

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125477.10 - phase: 0 /pseudo
(355 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
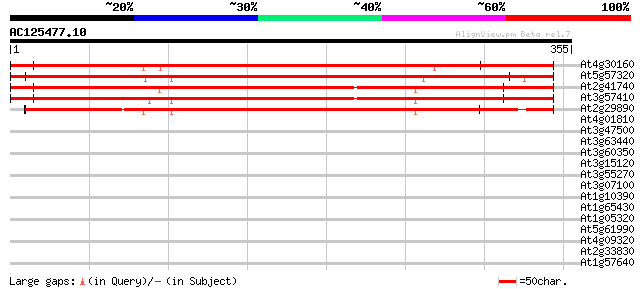
Score E
Sequences producing significant alignments: (bits) Value
At4g30160 putative villin 570 e-163
At5g57320 villin 528 e-150
At2g41740 putative villin 2 protein 452 e-127
At3g57410 villin 3 435 e-122
At2g29890 villin 1 (VLN1) 339 1e-93
At4g01810 protein transport factor like protein 36 0.037
At3g47500 H-protein promoter binding factor-2a 31 1.2
At3g63440 cytokinin oxidase -like protein 30 2.0
At3g60350 Arm repeat containing protein - like 30 2.0
At3g15120 chaperone-like ATPase 30 2.0
At3g55270 MAP kinase phosphatase (MKP1) 30 2.6
At3g07100 putative Sec24-like COPII protein 30 2.6
At1g10390 unknown protein 29 3.4
At1g65430 unknown protein 29 4.5
At1g05320 unknown protein 28 7.6
At5g61990 unknown protein 28 10.0
At4g09320 nucleoside-diphosphate kinase 28 10.0
At2g33830 putative auxin-regulated protein 28 10.0
At1g57640 28 10.0
>At4g30160 putative villin
Length = 974
Score = 570 bits (1468), Expect = e-163
Identities = 270/344 (78%), Positives = 311/344 (89%)
Query: 1 TTASKSGALRHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLS 60
TTA K+GALRHDIHYWLGKDTSQDEAG AA+KTVELDA LGGRAVQYREVQGHET+KFLS
Sbjct: 53 TTALKTGALRHDIHYWLGKDTSQDEAGTAAVKTVELDAALGGRAVQYREVQGHETEKFLS 112
Query: 61 YFKPCIIPQEGGAASGFKHVEAEEHKTRLFVCKGKHVVYVKEVPFARSSLNHDDIFILDT 120
YFKPCIIPQEGG ASGFKHV AEEH TRLFVC+GKHVV+VKEVPFARSSLNHDDI+ILDT
Sbjct: 113 YFKPCIIPQEGGVASGFKHVVAEEHITRLFVCRGKHVVHVKEVPFARSSLNHDDIYILDT 172
Query: 121 ESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFG 180
+SKIFQFNGSNSSIQERAKALEVVQYIKDTYHDG CEVA++EDG+LMAD++SGEFWG FG
Sbjct: 173 KSKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGTCEVATVEDGKLMADADSGEFWGFFG 232
Query: 181 GFAPLPRKTVSDDDKTIDSHPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLEVF 240
GFAPLPRKT +D+DKT +S +L CVEKG+A P E D+L +E+LDTNKCYILDCG+EVF
Sbjct: 233 GFAPLPRKTANDEDKTYNSDITRLFCVEKGQANPVEGDTLKREMLDTNKCYILDCGIEVF 292
Query: 241 VWIGRNTSLDERKSASGSTDELVSSTNRPKSQIIRVMEGFETVMFRSKFDSWPQTTNAAM 300
VW+GR TSLD+RK AS + +E++ S+ RPKSQ+IR++EGFETV FRSKF+SW Q TN +
Sbjct: 293 VWMGRTTSLDDRKIASKAAEEMIRSSERPKSQMIRIIEGFETVPFRSKFESWTQETNTTV 352
Query: 301 PEDGRGKVAALLKRQGLDVKGLVKADPVKEEPQPYIDCTGHLQV 344
EDGRG+VAALL+RQG++V+GL+KA P KEEPQ +IDCTG+LQV
Sbjct: 353 SEDGRGRVAALLQRQGVNVRGLMKAAPPKEEPQVFIDCTGNLQV 396
Score = 75.9 bits (185), Expect = 3e-14
Identities = 75/301 (24%), Positives = 133/301 (43%), Gaps = 33/301 (10%)
