

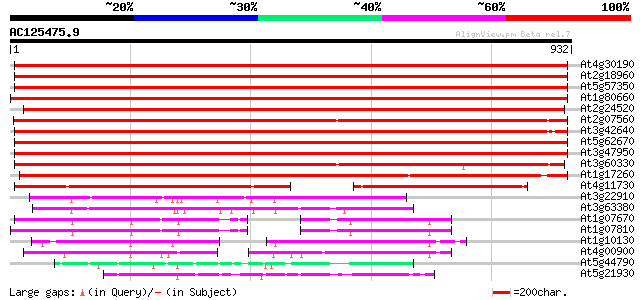
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125475.9 - phase: 0 /pseudo
(932 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g30190 H+-transporting ATPase type 2, plasma membrane 1597 0.0
At2g18960 plasma membrane proton ATPase (PMA) 1590 0.0
At5g57350 plasma membrane ATPase 3 (proton pump) (sp|P20431) 1575 0.0
At1g80660 H+-transporting ATPase 9 (aha9) 1555 0.0
At2g24520 putative plasma membrane proton ATPase 1534 0.0
At2g07560 pseudogene 1527 0.0
At3g42640 plasma membrane H+-ATPase-like protein 1518 0.0
At5g62670 plasma membrane proton ATPase-like 1502 0.0
At3g47950 H+-transporting ATPase - like protein 1486 0.0
At3g60330 plasma membrane H+-ATPase - like 1362 0.0
At1g17260 H+-transporting ATPase AHA10 1318 0.0
At4g11730 H+-transporting ATPase - like protein 607 e-173
At3g22910 calmodulin-stimulated calcium-ATPase, putative 199 5e-51
At3g63380 Ca2+-transporting ATPase -like protein 175 9e-44
At1g07670 endoplasmic reticulum-type calcium-transporting ATPase... 141 1e-33
At1g07810 ER-type Ca2+-pump protein 141 2e-33
At1g10130 putative calcium ATPase 137 3e-32
At4g00900 Ca2+-transporting ATPase - like protein 127 4e-29
At5g44790 COPPER-TRANSPORTING ATPASE RAN1 (RESPONSIVE-TO-ANTAGON... 121 2e-27
At5g21930 metal-transporting ATPase - like protein 111 2e-24
>At4g30190 H+-transporting ATPase type 2, plasma membrane
Length = 948
Score = 1597 bits (4136), Expect = 0.0
Identities = 797/921 (86%), Positives = 860/921 (92%), Gaps = 2/921 (0%)
Query: 8 SLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKILKFL 67
SLE IKNETVDLE+IP+EEVF+QLKC++EGL+++EG +R+QIFGPNKLEEKK+SK+LKFL
Sbjct: 3 SLEDIKNETVDLEKIPIEEVFQQLKCSREGLTTQEGEDRIQIFGPNKLEEKKESKLLKFL 62
Query: 68 GFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNAAA 127
GFMWNPLSWVME AA+MAI LANG+G+PPDWQDFVGIICLLVINSTISFIEENNAGNAAA
Sbjct: 63 GFMWNPLSWVMEMAAIMAIALANGDGRPPDWQDFVGIICLLVINSTISFIEENNAGNAAA 122
Query: 128 ALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSALT 187
ALMAGLAPKTKVLRDGKWSEQEAAILVPGDI+SIKLGDI+PADARLLEGDPLK+DQSALT
Sbjct: 123 ALMAGLAPKTKVLRDGKWSEQEAAILVPGDIVSIKLGDIIPADARLLEGDPLKVDQSALT 182
Query: 188 GESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLTA 247
GESLPVT++PG EV+SGSTCKQGE+EAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLTA
Sbjct: 183 GESLPVTKHPGQEVFSGSTCKQGEIEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLTA 242
Query: 248 IGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGSH 307
IGNFCICSIA+GM+ EIIVMYPIQ RKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGSH
Sbjct: 243 IGNFCICSIAIGMVIEIIVMYPIQRRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGSH 302
Query: 308 RLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLLA 367
RLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNL+EVF KGV+K+ V+L A
Sbjct: 303 RLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLVEVFCKGVEKDQVLLFA 362
Query: 368 ARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGNGNWHRAS 427
A ASRVENQDAIDAA+VG LADPKEARAG+RE+HFLPFNPVDKRTALTYIDG+GNWHR S
Sbjct: 363 AMASRVENQDAIDAAMVGMLADPKEARAGIREVHFLPFNPVDKRTALTYIDGSGNWHRVS 422
Query: 428 KGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQFVGL 487
KGAPEQI++L K D + + +IIDK+AERGLRSLAVARQ VPEKTKESPGAPW+FVGL
Sbjct: 423 KGAPEQILELAKASNDLSKKVLSIIDKYAERGLRSLAVARQVVPEKTKESPGAPWEFVGL 482
Query: 488 LSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLGQDK 547
L LFDPPRHDSAETIRRAL+LGVNVKMITGDQLAI KETGRRLGMGTNMYPS+ LLG K
Sbjct: 483 LPLFDPPRHDSAETIRRALNLGVNVKMITGDQLAIGKETGRRLGMGTNMYPSSALLGTHK 542
Query: 548 DANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRADIG 607
DAN+A++PVEELIEKADGFAGVFPEHKYEIV+KLQERKHI GMTGDGVNDAPALK+ADIG
Sbjct: 543 DANLASIPVEELIEKADGFAGVFPEHKYEIVKKLQERKHIVGMTGDGVNDAPALKKADIG 602
Query: 608 IAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFMF 667
IAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFM
Sbjct: 603 IAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFML 662
Query: 668 IALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGGYLAL 727
IALIW+FDFS FMVLIIAILNDGTIMTISKDRV PSP PDSWKLKEIFATG+VLGGY A+
Sbjct: 663 IALIWEFDFSAFMVLIIAILNDGTIMTISKDRVKPSPTPDSWKLKEIFATGVVLGGYQAI 722
Query: 728 MTVIFFWAMKENDFFPDKFGVRKL--NHDEMMSALYLQVSIVSQALIFVTRSRGWSFLER 785
MTVIFFWA + DFF D FGVR + N+ E+M A+YLQVSI+SQALIFVTRSR WSF+ER
Sbjct: 723 MTVIFFWAAHKTDFFSDTFGVRSIRDNNHELMGAVYLQVSIISQALIFVTRSRSWSFVER 782
Query: 786 PGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKFAIR 845
PGALL+IAF IAQLIAT+IAVYANW FAK++GIGWGWAGVIWLYSIV Y PLDV KFAIR
Sbjct: 783 PGALLMIAFLIAQLIATLIAVYANWEFAKIRGIGWGWAGVIWLYSIVTYFPLDVFKFAIR 842
