

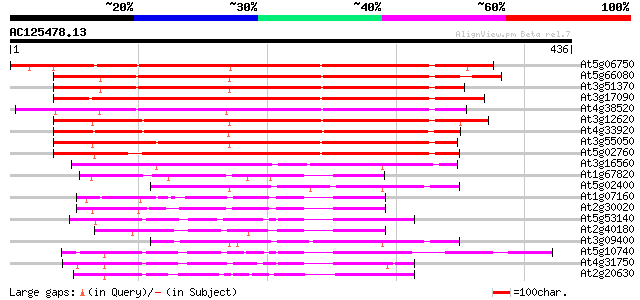
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125478.13 - phase: 0
(436 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g06750 protein phosphatase 2C-like 329 2e-90
At5g66080 protein phosphatase 2C-like protein 288 4e-78
At3g51370 protein phosphatase 2C like protein 286 2e-77
At3g17090 protein phosphatase-2c, putative 282 2e-76
At4g38520 putative protein phosphatase-2c 280 1e-75
At3g12620 unknown protein 273 1e-73
At4g33920 unknown protein 269 3e-72
At3g55050 protein phosphatase 2C - like protein 265 4e-71
At5g02760 protein phosphatase - like protein 261 7e-70
At3g16560 unknown protein 148 5e-36
At1g67820 protein phosphatase-like protein 110 2e-24
At5g02400 putative protein 105 4e-23
At1g07160 unknown protein 103 1e-22
At2g30020 putative protein phosphatase 2C 100 3e-21
At5g53140 protein phosphatase 2C-like 99 4e-21
At2g40180 protein phosphatase 2C (AthPP2C5) 99 5e-21
At3g09400 unknown protein 99 6e-21
At5g10740 protein phosphatase 2C -like protein 98 8e-21
At4g31750 unknown protein 96 4e-20
At2g20630 putative protein phosphatase 2C 95 7e-20
>At5g06750 protein phosphatase 2C-like
Length = 393
Score = 329 bits (843), Expect = 2e-90
Identities = 193/396 (48%), Positives = 249/396 (62%), Gaps = 27/396 (6%)
Query: 1 MHPWLENIVNLCRK--RKNDNINMALFDQDPHAS-----------SIDLKRHCYGQFSSA 47
M WL + C + R+ +N D D H S +L+RH +G FS A
Sbjct: 1 MFSWLARMALFCLRPMRRYGRMNRDDDDDDDHDGDSSSSGDSLLWSRELERHSFGDFSIA 60
Query: 48 FVQANEAMEDHSQVEVASRKALFLGVYDGHAGFEASVFITQHLFDHLLRAVRANENKITE 107
VQANE +EDHSQVE + A+F+GVYDGH G EAS +I+ HLF HL+R R + I+E
Sbjct: 61 VVQANEVIEDHSQVETGNG-AVFVGVYDGHGGPEASRYISDHLFSHLMRVSR-ERSCISE 118
Query: 108 PTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWKKTLHVANLGDSRAVIGTM 167
LR A SATE GFL V + K + VGSCCL G+IWK TL +AN+GDSRAV+G+M
Sbjct: 119 EALRAAFSATEEGFLTLVRRTCGLKPLIAAVGSCCLVGVIWKGTLLIANVGDSRAVLGSM 178
Query: 168 VNN-----KIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMYEREVWRVKGIITVSRSI 222
+N KI AEQLT DHN E +R+EL S HPDD+ IV+ + VWR+KGII VSRSI
Sbjct: 179 GSNNNRSNKIVAEQLTSDHNAALEEVRQELRSLHPDDSHIVVLKHGVWRIKGIIQVSRSI 238
Query: 223 GDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLTENDKFLIFASDGLWDFL 282
GD YLKRPEFSLD SFP+F + E R VLSAEP + +R L +DKF+IFASDGLW+ +
Sbjct: 239 GDAYLKRPEFSLDPSFPRF-HLAEELQRPVLSAEPCVYTRVLQTSDKFVIFASDGLWEQM 297
Query: 283 SNEQAVEIVQNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNANLGRGDGNRRYFHDDIS 342
+N+QAVEIV + R GIA+RLV + AA R Y+ +K G RR+FHDDI+
Sbjct: 298 TNQQAVEIVNKHPRPGIARRLVRRAITIAAKKREMNYDDLKKVE----RGVRRFFHDDIT 353
Query: 343 VIVVFLDKKSIL--RMPLHNLSYKSSSARPTTSAFA 376
V+V+F+D + ++ + + LS K S S F+
Sbjct: 354 VVVIFIDNELLMVEKATVPELSIKGFSHTVGPSKFS 389
>At5g66080 protein phosphatase 2C-like protein
Length = 385
Score = 288 bits (737), Expect = 4e-78
Identities = 162/358 (45%), Positives = 221/358 (61%), Gaps = 25/358 (6%)
Query: 35 DLKRHCYGQFSSAFVQANEAMEDHSQVEVASRKAL--------FLGVYDGHAGFEASVFI 86
D H +G FS A VQAN +ED SQVE L F+GVYDGH G E S F+
Sbjct: 39 DSAHHLFGDFSMAVVQANNLLEDQSQVESGPLTTLSSSGPYGTFVGVYDGHGGPETSRFV 98
Query: 87 TQHLFDHLLRAVRANENKITEPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGI 146
HLF HL R A ++ ++ +R A ATE GFL V K + K ++ VGSCCL G+
Sbjct: 99 NDHLFHHLKRFA-AEQDSMSVDVIRKAYEATEEGFLGVVAKQWAVKPHIAAVGSCCLIGV 157
Query: 147 IWKKTLHVANLGDSRAVIGTMVN--NKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVM 204