Query: 16 WLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYFKPCIIPQEGGAAS 75
W GK + ++E G+A ++ + Q R +G E +F + I+ +GG +S
Sbjct: 440 WFGKQSVEEERGSAVSMASKMVESMKFVPAQARIYEGKEPIQFFVIMQSFIV-FKGGISS 498
Query: 76 GFKHVEAE---------EHKTRLFVCKG---KHVVYVKEVPFARSSLNHDDIFILDTESK 123
G+K AE E+ LF +G +++ ++ P A +SLN +IL +S
Sbjct: 499 GYKKYIAEKEVDDDTYNENGVALFRIQGSGPENMQAIQVDPVA-ASLNSSYYYILHNDSS 557
Query: 124 IFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFGGFA 183
+F + G+ S+ ++ A + IK + ++G SES +FW L GG A
Sbjct: 558 VFTWAGNLSTATDQELAERQLDLIKPNQQS-----RAQKEG-----SESEQFWELLGGKA 607
Query: 184 PLPRKTVSDDDKTIDSHPPKLLCV-EKGKAEPFETDSLTKELLDTNKCYILDCGLEVFVW 242
+ ++ K + P C K + E + T++ L T +I+DC E+FVW
Sbjct: 608 EYSSQKLT---KEPERDPHLFSCTFTKEVLKVTEIYNFTQDDLMTEDIFIIDCHSEIFVW 664
Query: 243 IGRNTSLDERKSASGSTDELVSSTN-----RPKSQIIRVMEGFETVMFRSKFDSWPQTTN 297
+G+ + A ++ + + P++ I +MEG E F F SW + +
Sbjct: 665 VGQEVVPKNKLLALTIGEKFIEKDSLLEKLSPEAPIYVIMEGGEPSFFTRFFTSWDSSKS 724
Query: 298 A 298
A
Sbjct: 725 A 725
>At5g57320 villin
Length = 962
Score = 528 bits (1361), Expect = e-150
Identities = 251/346 (72%), Positives = 300/346 (86%), Gaps = 2/346 (0%)
Query: 1 TTASKSGALRHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLS 60
TTAS+SG+L HDIHYWLGKD+SQDEAGA A+ TVELD+ LGGRAVQYREVQGHET+KFLS
Sbjct: 53 TTASRSGSLHHDIHYWLGKDSSQDEAGAVAVMTVELDSALGGRAVQYREVQGHETEKFLS 112
Query: 61 YFKPCIIPQEGGAASGFKHVEAEEHKTRLFVCKGKHVVYVKEVPFARSSLNHDDIFILDT 120
YFKPCIIPQEGG ASGF HV+ EEH+TRL++CKGKHVV VKEVPF RS+LNH+D+FILDT
Sbjct: 113 YFKPCIIPQEGGVASGFNHVKPEEHQTRLYICKGKHVVRVKEVPFVRSTLNHEDVFILDT 172
Query: 121 ESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFG 180
ESKIFQF+GS SSIQERAKALEVVQYIKDTYHDGKC++A++EDGR+MAD+E+GEFWGLFG
Sbjct: 173 ESKIFQFSGSKSSIQERAKALEVVQYIKDTYHDGKCDIAAVEDGRMMADAEAGEFWGLFG 232
Query: 181 GFAPLPRKTVSDDDKTIDSHPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLEVF 240
GFAPLP+K +DD+T S KL VEKG+ + E + LTKELLDTNKCYILDCGLE+F
Sbjct: 233 GFAPLPKKPAVNDDETAASDGIKLFSVEKGQTDAVEAECLTKELLDTNKCYILDCGLELF 292
Query: 241 VWIGRNTSLDERKSASGSTDELVSSTNRPKSQIIRVMEGFETVMFRSKFDSWPQTTNAAM 300
VW GR+TS+D+RKSA+ + +E S+ PKS ++ VMEG+ETVMFRSKFDSWP ++ A
Sbjct: 293 VWKGRSTSIDQRKSATEAAEEFFRSSEPPKSNLVSVMEGYETVMFRSKFDSWPASSTIAE 352
Query: 301 PEDGRGKVAALLKRQGLDVKGLVK--ADPVKEEPQPYIDCTGHLQV 344
P+ GRGKVAALL+RQG++V+GLVK + K+EP+PYID TG+LQV
Sbjct: 353 PQQGRGKVAALLQRQGVNVQGLVKTSSSSSKDEPKPYIDGTGNLQV 398
Score = 66.2 bits (160), Expect = 3e-11
Identities = 74/323 (22%), Positives = 138/323 (41%), Gaps = 33/323 (10%)