Query: 846 YILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKSSYRE 905
YILSGKAW NL +NKTAFT KKDYGKEEREAQWA AQRTLHGLQP E+ IF EK SYRE
Sbjct: 843 YILSGKAWLNLFENKTAFTMKKDYGKEEREAQWALAQRTLHGLQPKEAVNIFPEKGSYRE 902
Query: 906 LSEIAEQAKRRAEVARLIPFH 926
LSEIAEQAKRRAE+ARL H
Sbjct: 903 LSEIAEQAKRRAEIARLRELH 923
>At2g18960 plasma membrane proton ATPase (PMA)
Length = 949
Score = 1590 bits (4117), Expect = 0.0
Identities = 792/920 (86%), Positives = 857/920 (93%), Gaps = 2/920 (0%)
Query: 9 LEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKILKFLG 68
LE IKNETVDLE+IP+EEVF+QLKCT+EGL+++EG +R+ IFGPNKLEEKK+SKILKFLG
Sbjct: 4 LEDIKNETVDLEKIPIEEVFQQLKCTREGLTTQEGEDRIVIFGPNKLEEKKESKILKFLG 63
Query: 69 FMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNAAAA 128
FMWNPLSWVMEAAALMAI LANG+ +PPDWQDFVGIICLLVINSTISFIEENNAGNAAAA
Sbjct: 64 FMWNPLSWVMEAAALMAIALANGDNRPPDWQDFVGIICLLVINSTISFIEENNAGNAAAA 123
Query: 129 LMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSALTG 188
LMAGLAPKTKVLRDGKWSEQEAAILVPGDI+SIKLGDI+PADARLLEGDPLK+DQSALTG
Sbjct: 124 LMAGLAPKTKVLRDGKWSEQEAAILVPGDIVSIKLGDIIPADARLLEGDPLKVDQSALTG 183
Query: 189 ESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLTAI 248
ESLPVT++PG EV+SGSTCKQGE+EAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLT+I
Sbjct: 184 ESLPVTKHPGQEVFSGSTCKQGEIEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLTSI 243
Query: 249 GNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGSHR 308
GNFCICSIA+G+ EI+VMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGSHR
Sbjct: 244 GNFCICSIAIGIAIEIVVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGSHR 303
Query: 309 LSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLLAA 368
LSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNL+EVF KGV+K+ V+L AA
Sbjct: 304 LSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLVEVFCKGVEKDQVLLFAA 363
Query: 369 RASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGNGNWHRASK 428
ASRVENQDAIDAA+VG LADPKEARAG+RE+HFLPFNPVDKRTALTYID +GNWHR SK
Sbjct: 364 MASRVENQDAIDAAMVGMLADPKEARAGIREVHFLPFNPVDKRTALTYIDSDGNWHRVSK 423
Query: 429 GAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQFVGLL 488
GAPEQI+DL R D ++ + + IDK+AERGLRSLAVARQ VPEKTKESPG PW+FVGLL
Sbjct: 424 GAPEQILDLANARPDLRKKVLSCIDKYAERGLRSLAVARQVVPEKTKESPGGPWEFVGLL 483
Query: 489 SLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLGQDKD 548
LFDPPRHDSAETIRRAL+LGVNVKMITGDQLAI KETGRRLGMGTNMYPSA LLG DKD
Sbjct: 484 PLFDPPRHDSAETIRRALNLGVNVKMITGDQLAIGKETGRRLGMGTNMYPSAALLGTDKD 543
Query: 549 ANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRADIGI 608
+NIA++PVEELIEKADGFAGVFPEHKYEIV+KLQERKHI GMTGDGVNDAPALK+ADIGI
Sbjct: 544 SNIASIPVEELIEKADGFAGVFPEHKYEIVKKLQERKHIVGMTGDGVNDAPALKKADIGI 603
Query: 609 AVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFMFI 668
AVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFM I
Sbjct: 604 AVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFMLI 663
Query: 669 ALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGGYLALM 728
ALIW+FDFS FMVLIIAILNDGTIMTISKDRV PSP PDSWKLKEIFATGIVLGGY A+M
Sbjct: 664 ALIWEFDFSAFMVLIIAILNDGTIMTISKDRVKPSPTPDSWKLKEIFATGIVLGGYQAIM 723
Query: 729 TVIFFWAMKENDFFPDKFGVRKL--NHDEMMSALYLQVSIVSQALIFVTRSRGWSFLERP 786
+VIFFWA + DFF DKFGVR + N+DE+M A+YLQVSI+SQALIFVTRSR WSF+ERP
Sbjct: 724 SVIFFWAAHKTDFFSDKFGVRSIRDNNDELMGAVYLQVSIISQALIFVTRSRSWSFVERP 783
Query: 787 GALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKFAIRY 846
GALL+IAF IAQL+AT+IAVYA+W FAKV+GIGWGWAGVIW+YSIV Y P D++KFAIRY
Sbjct: 784 GALLMIAFVIAQLVATLIAVYADWTFAKVKGIGWGWAGVIWIYSIVTYFPQDILKFAIRY 843
Query: 847 ILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKSSYREL 906
ILSGKAW +L DN+TAFTTKKDYG EREAQWA AQRTLHGLQP E IF EK SYREL
Sbjct: 844 ILSGKAWASLFDNRTAFTTKKDYGIGEREAQWAQAQRTLHGLQPKEDVNIFPEKGSYREL 903
Query: 907 SEIAEQAKRRAEVARLIPFH 926
SEIAEQAKRRAE+ARL H
Sbjct: 904 SEIAEQAKRRAEIARLRELH 923
>At5g57350 plasma membrane ATPase 3 (proton pump) (sp|P20431)
Length = 949
Score = 1575 bits (4077), Expect = 0.0
Identities = 779/920 (84%), Positives = 850/920 (91%), Gaps = 2/920 (0%)
Query: 9 LEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKILKFLG 68
LE I NE VDLE+IP+EEVF+QLKC++EGLS EG NRLQIFGPNKLEEKK+SK+LKFLG
Sbjct: 5 LEDIVNENVDLEKIPIEEVFQQLKCSREGLSGAEGENRLQIFGPNKLEEKKESKLLKFLG 64
Query: 69 FMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNAAAA 128
FMWNPLSWVMEAAA+MAI LANG GKPPDWQDFVGI+CLLVINSTISF+EENNAGNAAAA
Sbjct: 65 FMWNPLSWVMEAAAIMAIALANGGGKPPDWQDFVGIVCLLVINSTISFVEENNAGNAAAA 124
Query: 129 LMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSALTG 188