+ L+VAN+GDSRAV+G ++ ++ A QL+ +HN E++R+E+ S HPDD+ IV+
Sbjct: 158 VCDGKLYVANVGDSRAVLGKVIKATGEVNALQLSAEHNVSIESVRQEMHSLHPDDSHIVV 217
Query: 205 YEREVWRVKGIITVSRSIGDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDL 264
+ VWRVKGII VSRSIGD YLK+ EF+ + + K+ + EP R +LS EP + DL
Sbjct: 218 LKHNVWRVKGIIQVSRSIGDVYLKKSEFNKEPLYTKYR-LREPMKRPILSWEPSITVHDL 276
Query: 265 TENDKFLIFASDGLWDFLSNEQAVEIVQNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKN 324
+D+FLIFASDGLW+ LSN++AVEIVQN+ RNGIA+RLV L +AA R Y+ +
Sbjct: 277 QPDDQFLIFASDGLWEQLSNQEAVEIVQNHPRNGIARRLVKAALQEAAKKREMRYSDLNK 336
Query: 325 ANLGRGDGNRRYFHDDISVIVVFLDKKSILRMPLHNLSYKSSSARPTTSAFAESGLTM 382
G RR+FHDDI+V+V+FLD NL ++SS + + + G+T+
Sbjct: 337 IE----RGVRRHFHDDITVVVLFLDT---------NLLSRASSLKTPSVSIRGGGITL 381
>At3g51370 protein phosphatase 2C like protein
Length = 379
Score = 286 bits (731), Expect = 2e-77
Identities = 151/328 (46%), Positives = 208/328 (63%), Gaps = 15/328 (4%)
Query: 35 DLKRHCYGQFSSAFVQANEAMEDHSQVEVASRKAL-------FLGVYDGHAGFEASVFIT 87
D +H G+FS A VQAN +ED SQVE L F+G+YDGH G E S F+
Sbjct: 37 DFGQHLVGEFSMAVVQANNLLEDQSQVESGPLSTLDSGPYGTFIGIYDGHGGPETSRFVN 96
Query: 88 QHLFDHLLRAVRANENKITEPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGII 147
HLF HL R A + ++ ++ A ATE GFL V K + K + VGSCCL G+I
Sbjct: 97 DHLFQHLKRFA-AEQASMSVDVIKKAYEATEEGFLGVVTKQWPTKPQIAAVGSCCLVGVI 155
Query: 148 WKKTLHVANLGDSRAVIGTMVN--NKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMY 205
L++AN+GDSRAV+G + ++ A QL+ +HN E++R+E+ S HPDD+ IVM
Sbjct: 156 CGGMLYIANVGDSRAVLGRAMKATGEVIALQLSAEHNVSIESVRQEMHSLHPDDSHIVML 215
Query: 206 EREVWRVKGIITVSRSIGDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLT 265
+ VWRVKG+I +SRSIGD YLK+ EF+ + + K+ + EPF R +LS EP + ++
Sbjct: 216 KHNVWRVKGLIQISRSIGDVYLKKAEFNKEPLYTKYR-IREPFKRPILSGEPTITEHEIQ 274
Query: 266 ENDKFLIFASDGLWDFLSNEQAVEIVQNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNA 325
DKFLIFASDGLW+ +SN++AV+IVQN+ RNGIA+RLV L +AA R Y+ +K
Sbjct: 275 PQDKFLIFASDGLWEQMSNQEAVDIVQNHPRNGIARRLVKMALQEAAKKREMRYSDLKKI 334
Query: 326 NLGRGDGNRRYFHDDISVIVVFLDKKSI 353
G RR+FHDDI+V+++FLD +
Sbjct: 335 E----RGVRRHFHDDITVVIIFLDTNQV 358
>At3g17090 protein phosphatase-2c, putative
Length = 384
Score = 282 bits (722), Expect = 2e-76
Identities = 156/336 (46%), Positives = 210/336 (62%), Gaps = 7/336 (2%)
Query: 35 DLKRHCYGQFSSAFVQANEAMEDHSQVEVASRKALFLGVYDGHAGFEASVFITQHLFDHL 94
DL ++C G FS A +QAN+ +ED SQVE + F+GVYDGH G EA+ ++ HLF+H
Sbjct: 50 DLGKYCGGDFSMAVIQANQVLEDQSQVE-SGNFGTFVGVYDGHGGPEAARYVCDHLFNHF 108
Query: 95 LRAVRANENKITEPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWKKTLHV 154
+ +T T+ A ATE GF V + +++ NL VG+CCL G+I++ TL V
Sbjct: 109 REISAETQGVVTRETIERAFHATEEGFASIVSELWQEIPNLATVGTCCLVGVIYQNTLFV 168
Query: 155 ANLGDSRAVIGTMVN-NKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMYEREVWRVK 213
A+LGDSR V+G N + A QL+ +HN +E IR EL HPDD IV++ VWRVK
Sbjct: 169 ASLGDSRVVLGKKGNCGGLSAIQLSTEHNANNEDIRWELKDLHPDDPQIVVFRHGVWRVK 228
Query: 214 GIITVSRSIGDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLTENDKFLIF 273
GII VSRSIGD Y+KRPEF+ + KF + EP R ++SA P + S L ND FLIF
Sbjct: 229 GIIQVSRSIGDMYMKRPEFNKEPISQKF-RIAEPMKRPLMSATPTILSHPLHPNDSFLIF 287
Query: 274 ASDGLWDFLSNEQAVEIVQNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNANLGRGDGN 333
ASDGLW+ L+NE+AVEIV N+ R G AKRL+ L +AA R Y+ ++ +
Sbjct: 288 ASDGLWEHLTNEKAVEIVHNHPRAGSAKRLIKAALHEAARKREMRYSDLRKID----KKV 343