Query: 11 HDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYFKPCIIPQE 70
H + W GK + +++ +A ++ + Q R +G E +F + I +
Sbjct: 437 HLVGTWFGKQSVEEDRASAISLANKMVESMKFVPAQARINEGKEPIQFFVIMQS-FITFK 495
Query: 71 GGAASGFKHVEAEE---------HKTRLFVCKGKHVVYVK--EVPFARSSLNHDDIFILD 119
GG + FK AE LF +G ++ ++ A + LN +IL
Sbjct: 496 GGVSDAFKKYIAENDIPDTTYEAEGVALFRVQGSGPENMQAIQIEAASAGLNSSHCYILH 555
Query: 120 TESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLF 179
+S +F + G+ +S +++ E+++ + D + A E SES +FW L
Sbjct: 556 GDSTVFTWCGNLTSSEDQ----ELMERMLDLIKPNEPTKAQKEG------SESEQFWELL 605
Query: 180 GGFAPLPRKTVSDDDKTIDSHPPKLLCVEKGKA-EPFETDSLTKELLDTNKCYILDCGLE 238
GG + P + + D +S P C ++ + E + T++ L T +ILDC E
Sbjct: 606 GGKSEYPSQKIKRDG---ESDPHLFSCTYTNESLKATEIFNFTQDDLMTEDIFILDCHTE 662
Query: 239 VFVWIGRNTSLDERKSASGSTD-----ELVSSTNRPKSQIIRVMEGFETVMFRSKFDSWP 293
VFVW+G+ ++ A + + + ++ I V EG E F ++F +W
Sbjct: 663 VFVWVGQQVDPKKKPQALDIGENFLKHDFLLENLASETPIYIVTEGNEPPFF-TRFFTW- 720
Query: 294 QTTNAAMPEDGRGKVAALLKRQG 316
++ + M D + A+L +G
Sbjct: 721 DSSKSGMHGDSFQRKLAILTNKG 743
>At2g41740 putative villin 2 protein
Length = 976
Score = 452 bits (1163), Expect = e-127
Identities = 213/345 (61%), Positives = 273/345 (78%), Gaps = 2/345 (0%)
Query: 1 TTASKSGALRHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLS 60
TT +K GA DIH+W+GKDTSQDEAG AA+KTVELDAVLGGRAVQ+RE+QGHE+ KFLS
Sbjct: 51 TTQNKGGAYLFDIHFWIGKDTSQDEAGTAAVKTVELDAVLGGRAVQHREIQGHESDKFLS 110
Query: 61 YFKPCIIPQEGGAASGFKHVEAEEHKTRLFVCKGKHVVYVKEVPFARSSLNHDDIFILDT 120
YFKPCIIP EGG ASGFK VE E +TRL+ CKGK + +K+VPFARSSLNHDD+FILDT
Sbjct: 111 YFKPCIIPLEGGVASGFKTVEEEVFETRLYTCKGKRAIRLKQVPFARSSLNHDDVFILDT 170
Query: 121 ESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFG 180
E KI+QFNG+NS+IQERAKALEVVQY+KD YH+G C+VA ++DG+L +S+SG FW LFG
Sbjct: 171 EEKIYQFNGANSNIQERAKALEVVQYLKDKYHEGTCDVAIVDDGKLDTESDSGAFWVLFG 230
Query: 181 GFAPLPRKTVSDDDKTIDSHPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLEVF 240
GFAP+ RK +DDD +S PPKL C+ GK EP + D L+K +L+ KCY+LDCG E++
Sbjct: 231 GFAPIGRKVANDDDIVPESTPPKLYCITDGKMEPIDGD-LSKSMLENTKCYLLDCGAEIY 289
Query: 241 VWIGRNTSLDERKSASGSTDELVSSTNRPK-SQIIRVMEGFETVMFRSKFDSWPQTTNAA 299
+W+GR T +DERK+AS S +E ++S NRPK + + RV++G+E+ F+S FDSWP +
Sbjct: 290 IWVGRVTQVDERKAASQSAEEFLASENRPKATHVTRVIQGYESHSFKSNFDSWPSGSATP 349
Query: 300 MPEDGRGKVAALLKRQGLDVKGLVKADPVKEEPQPYIDCTGHLQV 344
E+GRGKVAALLK+QG+ +KG+ K+ PV E+ P ++ G L+V
Sbjct: 350 GNEEGRGKVAALLKQQGVGLKGIAKSAPVNEDIPPLLESGGKLEV 394
Score = 81.3 bits (199), Expect = 8e-16
Identities = 81/318 (25%), Positives = 132/318 (41%), Gaps = 41/318 (12%)