LMAGLAPKTKVLRDGKWSEQEA+ILVPGDI+SIKLGDI+PADARLLEGDPLK+DQSALTG
Sbjct: 125 LMAGLAPKTKVLRDGKWSEQEASILVPGDIVSIKLGDIIPADARLLEGDPLKVDQSALTG 184
Query: 189 ESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLTAI 248
ESLP T+ PG+EV+SGSTCKQGE+EAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLTAI
Sbjct: 185 ESLPATKGPGEEVFSGSTCKQGEIEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLTAI 244
Query: 249 GNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGSHR 308
GNFCICSIAVG+ EI+VMYPIQ R YRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGSH+
Sbjct: 245 GNFCICSIAVGIAIEIVVMYPIQRRHYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGSHK 304
Query: 309 LSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLLAA 368
LSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEV+ KGV+K+ V+L AA
Sbjct: 305 LSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVYCKGVEKDEVLLFAA 364
Query: 369 RASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGNGNWHRASK 428
RASRVENQDAIDAA+VG LADPKEARAG+REIHFLPFNPVDKRTALT+ID NGNWHR SK
Sbjct: 365 RASRVENQDAIDAAMVGMLADPKEARAGIREIHFLPFNPVDKRTALTFIDSNGNWHRVSK 424
Query: 429 GAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQFVGLL 488
GAPEQI+DLC R D ++ +H+ IDK+AERGLRSLAV+RQ VPEKTKES G+PW+FVG+L
Sbjct: 425 GAPEQILDLCNARADLRKRVHSTIDKYAERGLRSLAVSRQTVPEKTKESSGSPWEFVGVL 484
Query: 489 SLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLGQDKD 548
LFDPPRHDSAETIRRAL LGVNVKMITGDQLAIAKETGRRLGMG+NMYPS++LLG+ KD
Sbjct: 485 PLFDPPRHDSAETIRRALDLGVNVKMITGDQLAIAKETGRRLGMGSNMYPSSSLLGKHKD 544
Query: 549 ANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRADIGI 608
+A +PVE+LIEKADGFAGVFPEHKYEIV+KLQERKHICGMTGDGVNDAPALK+ADIGI
Sbjct: 545 EAMAHIPVEDLIEKADGFAGVFPEHKYEIVKKLQERKHICGMTGDGVNDAPALKKADIGI 604
Query: 609 AVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFMFI 668
AVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFM I
Sbjct: 605 AVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFMLI 664
Query: 669 ALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGGYLALM 728
ALIWKFDFSPFMVLIIAILNDGTIMTISKDRV PSP PDSWKLKEIFATG+VLGGY+A+M
Sbjct: 665 ALIWKFDFSPFMVLIIAILNDGTIMTISKDRVKPSPTPDSWKLKEIFATGVVLGGYMAIM 724
Query: 729 TVIFFWAMKENDFFPDKFGVRKL--NHDEMMSALYLQVSIVSQALIFVTRSRGWSFLERP 786
TV+FFWA + DFFP F VR L + EMMSALYLQVSIVSQALIFVTRSR WSF ERP
Sbjct: 725 TVVFFWAAYKTDFFPRTFHVRDLRGSEHEMMSALYLQVSIVSQALIFVTRSRSWSFTERP 784
Query: 787 GALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKFAIRY 846
G L+IAF++AQLIAT IAVY NW FA+++GIGWGWAGVIWLYSIVFY PLD+MKFAIRY
Sbjct: 785 GYFLLIAFWVAQLIATAIAVYGNWEFARIKGIGWGWAGVIWLYSIVFYFPLDIMKFAIRY 844
Query: 847 ILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKSSYREL 906
IL+G AW N++DN+TAFTTK++YG EEREAQWAHAQRTLHGLQ E++ + E+ YREL
Sbjct: 845 ILAGTAWKNIIDNRTAFTTKQNYGIEEREAQWAHAQRTLHGLQNTETANVVPERGGYREL 904
Query: 907 SEIAEQAKRRAEVARLIPFH 926
SEIA QAKRRAE+ARL H
Sbjct: 905 SEIANQAKRRAEIARLRELH 924
>At1g80660 H+-transporting ATPase 9 (aha9)
Length = 954
Score = 1555 bits (4027), Expect = 0.0
Identities = 769/928 (82%), Positives = 847/928 (90%), Gaps = 2/928 (0%)
Query: 1 MAESKSISLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKD 60
MA +K S + IKNE +DLE+IP+EEV QL+CT+EGL+S+EG RL+IFGPNKLEEKK+
Sbjct: 1 MAGNKDSSWDDIKNEGIDLEKIPIEEVLTQLRCTREGLTSDEGQTRLEIFGPNKLEEKKE 60
Query: 61 SKILKFLGFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEEN 120
+K+LKFLGFMWNPLSWVME AA+MAI LANG G+PPDWQDFVGI LL+INSTISFIEEN
Sbjct: 61 NKVLKFLGFMWNPLSWVMELAAIMAIALANGGGRPPDWQDFVGITVLLIINSTISFIEEN 120
Query: 121 NAGNAAAALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLK 180
NAGNAAAALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPAD RLL+GDPLK
Sbjct: 121 NAGNAAAALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADGRLLDGDPLK 180
Query: 181 IDQSALTGESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGH 240
IDQSALTGESLPVT++PG EVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQ GH
Sbjct: 181 IDQSALTGESLPVTKHPGQEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQEGH 240
Query: 241 FQKVLTAIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSV 300
FQKVLTAIGNFCICSIA+GML EI+VMYPIQ R YRDGIDNLLVLLIGGIPIAMPTVLSV
Sbjct: 241 FQKVLTAIGNFCICSIAIGMLIEIVVMYPIQKRAYRDGIDNLLVLLIGGIPIAMPTVLSV 300
Query: 301 TMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDK 360
TMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKL+VDK+++EVF K +DK
Sbjct: 301 TMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLTVDKSMVEVFVKDLDK 360
Query: 361 EHVMLLAARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGN 420
+ +++ AARASRVENQDAIDA IVG L DP+EAR G+ E+HF PFNPVDKRTA+TYID N