Query: 334 RRYFHDDISVIVVFLDKKSILRMPLHNLSYKSSSAR 369
RR+FHDDI+VIVVFL+ I R +++ + S R
Sbjct: 344 RRHFHDDITVIVVFLNHDLISRGHINSTQDTTVSIR 379
>At4g38520 putative protein phosphatase-2c
Length = 400
Score = 280 bits (716), Expect = 1e-75
Identities = 159/364 (43%), Positives = 216/364 (58%), Gaps = 19/364 (5%)
Query: 5 LENIVNLCRKRKNDNINMALFDQDPHASSI----DLKRHCYGQFSSAFVQANEAMEDHSQ 60
L N +N C ++D + D + D +H +G FS A VQAN +ED SQ
Sbjct: 5 LMNFLNACLWPRSDQQARSASDSGGRQEGLLWFRDSGQHVFGDFSMAVVQANSLLEDQSQ 64
Query: 61 VEVASRKA-------LFLGVYDGHAGFEASVFITQHLFDHLLRAVRANENKITEPTLRDA 113
+E S + F+GVYDGH G E S FI H+F HL R A + ++ ++ A
Sbjct: 65 LESGSLSSHDSGPFGTFVGVYDGHGGPETSRFINDHMFHHLKRFT-AEQQCMSSEVIKKA 123
Query: 114 VSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWKKTLHVANLGDSRAVIGTM--VNNK 171
ATE GFL V ++ + + VGSCCL +I L+VAN GDSRAV+G + V +
Sbjct: 124 FQATEEGFLSIVTNQFQTRPQIATVGSCCLVSVICDGKLYVANAGDSRAVLGQVMRVTGE 183
Query: 172 IQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMYEREVWRVKGIITVSRSIGDTYLKRPE 231
A QL+ +HN E++R+EL + HPD IV+ + VWRVKGII VSRSIGD YLKR E
Sbjct: 184 AHATQLSAEHNASIESVRRELQALHPDHPDIVVLKHNVWRVKGIIQVSRSIGDVYLKRSE 243
Query: 232 FSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLTENDKFLIFASDGLWDFLSNEQAVEIV 291
F+ + + KF + PF + +LSAEP + L +D+F+I ASDGLW+ +SN++AV+IV
Sbjct: 244 FNREPLYAKFR-LRSPFSKPLLSAEPAITVHTLEPHDQFIICASDGLWEHMSNQEAVDIV 302
Query: 292 QNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNANLGRGDGNRRYFHDDISVIVVFLDKK 351
QN+ RNGIAKRLV L +AA R Y+ +K + G RR+FHDDI+VIVVF D
Sbjct: 303 QNHPRNGIAKRLVKVALQEAAKKREMRYSDLKKID----RGVRRHFHDDITVIVVFFDTN 358
Query: 352 SILR 355
+ R
Sbjct: 359 LVSR 362
>At3g12620 unknown protein
Length = 385
Score = 273 bits (699), Expect = 1e-73
Identities = 154/351 (43%), Positives = 217/351 (60%), Gaps = 19/351 (5%)
Query: 35 DLKRHCYGQFSSAFVQANEAMEDHSQVEV-------ASRKALFLGVYDGHAGFEASVFIT 87
D H G+FS + +QAN +EDHS++E + +A F+GVYDGH G EA+ F+
Sbjct: 41 DSGNHVAGEFSMSVIQANNLLEDHSKLESGPVSMFDSGPQATFVGVYDGHGGPEAARFVN 100
Query: 88 QHLFDHLLRAVRANENKITEPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGII 147
+HLFD++ + N ++ + A ATE FL V + ++ K + VG+CCL GII
Sbjct: 101 KHLFDNIRKFTSENHG-MSANVITKAFLATEEDFLSLVRRQWQIKPQIASVGACCLVGII 159
Query: 148 WKKTLHVANLGDSRAVIGTMVN--NKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMY 205
L++AN GDSR V+G + ++A QL+ +HN E++R+EL S HP+D IV+
Sbjct: 160 CSGLLYIANAGDSRVVLGRLEKAFKIVKAVQLSSEHNASLESVREELRSLHPNDPQIVVL 219
Query: 206 EREVWRVKGIITVSRSIGDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLT 265
+ +VWRVKGII VSRSIGD YLK+ EF+ + KF VPE F + +L AEP + +
Sbjct: 220 KHKVWRVKGIIQVSRSIGDAYLKKAEFNREPLLAKFR-VPEVFHKPILRAEPAITVHKIH 278
Query: 266 ENDKFLIFASDGLWDFLSNEQAVEIVQNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNA 325
D+FLIFASDGLW+ LSN++AV+IV RNGIA++L+ T L +AA R Y+ +K
Sbjct: 279 PEDQFLIFASDGLWEHLSNQEAVDIVNTCPRNGIARKLIKTALREAAKKREMRYSDLKKI 338
Query: 326 NLGRGDGNRRYFHDDISVIVVFLD----KKSILRMPLHNLSYKSSSARPTT 372
+ G RR+FHDDI+VIVVFLD +S R PL ++S A P+T
Sbjct: 339 D----RGVRRHFHDDITVIVVFLDSHLVSRSTSRRPLLSISGGGDLAGPST 385
>At4g33920 unknown protein
Length = 380
Score = 269 bits (687), Expect = 3e-72
Identities = 153/321 (47%), Positives = 202/321 (62%), Gaps = 12/321 (3%)
Query: 35 DLKRHCYGQFSSAFVQANEAMEDHSQVEVASRKALFLGVYDGHAGFEASVFITQHLFDHL 94
+L+ H G +S A VQAN +ED SQV +S A ++GVYDGH G EAS F+ +HLF ++