Query: 16 WLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYFKPCIIPQEGGAAS 75
W GK + ++ A + L GR VQ R +G E +F++ F+P ++ +GG +S
Sbjct: 439 WFGKKSIPEDQDTAIRLANTMSNSLKGRPVQGRIYEGKEPPQFVALFQPMVV-LKGGLSS 497
Query: 76 GFKHVEAEEHKTRLFVCK-----------GKHVVYVKEVPFARSSLNHDDIFILDTESKI 124
G+K E T G H +V +SLN + F+L + + +
Sbjct: 498 GYKSSMGESESTDETYTPESIALVQVSGTGVHNNKAVQVETVATSLNSYECFLLQSGTSM 557
Query: 125 FQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFGGFAP 184
F ++G+ S+ ++ A +V +++K I +ES FW GG
Sbjct: 558 FLWHGNQSTHEQLELATKVAEFLKP----------GITLKHAKEGTESSTFWFALGGKQN 607
Query: 185 LPRKTVSDDDKTIDSHPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLEVFVWIG 244
K S + TI +GK + E + ++ L T Y LD EVFVW+G
Sbjct: 608 FTSKKASSE--TIRDPHLFSFAFNRGKFQVEEIYNFAQDDLLTEDIYFLDTHAEVFVWVG 665
Query: 245 RNTSLDERKSA----------SGSTDELVSSTNRPKSQIIRVMEGFETVMFRSKFDSWPQ 294
+ E+++ +GS + L PK I ++ EG E F + F SW
Sbjct: 666 QCVEPKEKQTVFEIGQKYIDLAGSLEGL-----HPKVPIYKINEGNEPCFFTTYF-SW-D 718
Query: 295 TTNAAMPEDGRGKVAALL 312
T A + + K A+LL
Sbjct: 719 ATKAIVQGNSFQKKASLL 736
>At3g57410 villin 3
Length = 965
Score = 435 bits (1118), Expect = e-122
Identities = 206/345 (59%), Positives = 270/345 (77%), Gaps = 2/345 (0%)
Query: 1 TTASKSGALRHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLS 60
TT +K GA DIH+W+GKDTSQDEAG AA+KTVELDA LGGRAVQYRE+QGHE+ KFLS
Sbjct: 53 TTQNKGGAYLFDIHFWIGKDTSQDEAGTAAVKTVELDAALGGRAVQYREIQGHESDKFLS 112
Query: 61 YFKPCIIPQEGGAASGFKHVEAEEHKTRLFVCKGKHVVYVKEVPFARSSLNHDDIFILDT 120
YFKPCIIP EGG ASGFK E EE +TRL+ CKGK V++K+VPFARSSLNHDD+FILDT
Sbjct: 113 YFKPCIIPLEGGVASGFKKPEEEEFETRLYTCKGKRAVHLKQVPFARSSLNHDDVFILDT 172
Query: 121 ESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFG 180
+ KI+QFNG+NS+IQERAKAL V+QY+KD +H+G +VA ++DG+L +S+SGEFW LFG
Sbjct: 173 KEKIYQFNGANSNIQERAKALVVIQYLKDKFHEGTSDVAIVDDGKLDTESDSGEFWVLFG 232
Query: 181 GFAPLPRKTVSDDDKTIDSHPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLEVF 240
GFAP+ RK S+D+ ++ PPKL + G+ E + D L+K +L+ NKCY+LDCG E+F
Sbjct: 233 GFAPIARKVASEDEIIPETTPPKLYSIADGQVESIDGD-LSKSMLENNKCYLLDCGSEIF 291
Query: 241 VWIGRNTSLDERKSASGSTDELVSSTNRPK-SQIIRVMEGFETVMFRSKFDSWPQTTNAA 299
+W+GR T ++ERK+A + ++ V+S NRPK ++I RV++G+E F+S FDSWP +
Sbjct: 292 IWVGRVTQVEERKTAIQAAEDFVASENRPKATRITRVIQGYEPHSFKSNFDSWPSGSATP 351
Query: 300 MPEDGRGKVAALLKRQGLDVKGLVKADPVKEEPQPYIDCTGHLQV 344
E+GRGKVAALLK+QG+ +KGL K+ PV E+ P ++ G L+V
Sbjct: 352 ANEEGRGKVAALLKQQGVGLKGLSKSTPVNEDIPPLLEGGGKLEV 396
Score = 87.8 bits (216), Expect = 8e-18
Identities = 83/318 (26%), Positives = 147/318 (46%), Gaps = 41/318 (12%)