Sbjct: 361 DQLLVNAARASRVENQDAIDACIVGMLGDPREAREGITEVHFFPFNPVDKRTAITYIDAN 420
Query: 421 GNWHRASKGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGA 480
GNWHR SKGAPEQI++LC LRED + H IIDKFA+RGLRSLAV RQ V EK K SPG
Sbjct: 421 GNWHRVSKGAPEQIIELCNLREDASKRAHDIIDKFADRGLRSLAVGRQTVSEKDKNSPGE 480
Query: 481 PWQFVGLLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSA 540
PWQF+GLL LFDPPRHDSAETIRRAL LGVNVKMITGDQLAI KETGRRLGMGTNMYPS+
Sbjct: 481 PWQFLGLLPLFDPPRHDSAETIRRALDLGVNVKMITGDQLAIGKETGRRLGMGTNMYPSS 540
Query: 541 TLLGQDKDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPA 600
LLGQDKD +IA+LPV+ELIEKADGFAGVFPEHKYEIV++LQE KHICGMTGDGVNDAPA
Sbjct: 541 ALLGQDKDESIASLPVDELIEKADGFAGVFPEHKYEIVKRLQEMKHICGMTGDGVNDAPA 600
Query: 601 LKRADIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIR 660
LKRADIGIAVADATDAAR ASDIVLTEPGLSVI+SAVLTSRAIFQRMKNYTIYAVSITIR
Sbjct: 601 LKRADIGIAVADATDAARSASDIVLTEPGLSVIVSAVLTSRAIFQRMKNYTIYAVSITIR 660
Query: 661 IVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIV 720
IV GFM +ALIWKFDFSPFMVLI+AILNDGTIMTISKDRV PSPLPDSWKLKEIFATG+V
Sbjct: 661 IVMGFMLLALIWKFDFSPFMVLIVAILNDGTIMTISKDRVKPSPLPDSWKLKEIFATGVV 720
Query: 721 LGGYLALMTVIFFWAMKENDFFPDKFGVRKL--NHDEMMSALYLQVSIVSQALIFVTRSR 778
LG YLA+MTV+FFWA + DFF KFGVR + N E+ +A+YLQVSIVSQALIFVTRSR
Sbjct: 721 LGTYLAVMTVVFFWAAESTDFFSAKFGVRSISGNPHELTAAVYLQVSIVSQALIFVTRSR 780
Query: 779 GWSFLERPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLD 838
WS++ERPG L+ AFF+AQLIAT+IAVYANW FA+++GIGWGWAGVIWLYSIVFYIPLD
Sbjct: 781 SWSYVERPGFWLISAFFMAQLIATLIAVYANWNFARIRGIGWGWAGVIWLYSIVFYIPLD 840
Query: 839 VMKFAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFN 898
++KF IRY LSG+AW+N+++NKTAFT+KKDYGK EREAQWA AQRTLHGLQP ++S +FN
Sbjct: 841 ILKFIIRYSLSGRAWDNVIENKTAFTSKKDYGKGEREAQWAQAQRTLHGLQPAQTSDMFN 900
Query: 899 EKSSYRELSEIAEQAKRRAEVARLIPFH 926
+KS+YRELSEIA+QAKRRAEVARL H
Sbjct: 901 DKSTYRELSEIADQAKRRAEVARLRERH 928
>At2g24520 putative plasma membrane proton ATPase
Length = 931
Score = 1534 bits (3972), Expect = 0.0
Identities = 749/901 (83%), Positives = 835/901 (92%), Gaps = 2/901 (0%)
Query: 24 VEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKILKFLGFMWNPLSWVMEAAAL 83
+EEVFE+LKCTK+GL++ E ++RL +FGPNKLEEKK+SK+LKFLGFMWNPLSWVME AAL
Sbjct: 1 MEEVFEELKCTKQGLTANEASHRLDVFGPNKLEEKKESKLLKFLGFMWNPLSWVMEVAAL 60
Query: 84 MAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNAAAALMAGLAPKTKVLRDG 143
MAI LANG G+PPDWQDFVGI+CLL+INSTISFIEENNAGNAAAALMAGLAPKTKVLRD
Sbjct: 61 MAIALANGGGRPPDWQDFVGIVCLLLINSTISFIEENNAGNAAAALMAGLAPKTKVLRDN 120
Query: 144 KWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSALTGESLPVTRNPGDEVYS 203
+WSEQEA+ILVPGD+ISIKLGDI+PADARLL+GDPLKIDQS+LTGES+PVT+NP DEV+S
Sbjct: 121 QWSEQEASILVPGDVISIKLGDIIPADARLLDGDPLKIDQSSLTGESIPVTKNPSDEVFS 180
Query: 204 GSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLTAIGNFCICSIAVGMLAE 263
GS CKQGE+EA+VIATGVHTFFGKAAHLVD+TNQ+GHFQKVLT+IGNFCICSIA+G++ E
Sbjct: 181 GSICKQGEIEAIVIATGVHTFFGKAAHLVDNTNQIGHFQKVLTSIGNFCICSIALGIIVE 240
Query: 264 IIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGSHRLSQQGAITKRMTAIE 323
++VMYPIQ R+YRDGIDNLLVLLIGGIPIAMP+VLSVTMA GSHRL QQGAITKRMTAIE
Sbjct: 241 LLVMYPIQRRRYRDGIDNLLVLLIGGIPIAMPSVLSVTMATGSHRLFQQGAITKRMTAIE 300
Query: 324 EMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLLAARASRVENQDAIDAAI 383
EMAGMDVLC DKTGTLTLNKL+VDKNL+EVF KGV KEHV LLAARASR+ENQDAIDAAI
Sbjct: 301 EMAGMDVLCCDKTGTLTLNKLTVDKNLVEVFAKGVGKEHVFLLAARASRIENQDAIDAAI 360
Query: 384 VGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGNGNWHRASKGAPEQIMDLCKLRED 443
VG LADPKEARAGVRE+HF PFNPVDKRTALTY+D +GNWHRASKGAPEQI++LC +ED
Sbjct: 361 VGMLADPKEARAGVREVHFFPFNPVDKRTALTYVDSDGNWHRASKGAPEQILNLCNCKED 420
Query: 444 TKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQFVGLLSLFDPPRHDSAETIR 503
+R +H +IDKFAERGLRSLAVARQEV EK K++PG PWQ VGLL LFDPPRHDSAETIR
Sbjct: 421 VRRKVHGVIDKFAERGLRSLAVARQEVLEKKKDAPGGPWQLVGLLPLFDPPRHDSAETIR 480
Query: 504 RALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLGQDKDANIAALPVEELIEKA 563
RAL+LGVNVKMITGDQLAI KETGRRLGMGTNMYPS+ LLGQ KD+++ ALPV+ELIEKA
Sbjct: 481 RALNLGVNVKMITGDQLAIGKETGRRLGMGTNMYPSSALLGQVKDSSLGALPVDELIEKA 540
Query: 564 DGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRADIGIAVADATDAARGASDI 623
DGFAGVFPEHKYEIV +LQ+R HICGMTGDGVNDAPALK+ADIGIAV DATDAARGASDI
Sbjct: 541 DGFAGVFPEHKYEIVHRLQQRNHICGMTGDGVNDAPALKKADIGIAVVDATDAARGASDI 600
Query: 624 VLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFMFIALIWKFDFSPFMVLI 683
VLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFMFIALIW+FDFSPFMVLI