Sbjct: 27 ELRPHAGGDYSIAVVQANSRLEDQSQVFTSS-SATYVGVYDGHGGPEASRFVNRHLFPYM 85
Query: 95 LRAVRANENKITEPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWKKTLHV 154
+ R + ++ ++ A TE F V+++ K + VGSCCL G I TL+V
Sbjct: 86 HKFAREHGG-LSVDVIKKAFKETEEEFCGMVKRSLPMKPQMATVGSCCLVGAISNDTLYV 144
Query: 155 ANLGDSRAVIGTMV-----NNKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMYEREV 209
ANLGDSRAV+G++V N AE+L+ DHN E +RKE+ + +PDD+ IV+Y R V
Sbjct: 145 ANLGDSRAVLGSVVSGVDSNKGAVAERLSTDHNVAVEEVRKEVKALNPDDSQIVLYTRGV 204
Query: 210 WRVKGIITVSRSIGDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLTENDK 269
WR+KGII VSRSIGD YLK+PE+ D F + P P R ++AEP + R L D
Sbjct: 205 WRIKGIIQVSRSIGDVYLKKPEYYRDPIFQRHGN-PIPLRRPAMTAEPSIIVRKLKPQDL 263
Query: 270 FLIFASDGLWDFLSNEQAVEIVQNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNANLGR 329
FLIFASDGLW+ LS+E AVEIV + R GIA+RLV L +AA R Y +K
Sbjct: 264 FLIFASDGLWEHLSDETAVEIVLKHPRTGIARRLVRAALEEAAKKREMRYGDIKKI---- 319
Query: 330 GDGNRRYFHDDISVIVVFLDK 350
G RR+FHDDISVIVV+LD+
Sbjct: 320 AKGIRRHFHDDISVIVVYLDQ 340
>At3g55050 protein phosphatase 2C - like protein
Length = 409
Score = 265 bits (677), Expect = 4e-71
Identities = 147/323 (45%), Positives = 201/323 (61%), Gaps = 15/323 (4%)
Query: 35 DLKRHCYGQFSSAFVQANEAMEDHSQVEV-------ASRKALFLGVYDGHAGFEASVFIT 87
D H G+FS A VQAN +EDHSQ+E + +A F+GVYDGH G EA+ F+
Sbjct: 67 DSGNHITGEFSMAVVQANNLLEDHSQLESGPISLHESGPEATFVGVYDGHGGPEAARFVN 126
Query: 88 QHLFDHLLRAVRANENKITEPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGII 147
LF ++ R + R V ATE FL V++ ++ K + VG+CCL GI+
Sbjct: 127 DRLFYNIKRYTSEQRGMSPDVITRGFV-ATEEEFLGLVQEQWKTKPQIASVGACCLVGIV 185
Query: 148 WKKTLHVANLGDSRAVIGTMVN--NKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMY 205
L+VAN GDSR V+G + N +++A QL+ +HN E++R+EL HPDD IV+
Sbjct: 186 CNGLLYVANAGDSRVVLGKVANPFKELKAVQLSTEHNASIESVREELRLLHPDDPNIVVL 245
Query: 206 EREVWRVKGIITVSRSIGDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLT 265
+ +VWRVKGII VSRSIGD YLKR EF+ + PKF VPE F + ++ AEP + +
Sbjct: 246 KHKVWRVKGIIQVSRSIGDAYLKRAEFNQEPLLPKF-RVPERFEKPIMRAEPTITVHKIH 304
Query: 266 ENDKFLIFASDGLWDFLSNEQAVEIVQNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNA 325
D+FLIFASDGLW+ LSN++AV+IV + RNG+A++LV L +AA R Y+ ++
Sbjct: 305 PEDQFLIFASDGLWEHLSNQEAVDIVNSCPRNGVARKLVKAALQEAAKKREMRYSDLEKI 364
Query: 326 NLGRGDGNRRYFHDDISVIVVFL 348
G RR+FHDDI+VIVVFL
Sbjct: 365 E----RGIRRHFHDDITVIVVFL 383
>At5g02760 protein phosphatase - like protein
Length = 361
Score = 261 bits (666), Expect = 7e-70
Identities = 140/322 (43%), Positives = 200/322 (61%), Gaps = 22/322 (6%)
Query: 35 DLKRHCYGQFSSAFVQANEAMEDHSQVEVA-------SRKALFLGVYDGHAGFEASVFIT 87
DL H +G+FS A +QAN MED Q+E + + F+GVYDGH G EAS FI
Sbjct: 30 DLGLHTFGEFSMAMIQANSVMEDQCQIESGPLTFNNPTVQGTFVGVYDGHGGPEASRFIA 89
Query: 88 QHLFDHLLRAVRANENKITEPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGII 147
++F +I+E + A + T+ FL+ V K + + VGSCCLAG+I
Sbjct: 90 DNIFP----------KEISEQVISKAFAETDKDFLKTVTKQWPTNPQMASVGSCCLAGVI 139
Query: 148 WKKTLHVANLGDSRAVIGTMVNNKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMYER 207
+++AN GDSRAV+G ++A QL+ +HN E+ R+EL S HP+D TI++ +
Sbjct: 140 CNGLVYIANTGDSRAVLGRSERGGVRAVQLSVEHNANLESARQELWSLHPNDPTILVMKH 199
Query: 208 EVWRVKGIITVSRSIGDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLTEN 267
+WRVKG+I V+RSIGD YLKR EF+ + PKF +PE F + +LSA+P + L+
Sbjct: 200 RLWRVKGVIQVTRSIGDAYLKRAEFNREPLLPKF-RLPEHFTKPILSADPSVTITRLSPQ 258