Query: 16 WLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYFKPCIIPQEGGAAS 75
W GK+++Q++ A + L GR VQ R +G E +F++ F+ ++ +GG +S
Sbjct: 441 WFGKNSNQEDQETAVRLASTMTNSLKGRPVQARIFEGKEPPQFVALFQHMVV-LKGGLSS 499
Query: 76 GFKHVEAEEHKT---------RLFVCKGKHVVYVK--EVPFARSSLNHDDIFILDTESKI 124
G+K+ E+ + L G V K +V +SLN D F+L + + +
Sbjct: 500 GYKNSMTEKGSSGETYTPESIALIQVSGTGVHNNKALQVEAVATSLNSYDCFLLQSGTSM 559
Query: 125 FQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFGGFAP 184
F + G++S+ +++ A +V +++K + ++G +ES FW GG
Sbjct: 560 FLWVGNHSTHEQQELAAKVAEFLKPG-----TTIKHAKEG-----TESSSFWFALGGKQN 609
Query: 185 LPRKTVSDDDKTIDSHPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLEVFVWIG 244
K VS + T+ +GK + E + ++ L T + ++LD EVFVW+G
Sbjct: 610 FTSKKVSSE--TVRDPHLFSFSFNRGKFQVEEIHNFDQDDLLTEEMHLLDTHAEVFVWVG 667
Query: 245 RNTSLDERKSA----------SGSTDELVSSTNRPKSQIIRVMEGFETVMFRSKFDSWPQ 294
+ E+++A +GS + L PK + ++ EG E F + F SW
Sbjct: 668 QCVDPKEKQTAFEIGQRYINLAGSLEGL-----SPKVPLYKITEGNEPCFFTTYF-SW-D 720
Query: 295 TTNAAMPEDGRGKVAALL 312
+T A + + K AALL
Sbjct: 721 STKATVQGNSYQKKAALL 738
>At2g29890 villin 1 (VLN1)
Length = 909
Score = 339 bits (870), Expect = 1e-93
Identities = 173/337 (51%), Positives = 232/337 (68%), Gaps = 7/337 (2%)
Query: 10 RHDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYFKPCIIPQ 69
++DIHYWLG D ++ ++ A+ K ++LDA LG VQYREVQG ET+KFLSYFKPCIIP
Sbjct: 62 QYDIHYWLGIDANEVDSILASDKALDLDAALGCCTVQYREVQGQETEKFLSYFKPCIIPV 121
Query: 70 EGGAASGFKHVEAEEHKTRLFVCKGKHVVYVKEVPFARSSLNHDDIFILDTESKIFQFNG 129
EG S + E ++ L CKG HVV VKEVPF RSSLNHDD+FILDT SK+F F G
Sbjct: 122 EG-KYSPKTGIAGETYQVTLLRCKGDHVVRVKEVPFLRSSLNHDDVFILDTASKVFLFAG 180
Query: 130 SNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFGGFAPLPRKT 189
NSS QE+AKA+EVV+YIKD HDG+CEVA+IEDG+ DS++GEFW FGG+AP+P+ +
Sbjct: 181 CNSSTQEKAKAMEVVEYIKDNKHDGRCEVATIEDGKFSGDSDAGEFWSFFGGYAPIPKLS 240
Query: 190 VSDDDKTIDSHPPKLLCVE-KGKAEPFETDSLTKELLDTNKCYILDCGLEVFVWIGRNTS 248
S + + +L ++ KG P T SL K++L+ NKCY+LDC EVFVW+GRNTS
Sbjct: 241 SSTTQEQTQTPCAELFWIDTKGNLHPTGTSSLDKDMLEKNKCYMLDCHSEVFVWMGRNTS 300
Query: 249 LDERKSASGSTDELVSSTNR-PKSQIIRVMEGFETVMFRSKFDSWPQTTNAAMPEDGRGK 307
L ERK++ S++E + R + ++ + EG E FRS F+ WPQT +++ +GR K
Sbjct: 301 LTERKTSISSSEEFLRKEGRSTTTSLVLLTEGLENARFRSFFNKWPQTVESSLYNEGREK 360
Query: 308 VAALLKRQGLDVKGLVKADPVKEEPQPYIDCTGHLQV 344
VAAL K++G DV+ L P +E+ Y +C +L+V
Sbjct: 361 VAALFKQKGYDVEEL----PDEEDDPLYTNCRDNLKV 393
Score = 70.9 bits (172), Expect = 1e-12
Identities = 70/303 (23%), Positives = 128/303 (42%), Gaps = 29/303 (9%)