Sbjct: 601 VLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFMFIALIWQFDFSPFMVLI 660
Query: 684 IAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGGYLALMTVIFFWAMKENDFFP 743
IAILNDGTIMTISKDR+ PSP PDSWKL++IF+TG+VLGGY ALMTV+FFW MK++DFF
Sbjct: 661 IAILNDGTIMTISKDRMKPSPQPDSWKLRDIFSTGVVLGGYQALMTVVFFWVMKDSDFFS 720
Query: 744 DKFGVRKLNH--DEMMSALYLQVSIVSQALIFVTRSRGWSFLERPGALLVIAFFIAQLIA 801
+ FGVR L+ ++MM+ALYLQVSI+SQALIFVTRSR WS+ E PG LL+ AF IAQL+A
Sbjct: 721 NYFGVRPLSQRPEQMMAALYLQVSIISQALIFVTRSRSWSYAECPGLLLLGAFVIAQLVA 780
Query: 802 TIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKFAIRYILSGKAWNNLLDNKT 861
T IAVYANW FA+++G GWGWAGVIWLYS + YIPLD++KF IRY+LSGKAW NLL+NKT
Sbjct: 781 TFIAVYANWSFARIEGAGWGWAGVIWLYSFLTYIPLDLLKFGIRYVLSGKAWLNLLENKT 840
Query: 862 AFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKSSYRELSEIAEQAKRRAEVAR 921
AFTTKKDYGKEEREAQWA AQRTLHGLQP E + IFNEK+SY ELS+IAEQAKRRAEV R
Sbjct: 841 AFTTKKDYGKEEREAQWAAAQRTLHGLQPAEKNNIFNEKNSYSELSQIAEQAKRRAEVVR 900
Query: 922 L 922
L
Sbjct: 901 L 901
>At2g07560 pseudogene
Length = 949
Score = 1527 bits (3953), Expect = 0.0
Identities = 759/922 (82%), Positives = 837/922 (90%), Gaps = 5/922 (0%)
Query: 7 ISLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKILKF 66
IS ++IK E VDLE+IPV+EVF+QLKC++EGLSSEEG NRLQIFG NKLEEK ++K LKF
Sbjct: 5 ISWDEIKKENVDLEKIPVDEVFQQLKCSREGLSSEEGRNRLQIFGANKLEEKVENKFLKF 64
Query: 67 LGFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNAA 126
LGFMWNPLSWVMEAAA+MAI LANG G+PPDWQDFVGI CLL+INSTISFIEENNAGNAA
Sbjct: 65 LGFMWNPLSWVMEAAAIMAIVLANGGGRPPDWQDFVGITCLLIINSTISFIEENNAGNAA 124
Query: 127 AALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSAL 186
AALMA LAPKTKVLRDG+W EQEAAILVPGD+ISIKLGDIVPADARLLEGDPLKIDQSAL
Sbjct: 125 AALMANLAPKTKVLRDGRWGEQEAAILVPGDLISIKLGDIVPADARLLEGDPLKIDQSAL 184
Query: 187 TGESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLT 246
TGESLP T++ GDEV+SGSTCKQGE+EAVVIATGVHTFFGKAAHLVDSTN VGHFQKVLT
Sbjct: 185 TGESLPATKHQGDEVFSGSTCKQGEIEAVVIATGVHTFFGKAAHLVDSTNNVGHFQKVLT 244
Query: 247 AIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGS 306
AIGNFCICSI +GML EII+MYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGS
Sbjct: 245 AIGNFCICSIGIGMLIEIIIMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGS 304
Query: 307 HRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLL 366
HRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKL+VDKNLIEVF K VDK++V+LL
Sbjct: 305 HRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLTVDKNLIEVFSKDVDKDYVILL 364
Query: 367 AARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGNGNWHRA 426
+ARASRVENQDAID +IV L DPKEARAG+ E+HFLPFNPV+KRTA+TYID NG WHR
Sbjct: 365 SARASRVENQDAIDTSIVNMLGDPKEARAGITEVHFLPFNPVEKRTAITYIDTNGEWHRC 424
Query: 427 SKGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQFVG 486
SKGAPEQI++LC L+ +TKR H IIDKFAERGLRSL VARQ VPEK KES G PW+FVG
Sbjct: 425 SKGAPEQIIELCDLKGETKRRAHEIIDKFAERGLRSLGVARQRVPEKDKESAGTPWEFVG 484
Query: 487 LLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLGQD 546
LL LFDPPRHDSAETIRRAL LGVNVKMITGDQLAI KETGRRLGMGTNMYPS++LL ++
Sbjct: 485 LLPLFDPPRHDSAETIRRALDLGVNVKMITGDQLAIGKETGRRLGMGTNMYPSSSLL-EN 543
Query: 547 KDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRADI 606
KD +PV+ELIEKADGFAGVFPEHKYEIVRKLQERKHI GMTGDGVNDAPALK+ADI
Sbjct: 544 KDDTTGGVPVDELIEKADGFAGVFPEHKYEIVRKLQERKHIVGMTGDGVNDAPALKKADI 603
Query: 607 GIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFM 666
GIAV DATDAAR ASDIVLTEPGLSVI+SAVLTSRAIFQRMKNYTIYAVSITIRIV GFM
Sbjct: 604 GIAVDDATDAARSASDIVLTEPGLSVIVSAVLTSRAIFQRMKNYTIYAVSITIRIVLGFM 663
Query: 667 FIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGGYLA 726
+ALIW+FDFSPFMVLIIAILNDGTIMTISKDRV PSP+PDSWKLKEIFATG+VLG Y+A
Sbjct: 664 LVALIWEFDFSPFMVLIIAILNDGTIMTISKDRVKPSPIPDSWKLKEIFATGVVLGTYMA 723
Query: 727 LMTVIFFWAMKENDFFPDKFGVRKL--NHDEMMSALYLQVSIVSQALIFVTRSRGWSFLE 784
L+TV+FFW + FF DKFGVR L +E+++ LYLQVSI+SQALIFVTRSR WSF+E
Sbjct: 724 LVTVVFFWLAHDTTFFSDKFGVRSLQGKDEELIAVLYLQVSIISQALIFVTRSRSWSFVE 783
Query: 785 RPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKFAI 844
RPG LL+IAFF+AQLIAT+IA YA+W FA+++G GWGW GVIW+YSIV YIPLD++KF
Sbjct: 784 RPGLLLLIAFFVAQLIATLIATYAHWEFARIKGCGWGWCGVIWIYSIVTYIPLDILKFIT 843
Query: 845 RYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKSSYR 904
RY LSGKAWNN+++N+TAFTTKKDYG+ EREAQWA AQRTLHGL+PPES +F + ++Y
Sbjct: 844 RYTLSGKAWNNMIENRTAFTTKKDYGRGEREAQWALAQRTLHGLKPPES--MFEDTATYT 901
Query: 905 ELSEIAEQAKRRAEVARLIPFH 926
ELSEIAEQAK+RAEVARL H
Sbjct: 902 ELSEIAEQAKKRAEVARLREVH 923
>At3g42640 plasma membrane H+-ATPase-like protein
Length = 948
Score = 1518 bits (3931), Expect = 0.0