Query: 268 DKFLIFASDGLWDFLSNEQAVEIVQNNSRNGIAKRLVSTVLAQAAANRNSTYNTMKNANL 327
D+F+I ASDGLW+ LSN++AV+IV N+ R GIA+RL+ L +AA R Y+ + +
Sbjct: 259 DEFIILASDGLWEHLSNQEAVDIVHNSPRQGIARRLLKAALKEAAKKREMRYSDLTEIH- 317
Query: 328 GRGDGNRRYFHDDISVIVVFLD 349
G RR+FHDDI+VIVV+L+
Sbjct: 318 ---PGVRRHFHDDITVIVVYLN 336
>At3g16560 unknown protein
Length = 493
Score = 148 bits (374), Expect = 5e-36
Identities = 108/338 (31%), Positives = 166/338 (48%), Gaps = 48/338 (14%)
Query: 49 VQANEAMEDHSQVEVASRKA-LFLGVYDGHAGFEASVFITQHLFDHLLRAVRANENKITE 107
V A ED Q + LF +YDG G +A+ F+ L++ ++ ++ + ++ +
Sbjct: 153 VAGGAAGEDRVQAVCSEENGWLFCAIYDGFNGRDAADFLACTLYESIVFHLQLLDRQMKQ 212
Query: 108 PTLRD--------------------------------AVSATEAGFLEYVEKNYRQKNNL 135
D A+ E FL VE+ ++ +L
Sbjct: 213 TKSDDDGEKLELLSNISNVDYSSTDLFRQGVLDCLNRALFQAETDFLRMVEQEMEERPDL 272
Query: 136 GKVGSCCLAGIIWKKTLHVANLGDSRAVIGTM-VNNKIQAEQLTRDHNCKDEAIRKELMS 194
VGSC L ++ K L+V NLGDSRAV+ T N K+QA QLT DH +E L+S
Sbjct: 273 VSVGSCVLVTLLVGKDLYVLNLGDSRAVLATYNGNKKLQAVQLTEDHTVDNEVEEARLLS 332
Query: 195 EHPDDTTIVMYEREVWRVKGIITVSRSIGDTYLKRPEFSLDESFPKFEEVPEPFIRGVLS 254
EH DD IV+ ++KG + V+R++G YLK+ + L+++ V +S
Sbjct: 333 EHLDDPKIVI----GGKIKGKLKVTRALGVGYLKKEK--LNDALMGILRVRNLLSPPYVS 386
Query: 255 AEPEMRSRDLTENDKFLIFASDGLWDFLSNEQAV----EIVQNNSRNGIAKRLVSTVLAQ 310
EP MR +TE+D F+I ASDGL+DF SNE+A+ V +N AK L+ ++A+
Sbjct: 387 VEPSMRVHKITESDHFVIVASDGLFDFFSNEEAIGLVHSFVSSNPSGDPAKFLLERLVAK 446
Query: 311 AAANRNSTYNTMKNANLGRGDGNRRYFHDDISVIVVFL 348
AAA T + N GR RR +HDD++++V+ L
Sbjct: 447 AAARAGFTLEELTNVPAGR----RRRYHDDVTIMVITL 480
>At1g67820 protein phosphatase-like protein
Length = 447
Score = 110 bits (274), Expect = 2e-24
Identities = 86/252 (34%), Positives = 129/252 (51%), Gaps = 47/252 (18%)
Query: 55 MEDHSQVE---VASRKALFLGVYDGHAGFEASVFITQHLFDHLLRAVRANENKITEPTLR 111
MED ++ V + K F GVYDGH G +A+ F+ ++L +++ + + K
Sbjct: 116 MEDTHRIVPCLVGNSKKSFFGVYDGHGGAKAAEFVAENLHKYVVEMMENCKGK------E 169
Query: 112 DAVSATEAGFL----EYVEKNYRQKNNLGKV-GSCCLAGIIWKKTLHVANLGDSRAVIGT 166
+ V A +A FL +++EK ++++ G V G+CC+ +I + + V+NLGD RAV+
Sbjct: 170 EKVEAFKAAFLRTDRDFLEKVIKEQSLKGVVSGACCVTAVIQDQEMIVSNLGDCRAVLCR 229
Query: 167 MVNNKIQAEQLTRDHNC--KDEAIRKELMSEHPDDTT-----IVMYEREVWRVKGIITVS 219
AE LT DH DE R E S P T V + WRV+GI+ VS
Sbjct: 230 AG----VAEALTDDHKPGRDDEKERIESQSLIPFMTFGLQGGYVDNHQGAWRVQGILAVS 285
Query: 220 RSIGDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLTENDKFLIFASDGLW 279
RSIGD +LK+ + AEPE R +L ++ +FL+ ASDGLW
Sbjct: 286 RSIGDAHLKK----------------------WVVAEPETRVLELEQDMEFLVLASDGLW 323
Query: 280 DFLSNEQAVEIV 291
D +SN++AV V
Sbjct: 324 DVVSNQEAVYTV 335
>At5g02400 putative protein
Length = 674
Score = 105 bits (263), Expect = 4e-23
Identities = 81/278 (29%), Positives = 131/278 (46%), Gaps = 50/278 (17%)
Query: 110 LRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWKKTLHVANLGDSRAVIGTMVN 169
L A+ TE +LE ++ ++ L +GSC L ++ + ++V N+GDSRAV+G N
Sbjct: 401 LLQALRKTEDAYLELADQMVKENPELALMGSCVLVTLMKGEDVYVMNVGDSRAVLGRKPN 460
Query: 170 --------------------------------NKIQAEQLTRDHNCKDEAIRKELMSEHP 197
N + QL +H+ + E + + EHP
Sbjct: 461 LATGRKRQKELERIREDSSLEDKEILMNGAMRNTLVPLQLNMEHSTRIEEEVRRIKKEHP 520
Query: 198 DDTTIVMYEREVWRVKGIITVSRSIGDTYLKRPEF--SLDESFPKFEEVPEPFIRGVLSA 255
DD V E RVKG + V+R+ G +LK+P++ +L E F P+I +
Sbjct: 521 DDDCAV----ENDRVKGYLKVTRAFGAGFLKQPKWNDALLEMFRIDYIGTSPYI----TC 572