Query: 11 HDIHYWLGKDTSQDEAGAAAIKTVELDAVLGGRAVQYREVQGHETQKFLSYFKPCIIPQE 70
H ++ W+G ++ Q + A + G +V QG+E +F F+ ++ +
Sbjct: 432 HLLYVWIGCESIQQDRADAITNASAIVGTTKGESVLCHIYQGNEPSRFFPMFQSLVV-FK 490
Query: 71 GGAASGFKHVEAE---------EHKTRLFVCKGKHVVYVK--EVPFARSSLNHDDIFILD 119
GG + +K + AE E+K LF G ++ +V +SLN +IL
Sbjct: 491 GGLSRRYKVLLAEKEKIGEEYNENKASLFRVVGTSPRNMQAIQVNLVATSLNSSYSYILQ 550
Query: 120 TESKIFQFNGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLF 179
+ F + G SS + + L+ + Y DT C+ I +G +E+ FW L
Sbjct: 551 YGASAFTWIGKLSSDSDH-EVLDRMLYFLDT----SCQPIYIREG-----NETDTFWNLL 600
Query: 180 GGFAPLPRKTVSDDDKTIDSHPPKLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLEV 239
GG + P++ + K I+ + E + ++ L T ++LDC EV
Sbjct: 601 GGKSEYPKE--KEMRKQIEEPHLFTCSCSSDVLKVKEIYNFVQDDLTTEDVFLLDCQSEV 658
Query: 240 FVWIGRNTSLDERKSA-----SGSTDELVSSTNRPKSQIIRVMEGFETVMFRSKFDSWPQ 294
+VWIG N+++ ++ A +++ ++ + V EG E F F+ P+
Sbjct: 659 YVWIGSNSNIKSKEEALTLGLKFLEMDILEEGLTMRTPVYVVTEGHEPPFFTRFFEWVPE 718
Query: 295 TTN 297
N
Sbjct: 719 KAN 721
>At4g01810 protein transport factor like protein
Length = 880
Score = 35.8 bits (81), Expect = 0.037
Identities = 15/32 (46%), Positives = 22/32 (67%)
Query: 225 LDTNKCYILDCGLEVFVWIGRNTSLDERKSAS 256
+ ++K ILD G +VF+W+G S DE KSA+
Sbjct: 754 MQSDKAVILDHGTDVFIWLGAELSADEVKSAA 785
>At3g47500 H-protein promoter binding factor-2a
Length = 448
Score = 30.8 bits (68), Expect = 1.2
Identities = 22/71 (30%), Positives = 29/71 (39%), Gaps = 9/71 (12%)
Query: 234 DCGLEVFVWIGRNTSLDERKSASGSTDELVSSTNRPKSQIIRVMEGFETVMFRSKFDSWP 293
DC V N S+DE ++ SGS E + N + M G+ + WP
Sbjct: 246 DCSSGSSVTTSNNHSVDESRAQSGSVVEAQMNNNNNNN-----MNGYACI----PGVPWP 296
Query: 294 QTTNAAMPEDG 304
T N AMP G
Sbjct: 297 YTWNPAMPPPG 307
>At3g63440 cytokinin oxidase -like protein
Length = 504
Score = 30.0 bits (66), Expect = 2.0
Identities = 20/81 (24%), Positives = 38/81 (46%), Gaps = 3/81 (3%)
Query: 97 VVYVKEVPFARSSLNHDDIFILDTESKI-FQFNGSNSSIQERAKALEVVQYIKDTYHDGK 155
V++ K V S++ H I+++ T S++ G S+Q +A+ + ++ H K
Sbjct: 62 VLHPKSVSDIASTIRH--IWMMGTHSQLTVAARGRGHSLQGQAQTRHGIVIHMESLHPQK 119
Query: 156 CEVASIEDGRLMADSESGEFW 176
+V S++ D GE W
Sbjct: 120 LQVYSVDSPAPYVDVSGGELW 140
>At3g60350 Arm repeat containing protein - like
Length = 928
Score = 30.0 bits (66), Expect = 2.0
Identities = 17/58 (29%), Positives = 30/58 (51%)
Query: 128 NGSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFWGLFGGFAPL 185
NG+N+++ + A ALE + + + H+G + A+ L D ++ E FGG L
Sbjct: 590 NGNNAAVGQEAGALEALVQLTQSPHEGVKQEAAGALWNLAFDDKNRESIAAFGGVEAL 647