Identities = 753/921 (81%), Positives = 836/921 (90%), Gaps = 6/921 (0%)
Query: 8 SLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKILKFL 67
S ++IK E VDLERIPVEEVFEQLKC+KEGLSS+EGA RL+IFG NKLEEK ++K LKFL
Sbjct: 6 SWDEIKKENVDLERIPVEEVFEQLKCSKEGLSSDEGAKRLEIFGANKLEEKSENKFLKFL 65
Query: 68 GFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNAAA 127
GFMWNPLSWVME+AA+MAI LANG GK PDWQDF+GI+ LL+INSTISFIEENNAGNAAA
Sbjct: 66 GFMWNPLSWVMESAAIMAIVLANGGGKAPDWQDFIGIMVLLIINSTISFIEENNAGNAAA 125
Query: 128 ALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSALT 187
ALMA LAPKTKVLRDGKW EQEA+ILVPGD+ISIKLGDIVPADARLLEGDPLKIDQSALT
Sbjct: 126 ALMANLAPKTKVLRDGKWGEQEASILVPGDLISIKLGDIVPADARLLEGDPLKIDQSALT 185
Query: 188 GESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLTA 247
GESLP T++PGDEV+SGSTCKQGE+EAVVIATGVHTFFGKAAHLVDSTN VGHFQKVLT+
Sbjct: 186 GESLPTTKHPGDEVFSGSTCKQGEIEAVVIATGVHTFFGKAAHLVDSTNNVGHFQKVLTS 245
Query: 248 IGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGSH 307
IGNFCICSI +GML EI++MYPIQHR YRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGSH
Sbjct: 246 IGNFCICSIGLGMLIEILIMYPIQHRTYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGSH 305
Query: 308 RLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLLA 367
RLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDK+LIEVF K +D + V+L+A
Sbjct: 306 RLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKSLIEVFPKNMDSDSVVLMA 365
Query: 368 ARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGNGNWHRAS 427
ARASR+ENQDAIDA+IVG L DPKEARAG+ E+HFLPFNPVDKRTA+TYID +G+WHR+S
Sbjct: 366 ARASRIENQDAIDASIVGMLGDPKEARAGITEVHFLPFNPVDKRTAITYIDESGDWHRSS 425
Query: 428 KGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQFVGL 487
KGAPEQI++LC L+ +TKR H +ID FAERGLRSL VA+Q VPEKTKES G+PW+FVGL
Sbjct: 426 KGAPEQIIELCNLQGETKRKAHEVIDGFAERGLRSLGVAQQTVPEKTKESDGSPWEFVGL 485
Query: 488 LSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLGQDK 547
L LFDPPRHDSAETIRRAL LGVNVKMITGDQLAI ETGRRLGMGTNMYPS +LLG K
Sbjct: 486 LPLFDPPRHDSAETIRRALELGVNVKMITGDQLAIGIETGRRLGMGTNMYPSTSLLGNSK 545
Query: 548 DANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRADIG 607
D ++ +P++ELIEKADGFAGVFPEHKYEIV+KLQERKHICGMTGDGVNDAPALK+ADIG
Sbjct: 546 DESLVGIPIDELIEKADGFAGVFPEHKYEIVKKLQERKHICGMTGDGVNDAPALKKADIG 605
Query: 608 IAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFMF 667
IAVADATDAAR ASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIV GFM
Sbjct: 606 IAVADATDAARSASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVLGFML 665
Query: 668 IALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGGYLAL 727
+ALIW+FDF+PFMVLIIAILNDGTIMTISKDRV PSP+PDSWKL EIFATG+VLG Y+AL
Sbjct: 666 VALIWRFDFAPFMVLIIAILNDGTIMTISKDRVKPSPVPDSWKLNEIFATGVVLGTYMAL 725
Query: 728 MTVIFFWAMKENDFFPDKFGVRKL--NHDEMMSALYLQVSIVSQALIFVTRSRGWSFLER 785
TV+FFW + DFF FGVR + N +E+M+ALYLQVSI+SQALIFVTRSR WSF+ER
Sbjct: 726 TTVLFFWLAHDTDFFSKTFGVRSIQGNEEELMAALYLQVSIISQALIFVTRSRSWSFVER 785
Query: 786 PGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKFAIR 845
PG LL+IAF IAQL+AT+IAVYANWGFA++ G GWGWAG IW+YSI+ YIPLD++KF IR
Sbjct: 786 PGFLLLIAFVIAQLVATLIAVYANWGFARIVGCGWGWAGGIWVYSIITYIPLDILKFIIR 845
Query: 846 YILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKSSYRE 905
Y L+GKAW+N+++ KTAFTTKKDYGK EREAQWA AQRTLHGL PPE+ +FN+ + E
Sbjct: 846 YALTGKAWDNMINQKTAFTTKKDYGKGEREAQWALAQRTLHGLPPPEA--MFNDNKN--E 901
Query: 906 LSEIAEQAKRRAEVARLIPFH 926
LSEIAEQAKRRAEVARL H
Sbjct: 902 LSEIAEQAKRRAEVARLRELH 922
>At5g62670 plasma membrane proton ATPase-like
Length = 956
Score = 1502 bits (3888), Expect = 0.0
Identities = 747/924 (80%), Positives = 830/924 (88%), Gaps = 7/924 (0%)
Query: 9 LEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKILKFLG 68
LE + ETVDLE +P+EEVFE L+C++EGL++E RL +FG NKLEEKK+SK LKFLG
Sbjct: 8 LEAVLKETVDLENVPIEEVFESLRCSREGLTTEAADERLALFGHNKLEEKKESKFLKFLG 67
Query: 69 FMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNAAAA 128
FMWNPLSWVMEAAA+MAI LANG GKPPDWQDFVGII LLVINSTISFIEENNAGNAAAA
Sbjct: 68 FMWNPLSWVMEAAAIMAIALANGGGKPPDWQDFVGIITLLVINSTISFIEENNAGNAAAA 127
Query: 129 LMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSALTG 188
LMA LAPK KVLRDG+W EQ+AAILVPGDIISIKLGDIVPADARLLEGDPLKIDQS+LTG
Sbjct: 128 LMARLAPKAKVLRDGRWGEQDAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSSLTG 187
Query: 189 ESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLTAI 248
ESLPVT+ PGD VYSGSTCKQGELEAVVIATGVHTFFGKAAHLVD+TN VGHFQ+VLTAI
Sbjct: 188 ESLPVTKGPGDGVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDTTNHVGHFQQVLTAI 247
Query: 249 GNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGSHR 308