Query: 256 EPEMRSRDLTENDKFLIFASDGLWDFLSNEQAV----EIVQNNSRNGIAKRLVSTVLAQA 311
P + LT DKFLI +SDGL+++ SN++A+ + A+ L+ VL +A
Sbjct: 573 SPSLCHHKLTSRDKFLILSSDGLYEYFSNQEAIFEVESFISAFPEGDPAQHLIQEVLLRA 632
Query: 312 AANRNSTYNTMKNANLGRGDGNRRYFHDDISVIVVFLD 349
A ++ + L G+RR +HDD+SVIV+ L+
Sbjct: 633 ANKFGMDFHEL----LEIPQGDRRRYHDDVSVIVISLE 666
>At1g07160 unknown protein
Length = 380
Score = 103 bits (258), Expect = 1e-22
Identities = 85/246 (34%), Positives = 122/246 (49%), Gaps = 47/246 (19%)
Query: 53 EAMEDH----SQVEVASRKALFLGVYDGHAGFEASVFITQHLFDHLLRAVRA--NENKIT 106
EAMED + ++ ++A+F GVYDGH G A+ F ++L ++L + NE+KI
Sbjct: 133 EAMEDRFSAITNLQGDPKQAIF-GVYDGHGGPTAAEFAAKNLCSNILGEIVGGRNESKIE 191
Query: 107 EPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWKKTLHVANLGDSRAVIGT 166
E R + AT++ FL+ EKN K GSCC+ +I L VAN GD RAV+
Sbjct: 192 EAVKRGYL-ATDSEFLK--EKNV-------KGGSCCVTALISDGNLVVANAGDCRAVLSV 241
Query: 167 MVNNKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMYEREVWRVKGIITVSRSIGDTY 226
AE LT DH + R + S T VWR++G + VSR IGD +
Sbjct: 242 GGF----AEALTSDHRPSRDDERNRIESSGGYVDTF----NSVWRIQGSLAVSRGIGDAH 293
Query: 227 LKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLTENDKFLIFASDGLWDFLSNEQ 286
LK+ + +EPE+ + +FLI ASDGLWD +SN++
Sbjct: 294 LKQ----------------------WIISEPEINILRINPQHEFLILASDGLWDKVSNQE 331
Query: 287 AVEIVQ 292
AV+I +
Sbjct: 332 AVDIAR 337
>At2g30020 putative protein phosphatase 2C
Length = 396
Score = 99.8 bits (247), Expect = 3e-21
Identities = 78/245 (31%), Positives = 117/245 (46%), Gaps = 45/245 (18%)
Query: 53 EAMEDHSQVEV---ASRKALFLGVYDGHAGFEASVFITQHLFDHLLRAV--RANENKITE 107
EAMED RK GVYDGH G +A+ F ++L +++ V + +E++I E
Sbjct: 150 EAMEDRFSAITNLHGDRKQAIFGVYDGHGGVKAAEFAAKNLDKNIVEEVVGKRDESEIAE 209
Query: 108 PTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWKKTLHVANLGDSRAVIGTM 167
++ AT+A FL K K GSCC+ ++ + L V+N GD RAV+
Sbjct: 210 -AVKHGYLATDASFL---------KEEDVKGGSCCVTALVNEGNLVVSNAGDCRAVMSVG 259
Query: 168 VNNKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMYEREVWRVKGIITVSRSIGDTYL 227
A+ L+ DH + RK + + T VWR++G + VSR IGD L
Sbjct: 260 G----VAKALSSDHRPSRDDERKRIETTGGYVDTF----HGVWRIQGSLAVSRGIGDAQL 311
Query: 228 KRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLTENDKFLIFASDGLWDFLSNEQA 287
K+ + AEPE + + + +FLI ASDGLWD +SN++A
Sbjct: 312 KK----------------------WVIAEPETKISRIEHDHEFLILASDGLWDKVSNQEA 349
Query: 288 VEIVQ 292
V+I +
Sbjct: 350 VDIAR 354
>At5g53140 protein phosphatase 2C-like
Length = 420
Score = 99.4 bits (246), Expect = 4e-21
Identities = 73/272 (26%), Positives = 129/272 (46%), Gaps = 43/272 (15%)
Query: 47 AFVQANEAMEDHSQVEVAS---RKALFLGVYDGHAGFEASVFITQHLFDHLLRAVRANEN 103
+F MED ++ ++ + G++DGH G A+ ++ +HLF++L++ + +
Sbjct: 106 SFRGKRSTMEDFYDIKASTIEGQAVCMFGIFDGHGGSRAAEYLKEHLFNNLMKHPQFLTD 165
Query: 104 KITEPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWKKTLHVANLGDSRAV 163
T+ L + T+ FLE + YR GS A ++ L+VAN+GDSR
Sbjct: 166 --TKLALNETYKQTDVAFLESEKDTYRDD------GSTASAAVLVGNHLYVANVGDSR-- 215
Query: 164 IGTMVNNKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMYEREVWRVKGIITVSRSIG 223
T+V+ +A L+ DH RK + S ++M+ WRV G++ +SR+ G
Sbjct: 216 --TIVSKAGKAIALSDDHKPNRSDERKRIESAGG----VIMWAG-TWRVGGVLAMSRAFG 268
Query: 224 DTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLTENDKFLIFASDGLWDFLS 283
+ LK+ + AEPE++ ++ + L+ ASDGLWD +
Sbjct: 269 NRMLKQ----------------------FVVAEPEIQDLEIDHEAELLVLASDGLWDVVP 306