>At3g15120 chaperone-like ATPase
Length = 1954
Score = 30.0 bits (66), Expect = 2.0
Identities = 24/84 (28%), Positives = 38/84 (44%), Gaps = 11/84 (13%)
Query: 129 GSNSSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGEFW-GLFG----GFA 183
G+ S+QE+A +L+ + G S+ED ADS +G+ W G+ G
Sbjct: 1714 GTVISLQEKADSLD------NPNGSGDSNSISLEDPHKSADSNNGKAWDGVHGLESANNM 1767
Query: 184 PLPRKTVSDDDKTIDSHPPKLLCV 207
P P + V +T P L+C+
Sbjct: 1768 PEPVEQVETTGRTNPQDDPSLVCL 1791
>At3g55270 MAP kinase phosphatase (MKP1)
Length = 732
Score = 29.6 bits (65), Expect = 2.6
Identities = 16/67 (23%), Positives = 32/67 (46%), Gaps = 1/67 (1%)
Query: 225 LDTNKCYILDCGLEVFVWIGRNTSLDERKSASGSTDELVSSTNRPKSQIIRVMEGFETVM 284
LD+ +I+ +++W+GR K A + + ++ + ++ I+ V EG E V
Sbjct: 277 LDSRGAFIIQLPSAIYIWVGRQCETIMEKDAKAAVCQ-IARYEKVEAPIMVVREGDEPVY 335
Query: 285 FRSKFDS 291
+ F S
Sbjct: 336 YWDAFAS 342
>At3g07100 putative Sec24-like COPII protein
Length = 1054
Score = 29.6 bits (65), Expect = 2.6
Identities = 15/38 (39%), Positives = 19/38 (49%)
Query: 220 LTKELLDTNKCYILDCGLEVFVWIGRNTSLDERKSASG 257
L E LD+ YI D G + +W GR S D K+ G
Sbjct: 932 LAAESLDSRGLYIYDDGFRLVLWFGRMLSPDIAKNLLG 969
>At1g10390 unknown protein
Length = 1041
Score = 29.3 bits (64), Expect = 3.4
Identities = 25/122 (20%), Positives = 52/122 (42%), Gaps = 20/122 (16%)
Query: 250 DERKSASGSTDELVSSTNRPKSQIIRVMEGF----ETVMFRSKFDSWPQTTNAAMPE--- 302
DE S++ D L P++ +IR ++ + ++++ + + + P N P+
Sbjct: 750 DEESSSTPKADALFIPRENPRALVIRPVQQWSSRDKSILPKEQRPTAPLHDNGKSPDMAT 809
Query: 303 -------DGRGKVAALLKRQGLDVK------GLVKADPVKEEPQPYIDCTGHLQVLEYIL 349
+G G++ A +R V G ++D E+ +PY +GH I+
Sbjct: 810 DAANHDRNGNGELGATGERIHTSVNANQKPNGTTRSDQASEKERPYKTLSGHRAGEAAIV 869
Query: 350 YQ 351
Y+
Sbjct: 870 YE 871
>At1g65430 unknown protein
Length = 567
Score = 28.9 bits (63), Expect = 4.5
Identities = 22/93 (23%), Positives = 39/93 (41%), Gaps = 1/93 (1%)
Query: 249 LDERKSASGSTDELVSSTNRPKSQIIRVMEGFETVMFRSKFDSWPQTTNAAMPEDGRGKV 308
LD +K + ++L +P+SQ+ ++E + ++ + W +P+ GK
Sbjct: 381 LDLKKMQTDDIEKLSDIQCQPESQLKFIIEAWLQIVECRRVLKWTYAYGFYIPDQEHGKR 440
Query: 309 AALLKRQGLDVKGLVKADPVKE-EPQPYIDCTG 340
QG GL + E E PY+D G
Sbjct: 441 VFFEYLQGEAESGLERLHQCAEKELLPYLDAKG 473
>At1g05320 unknown protein
Length = 841
Score = 28.1 bits (61), Expect = 7.6
Identities = 51/239 (21%), Positives = 98/239 (40%), Gaps = 30/239 (12%)
Query: 66 IIPQEGGAASGFKHVEAEEHKTRLFVCKGKHVVYVKEVP-----FARSSLNHDDIFILDT 120
I+ +E G S + E +E ++L + E+ + H+DI + T
Sbjct: 190 IVAEEEGKKSSIQMQEYQEKVSKLESSLNQSSARNSELEEDLRIALQKGAEHEDIGNVST 249