GNFCICSIAVGM+ EI+VMYPIQHR YR GIDNLLVLLIGGIPIAMPTVLSVTMAIGSHR
Sbjct: 248 GNFCICSIAVGMIIEIVVMYPIQHRAYRPGIDNLLVLLIGGIPIAMPTVLSVTMAIGSHR 307
Query: 309 LSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLLAA 368
LSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKL+VDKNLIEVF KGVD + V+L+AA
Sbjct: 308 LSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLTVDKNLIEVFTKGVDADTVVLMAA 367
Query: 369 RASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGNGNWHRASK 428
+ASR+ENQDAIDAAIVG LADPKEARAGVRE+HFLPFNP DKRTALTYID +G HR SK
Sbjct: 368 QASRLENQDAIDAAIVGMLADPKEARAGVREVHFLPFNPTDKRTALTYIDSDGKMHRVSK 427
Query: 429 GAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQFVGLL 488
GAPEQI++L R + +R +HA+IDKFAERGLRSLAVA QEVPE TKES G PWQF+GL+
Sbjct: 428 GAPEQILNLAHNRAEIERRVHAVIDKFAERGLRSLAVAYQEVPEGTKESAGGPWQFMGLM 487
Query: 489 SLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLGQDKD 548
LFDPPRHDSAETIRRAL+LGVNVKMITGDQLAI KETGRRLGMGTNMYPS+ LLGQ KD
Sbjct: 488 PLFDPPRHDSAETIRRALNLGVNVKMITGDQLAIGKETGRRLGMGTNMYPSSALLGQHKD 547
Query: 549 ANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRADIGI 608
+I ALP+++LIEKADGFAGVFPEHKYEIV++LQ RKHICGMTGDGVNDAPALK+ADIGI
Sbjct: 548 ESIGALPIDDLIEKADGFAGVFPEHKYEIVKRLQARKHICGMTGDGVNDAPALKKADIGI 607
Query: 609 AVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFMFI 668
AVADATDAAR ASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIV GFM +
Sbjct: 608 AVADATDAARSASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVLGFMLL 667
Query: 669 ALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGGYLALM 728
ALIWKFDF PFMVLIIAILNDGTIMTISKDRV PSPLPDSWKL EIFATG+V G Y+A+M
Sbjct: 668 ALIWKFDFPPFMVLIIAILNDGTIMTISKDRVKPSPLPDSWKLSEIFATGVVFGSYMAMM 727
Query: 729 TVIFFWAMKENDFFPDKFGV---RKLNHDE---MMSALYLQVSIVSQALIFVTRSRGWSF 782
TVIFFWA + DFFP FGV K HD+ + SA+YLQVSI+SQALIFVTRSR WS+
Sbjct: 728 TVIFFWAAYKTDFFPRTFGVSTLEKTAHDDFRKLASAIYLQVSIISQALIFVTRSRSWSY 787
Query: 783 LERPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKF 842
+ERPG LLV+AF +AQL+AT+IAVYANW FA ++GIGWGWAGVIWLY+IVFYIPLD++KF
Sbjct: 788 VERPGMLLVVAFILAQLVATLIAVYANWSFAAIEGIGWGWAGVIWLYNIVFYIPLDIIKF 847
Query: 843 AIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKSS 902
IRY LSG+AW+ +++ + AFT +KD+GKE+RE QWAHAQRTLHGLQ P++ +F E++
Sbjct: 848 LIRYALSGRAWDLVIEQRVAFTRQKDFGKEQRELQWAHAQRTLHGLQAPDAK-MFPERTH 906
Query: 903 YRELSEIAEQAKRRAEVARLIPFH 926
+ ELS++AE+AKRRAE+ARL H
Sbjct: 907 FNELSQMAEEAKRRAEIARLRELH 930
>At3g47950 H+-transporting ATPase - like protein
Length = 960
Score = 1486 bits (3846), Expect = 0.0
Identities = 738/924 (79%), Positives = 826/924 (88%), Gaps = 7/924 (0%)
Query: 9 LEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKILKFLG 68
LE + E VDLE +P+EEVFE L+C+KEGL+++ RL +FG NKLEEKK+SK LKFLG
Sbjct: 12 LEAVLKEAVDLENVPIEEVFENLRCSKEGLTTQAADERLALFGHNKLEEKKESKFLKFLG 71
Query: 69 FMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNAAAA 128
FMWNPLSWVMEAAA+MAI LANG GKPPDWQDFVGII LLVINSTISFIEENNAGNAAAA
Sbjct: 72 FMWNPLSWVMEAAAIMAIALANGGGKPPDWQDFVGIITLLVINSTISFIEENNAGNAAAA 131
Query: 129 LMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSALTG 188
LMA LAPK KVLRDG+W EQ+AAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSALTG
Sbjct: 132 LMARLAPKAKVLRDGRWGEQDAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSALTG 191
Query: 189 ESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLTAI 248
ESLPVT++ GD VYSGSTCKQGE+EAVVIATGVHTFFGKAAHLVD+TNQ+GHFQ+VLTAI
Sbjct: 192 ESLPVTKSSGDGVYSGSTCKQGEIEAVVIATGVHTFFGKAAHLVDTTNQIGHFQQVLTAI 251
Query: 249 GNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGSHR 308
GNFCICSIAVGML EI+VMYPIQHR YR GIDNLLVLLIGGIPIAMPTVLSVTMAIGSHR
Sbjct: 252 GNFCICSIAVGMLIEIVVMYPIQHRAYRPGIDNLLVLLIGGIPIAMPTVLSVTMAIGSHR 311
Query: 309 LSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLLAA 368
LSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKL+VDKNLIEVF KGVD + V+L+AA
Sbjct: 312 LSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLTVDKNLIEVFMKGVDADTVVLMAA 371
Query: 369 RASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGNGNWHRASK 428
RASR+ENQDAIDAAIVG LADPK+ARAG++E+HFLPFNP DKRTALTYID GN HR SK
Sbjct: 372 RASRLENQDAIDAAIVGMLADPKDARAGIQEVHFLPFNPTDKRTALTYIDNEGNTHRVSK 431
Query: 429 GAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQFVGLL 488
GAPEQI++L + + +R +HA+IDKFAERGLRSLAVA Q+VPE K+S G PWQFVGL+
Sbjct: 432 GAPEQILNLAHNKSEIERRVHAVIDKFAERGLRSLAVAYQDVPEGRKDSAGGPWQFVGLM 491
Query: 489 SLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLGQDKD 548
LFDPPRHDSAETIRRAL+LGV+VKMITGDQLAI KETGRRLGMGTNMYPS+ LLGQ+KD