Query: 284 NEQAVEIVQNNSR-NGIAKRLVSTVLAQAAAN 314
NE AV + Q+ A++L T ++ +A+
Sbjct: 307 NEDAVALAQSEEEPEAAARKLTDTAFSRGSAD 338
>At2g40180 protein phosphatase 2C (AthPP2C5)
Length = 390
Score = 99.0 bits (245), Expect = 5e-21
Identities = 74/232 (31%), Positives = 114/232 (48%), Gaps = 48/232 (20%)
Query: 67 KALFLGVYDGHAGFEASVFITQHLFDHL---LRAVRANENKIT-EPTLRDAVSATEAGFL 122
K F GV+DGH G +A+ F +L +++ + + R+ E+ + E +R+ T+ FL
Sbjct: 158 KNAFFGVFDGHGGSKAAEFAAMNLGNNIEAAMASARSGEDGCSMESAIREGYIKTDEDFL 217
Query: 123 EYVEKNYRQKNNLGKVGSCCLAGIIWKKTLHVANLGDSRAVIGTMVNNKIQAEQLTRDHN 182
+ + G+CC+ +I K L V+N GD RAV+ + AE LT DHN
Sbjct: 218 KEGSRG----------GACCVTALISKGELAVSNAGDCRAVM----SRGGTAEALTSDHN 263
Query: 183 CK--DEAIRKELMSEHPDDTTIVMYEREVWRVKGIITVSRSIGDTYLKRPEFSLDESFPK 240
+E R E + + D VWR++G + VSR IGD YLK
Sbjct: 264 PSQANELKRIEALGGYVDCCN------GVWRIQGTLAVSRGIGDRYLKE----------- 306
Query: 241 FEEVPEPFIRGVLSAEPEMRSRDLTENDKFLIFASDGLWDFLSNEQAVEIVQ 292
+ AEPE R+ + +FLI ASDGLWD ++N++AV++V+
Sbjct: 307 -----------WVIAEPETRTLRIKPEFEFLILASDGLWDKVTNQEAVDVVR 347
>At3g09400 unknown protein
Length = 650
Score = 98.6 bits (244), Expect = 6e-21
Identities = 77/273 (28%), Positives = 129/273 (47%), Gaps = 47/273 (17%)
Query: 110 LRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWKKTLHVANLGDSRAVIGTMVN 169
L+ A+ TE F V +N L +GSC L ++ + ++V ++GDSRAV+ N
Sbjct: 384 LQQALEKTEESFDLMVNEN----PELALMGSCVLVTLMKGEDVYVMSVGDSRAVLARRPN 439
Query: 170 ---NKIQAE--------------------------QLTRDHNCKDEAIRKELMSEHPDDT 200
K+Q E QL ++H+ E + + EHPDD
Sbjct: 440 VEKMKMQKELERVKEESPLETLFITERGLSLLVPVQLNKEHSTSVEEEVRRIKKEHPDDI 499
Query: 201 TIVMYEREVWRVKGIITVSRSIGDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMR 260
+ E RVKG + V+R+ G +LK+P+++ E+ + + ++ P +
Sbjct: 500 LAI----ENNRVKGYLKVTRAFGAGFLKQPKWN--EALLEMFRIDYVGTSPYITCSPSLH 553
Query: 261 SRDLTENDKFLIFASDGLWDFLSNEQAV----EIVQNNSRNGIAKRLVSTVLAQAAANRN 316
L+ DKFLI +SDGL+++ SNE+A+ + A+ L+ VL +AA
Sbjct: 554 HHRLSSRDKFLILSSDGLYEYFSNEEAIFEVDSFISAFPEGDPAQHLIQEVLLRAAKKYG 613
Query: 317 STYNTMKNANLGRGDGNRRYFHDDISVIVVFLD 349
++ + L G+RR +HDD+SVIV+ L+
Sbjct: 614 MDFHEL----LEIPQGDRRRYHDDVSVIVISLE 642
Score = 28.5 bits (62), Expect = 7.6
Identities = 14/48 (29%), Positives = 29/48 (60%), Gaps = 1/48 (2%)
Query: 53 EAMEDHSQVEVASRKA-LFLGVYDGHAGFEASVFITQHLFDHLLRAVR 99
+A ED V ++ LF+G+YDG +G + ++ ++L+ +LR ++
Sbjct: 253 KAGEDRVHVILSEENGWLFVGIYDGFSGPDPPDYLIKNLYTAVLRELK 300
>At5g10740 protein phosphatase 2C -like protein
Length = 354
Score = 98.2 bits (243), Expect = 8e-21
Identities = 102/387 (26%), Positives = 166/387 (42%), Gaps = 82/387 (21%)
Query: 41 YGQFSSAFVQANEAMEDHSQVEVASRKALFLG---VYDGHAGFEASVFITQHLFDHLLRA 97
YG SSA +++ MED + + +G V+DGH G A+ ++ +HLF +L+
Sbjct: 34 YGYASSAGKRSS--MEDFFETRIDGINGEIVGLFGVFDGHGGARAAEYVKRHLFSNLITH 91
Query: 98 VRANENKITEPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWKKTLHVANL 157
+ + T+ + DA + T++ L+ + R GS I+ L VAN+
Sbjct: 92 PKFISD--TKSAITDAYNHTDSELLKSENSHNRD------AGSTASTAILVGDRLVVANV 143
Query: 158 GDSRAVIGTMVNNKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMYEREVWRVKGIIT 217
GDSRAVI + +A ++RDH D++ +E + VM+ WRV G++
Sbjct: 144 GDSRAVI----SRGGKAIAVSRDHK-PDQSDERERIENAGG---FVMWAG-TWRVGGVLA 194
Query: 218 VSRSIGDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLTENDKFLIFASDG 277
VSR+ GD LK+ + A+PE++ + + +FLI ASDG
Sbjct: 195 VSRAFGDRLLKQ----------------------YVVADPEIQEEKIDDTLEFLILASDG 232