Query: 121 ESKI-----FQFNGSN-SSIQERAKALEVVQYIKDTYHDGKCEVASIEDGRLMADSESGE 174
+ + FQ + +E+ K LE +Q +K++ + VA +E R ++++ +
Sbjct: 250 KRSVELQGLFQTSQLKLEKAEEKLKDLEAIQ-VKNSSLEATLSVA-MEKERDLSENLNAV 307
Query: 175 FWGLFGGFAPLPRKTVSDDDKTIDS------HPPKLLCVEKGKAEPFETDSLTKELLDTN 228
L L ++ D+ T S H L V+K + D+ K L + +
Sbjct: 308 MEKLKSSEERLEKQAREIDEATTRSIELEALHKHSELKVQKTMEDFSSRDTEAKSLTEKS 367
Query: 229 KCYILDCGLEVFVW-------IGRNTSLDERKSASGSTDELVSSTNRPKSQIIRVMEGF 280
K D ++ V+ G++ SL E S + +EL++ TN I+ +EG+
Sbjct: 368 K----DLEEKIRVYEGKLAEACGQSLSLQEELDQSSAENELLADTNNQLKIKIQELEGY 422
>At5g61990 unknown protein
Length = 974
Score = 27.7 bits (60), Expect = 10.0
Identities = 23/80 (28%), Positives = 36/80 (44%), Gaps = 15/80 (18%)
Query: 94 GKHVVYVKEVPFARSSLNHDDIFILDTESKIFQFNGSNSSIQERAKALEVVQYIKDTYHD 153
GK V++ E F ++LN D L ES I K L ++Y D D
Sbjct: 240 GKDVLFKTEKEFRTATLNVDGALKLK-ESMI-------------CKGLVPLKYTYDVLID 285
Query: 154 GKCEVASIEDGR-LMADSES 172
G C++ +ED + L+ + +S
Sbjct: 286 GLCKIKRLEDAKSLLVEMDS 305
>At4g09320 nucleoside-diphosphate kinase
Length = 169
Score = 27.7 bits (60), Expect = 10.0
Identities = 16/51 (31%), Positives = 23/51 (44%)
Query: 203 KLLCVEKGKAEPFETDSLTKELLDTNKCYILDCGLEVFVWIGRNTSLDERK 253
KL+ VE+ AE D +K YI+ + +W G+N L RK
Sbjct: 56 KLISVERSFAEKHYEDLSSKSFFSGLVDYIVSGPVVAMIWEGKNVVLTGRK 106
>At2g33830 putative auxin-regulated protein
Length = 108
Score = 27.7 bits (60), Expect = 10.0
Identities = 11/31 (35%), Positives = 17/31 (54%)
Query: 292 WPQTTNAAMPEDGRGKVAALLKRQGLDVKGL 322
W +T PE G G++ + Q LD+KG+
Sbjct: 2 WDETVAGPKPEHGLGRLRNKITTQPLDIKGV 32
>At1g57640
Length = 1444
Score = 27.7 bits (60), Expect = 10.0
Identities = 17/72 (23%), Positives = 30/72 (41%), Gaps = 3/72 (4%)
Query: 47 YREVQGHETQKFLSYFKPCIIPQEGGAASGFKHVEAEEHKTRLFVCKGKHVVYVKEVPFA 106
Y Q H+ KF + + C+ G++ + EE K F + E P++
Sbjct: 720 YAHNQNHKGDKFAARSRRCVFVGYPHGQKGWRLFDLEEQK---FFVSRDVIFQETEFPYS 776
Query: 107 RSSLNHDDIFIL 118
+ S N +D +L
Sbjct: 777 KMSCNEEDERVL 788
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.135 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,112,354
Number of Sequences: 26719
Number of extensions: 354544
Number of successful extensions: 903
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 847
Number of HSP's gapped (non-prelim): 30
length of query: 355
length of database: 11,318,596
effective HSP length: 100
effective length of query: 255
effective length of database: 8,646,696
effective search space: 2204907480
effective search space used: 2204907480
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC125477.10