Query: 278 LWDFLSNEQAVEIVQN-NSRNGIAKRLVSTVLAQAAANRNSTYNTMKNANLGRGDGNRRY 336
LWD SNE AV +V+ AK+LV + + +A
Sbjct: 233 LWDVFSNEAAVAMVKEVEDPEDSAKKLVGEAIKRGSA----------------------- 269
Query: 337 FHDDISVIVV-FLDKKSILRMPLHNLSYKSSSARPTTSAFAESGLTMHGLQRLQKTIKNR 395
D+I+ +VV FL+KKS + + S K + P A + +
Sbjct: 270 --DNITCVVVRFLEKKSASSSHISSSSSKEAKEMPPLGDLA-----------ISSNEAKQ 316
Query: 396 FQASSSQEGESSQTQDPEGESSQTQSL 422
Q S + E+ + P+ S T +L
Sbjct: 317 VQIGSGNKPENVTNRKPDTASRSTDTL 343
>At4g31750 unknown protein
Length = 311
Score = 95.9 bits (237), Expect = 4e-20
Identities = 83/281 (29%), Positives = 133/281 (46%), Gaps = 48/281 (17%)
Query: 42 GQFSSAFVQA---NEAMEDHSQVEVASRKALFLG---VYDGHAGFEASVFITQHLFDHLL 95
G+FS + + +MED + + + +G V+DGH G A+ ++ Q+LF +L+
Sbjct: 30 GKFSYGYASSPGKRSSMEDFYETRIDGVEGEIVGLFGVFDGHGGARAAEYVKQNLFSNLI 89
Query: 96 RAVRANENKITEPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWKKTLHVA 155
R + + T + DA + T++ FL+ R GS I+ L VA
Sbjct: 90 RHPKFISD--TTAAIADAYNQTDSEFLKSENSQNRD------AGSTASTAILVGDRLLVA 141
Query: 156 NLGDSRAVIGTMVNNKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMYEREVWRVKGI 215
N+GDSRAVI N A ++RDH R+ + D VM+ WRV G+
Sbjct: 142 NVGDSRAVICRGGN----AIAVSRDHKPDQSDERQRI----EDAGGFVMWAG-TWRVGGV 192
Query: 216 ITVSRSIGDTYLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLTENDKFLIFAS 275
+ VSR+ GD LK+ + A+PE++ + + +FLI AS
Sbjct: 193 LAVSRAFGDRLLKQ----------------------YVVADPEIQEEKVDSSLEFLILAS 230
Query: 276 DGLWDFLSNEQAVEIVQ--NNSRNGIAKRLVSTVLAQAAAN 314
DGLWD +SNE+AV +++ + G AKRL+ + +A+
Sbjct: 231 DGLWDVVSNEEAVGMIKAIEDPEEG-AKRLMMEAYQRGSAD 270
>At2g20630 putative protein phosphatase 2C
Length = 279
Score = 95.1 bits (235), Expect = 7e-20
Identities = 81/270 (30%), Positives = 136/270 (50%), Gaps = 45/270 (16%)
Query: 50 QANEAMEDHSQVEVASRKALFLG---VYDGHAGFEASVFITQHLFDHLLRAVRANENKIT 106
+A MED+ E LG ++DGH G + + ++ +LFD++L+ + T
Sbjct: 40 KAGHPMEDYVVSEFKKVDGHDLGLFAIFDGHLGHDVAKYLQTNLFDNILKEKDFWTD--T 97
Query: 107 EPTLRDAVSATEAGFLEYVEKNYRQKNNLGKVGSCCLAGIIWK-KTLHVANLGDSRAVIG 165
+ +R+A +T+A LE Q LGK GS + GI+ KTL +AN+GDSRAV
Sbjct: 98 KNAIRNAYISTDAVILE-------QSLKLGKGGSTAVTGILIDGKTLVIANVGDSRAV-- 148
Query: 166 TMVNNKIQAEQLTRDHNCKDEAIRKELMSEHPDDTTIVMYEREVWRVKGIITVSRSIGDT 225
M N + A QL+ DH E +KE+ S + I +V RV G + V+R+ GD
Sbjct: 149 -MSKNGV-ASQLSVDHEPSKE--QKEIESRGGFVSNI---PGDVPRVDGQLAVARAFGDK 201
Query: 226 YLKRPEFSLDESFPKFEEVPEPFIRGVLSAEPEMRSRDLTENDKFLIFASDGLWDFLSNE 285
LK LS++P++R ++ +F++FASDG+W +SN+
Sbjct: 202 SLKIH----------------------LSSDPDIRDENIDHETEFILFASDGVWKVMSNQ 239
Query: 286 QAVEIVQN-NSRNGIAKRLVSTVLAQAAAN 314
+AV+++++ AK L+ +++ + +
Sbjct: 240 EAVDLIKSIKDPQAAAKELIEEAVSKQSTD 269
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.131 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,167,992
Number of Sequences: 26719
Number of extensions: 369349
Number of successful extensions: 1381
Number of sequences better than 10.0: 90
Number of HSP's better than 10.0 without gapping: 79
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 1095
Number of HSP's gapped (non-prelim): 124
length of query: 436
length of database: 11,318,596
effective HSP length: 102
effective length of query: 334
effective length of database: 8,593,258
effective search space: 2870148172
effective search space used: 2870148172
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC125478.13