Sbjct: 492 PLFDPPRHDSAETIRRALNLGVSVKMITGDQLAIGKETGRRLGMGTNMYPSSALLGQNKD 551
Query: 549 ANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRADIGI 608
+I ALPV+ELIEKADGFAGVFPEHKYEIV++LQ RKHICGMTGDGVNDAPALK+ADIGI
Sbjct: 552 ESIVALPVDELIEKADGFAGVFPEHKYEIVKRLQARKHICGMTGDGVNDAPALKKADIGI 611
Query: 609 AVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFMFI 668
AVADATDAAR ASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIV GFM +
Sbjct: 612 AVADATDAARSASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVLGFMLL 671
Query: 669 ALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGGYLALM 728
ALIW+FDF PFMVLIIAILNDGTIMTISKDRV PSPLPDSWKL EIFATG+V G Y+A+M
Sbjct: 672 ALIWQFDFPPFMVLIIAILNDGTIMTISKDRVKPSPLPDSWKLSEIFATGVVFGSYMAMM 731
Query: 729 TVIFFWAMKENDFFPDKFGV---RKLNHDE---MMSALYLQVSIVSQALIFVTRSRGWSF 782
TVIFFW + DFFP FGV K HD+ + SA+YLQVSI+SQALIFVTRSR WSF
Sbjct: 732 TVIFFWVSYKTDFFPRTFGVATLEKTAHDDFRKLASAIYLQVSIISQALIFVTRSRSWSF 791
Query: 783 LERPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKF 842
+ERPG L+IAF +AQL+AT+IAVYANW FA ++GIGWGWAGVIWLY+I+FYIPLD +KF
Sbjct: 792 VERPGIFLMIAFILAQLVATLIAVYANWSFAAIEGIGWGWAGVIWLYNIIFYIPLDFIKF 851
Query: 843 AIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKSS 902
IRY LSG+AW+ +++ + AFT +KD+GKE+RE QWAHAQRTLHGLQ P++ +F +++
Sbjct: 852 FIRYALSGRAWDLVIEQRVAFTRQKDFGKEQRELQWAHAQRTLHGLQAPDTK-MFTDRTH 910
Query: 903 YRELSEIAEQAKRRAEVARLIPFH 926
EL+++AE+AKRRAE+ARL H
Sbjct: 911 VSELNQMAEEAKRRAEIARLRELH 934
>At3g60330 plasma membrane H+-ATPase - like
Length = 961
Score = 1362 bits (3524), Expect = 0.0
Identities = 681/930 (73%), Positives = 789/930 (84%), Gaps = 20/930 (2%)
Query: 8 SLEQIKNETVDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKILKFL 67
+L+ I E++DLE +PVEEVF+ LKCTKEGL+S E RL +FG NKLEEKK+SKILKFL
Sbjct: 6 ALKAITTESIDLENVPVEEVFQHLKCTKEGLTSNEVQERLTLFGYNKLEEKKESKILKFL 65
Query: 68 GFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNAAA 127
GFMWNPLSWVMEAAALMAIGLA+G GKP D+ DFVGI+ LL+INSTISF+EENNAGNAAA
Sbjct: 66 GFMWNPLSWVMEAAALMAIGLAHGGGKPADYHDFVGIVVLLLINSTISFVEENNAGNAAA 125
Query: 128 ALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSALT 187
ALMA LAPK K +RDGKW+E +AA LVPGDI+SIKLGDI+PADARLLEGDPLKIDQ+ LT
Sbjct: 126 ALMAQLAPKAKAVRDGKWNEIDAAELVPGDIVSIKLGDIIPADARLLEGDPLKIDQATLT 185
Query: 188 GESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLTA 247
GESLPVT+NPG VYSGSTCKQGE+EAVVIATGVHTFFGKAAHLVDST VGHFQKVLTA
Sbjct: 186 GESLPVTKNPGASVYSGSTCKQGEIEAVVIATGVHTFFGKAAHLVDSTTHVGHFQKVLTA 245
Query: 248 IGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGSH 307
IGNFCICSIAVGM EI+V+Y +Q R YR GIDNLLVLLIGGIPIAMPTVLSVTMAIG+H
Sbjct: 246 IGNFCICSIAVGMAIEIVVIYGLQKRGYRVGIDNLLVLLIGGIPIAMPTVLSVTMAIGAH 305
Query: 308 RLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLLA 367
RL+QQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVF++G+D++ +L+A
Sbjct: 306 RLAQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFKRGIDRDMAVLMA 365
Query: 368 ARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGNGNWHRAS 427
ARA+R+ENQDAID AIV L+DPKEARAG++E+HFLPF+P ++RTALTY+DG G HR S
Sbjct: 366 ARAARLENQDAIDTAIVSMLSDPKEARAGIKELHFLPFSPANRRTALTYLDGEGKMHRVS 425
Query: 428 KGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQFVGL 487
KGAPE+I+D+ + + K +HA IDKFAERGLRSL +A QEVP+ + G PW FV L
Sbjct: 426 KGAPEEILDMAHNKLEIKEKVHATIDKFAERGLRSLGLAYQEVPDGDVKGEGGPWDFVAL 485
Query: 488 LSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLGQDK 547
L LFDPPRHDSA+TI RALHLGV+VKMITGDQLAIAKETGRRLGMGTNMYPS++LL
Sbjct: 486 LPLFDPPRHDSAQTIERALHLGVSVKMITGDQLAIAKETGRRLGMGTNMYPSSSLL---S 542
Query: 548 DANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRADIG 607
D N + V+ELIE ADGFAGVFPEHKYEIV++LQ RKHICGMTGDGVNDAPALK+ADIG
Sbjct: 543 DNNTEGVSVDELIENADGFAGVFPEHKYEIVKRLQSRKHICGMTGDGVNDAPALKKADIG 602
Query: 608 IAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFMF 667
IAV DATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIV GFM
Sbjct: 603 IAVDDATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVMGFML 662
Query: 668 IALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGGYLAL 727
+ + W+FDF PFMVL+IAILNDGTIMTISKDRV PSP PD WKLKEIFATG+VLG YLA+
Sbjct: 663 LCVFWEFDFPPFMVLVIAILNDGTIMTISKDRVKPSPTPDCWKLKEIFATGVVLGAYLAI 722
Query: 728 MTVIFFWAMKENDFFPDKFGVRKLN---------------HDEMMSALYLQVSIVSQALI 772
MTV+FFWA E +FF + F VR N +++M SA+YLQVS +SQALI
Sbjct: 723 MTVVFFWAAYETNFFHNIFHVRNFNQHHFKMKDKKVAAHLNEQMASAVYLQVSTISQALI 782
Query: 773 FVTRSRGWSFLERPGALLVIAFFIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIV 832
FVTRSR WSF+ERPG LLVIAF IAQL+A++I+ ANW FA ++ IGWGW GVIW+++IV
Sbjct: 783 FVTRSRSWSFVERPGFLLVIAFLIAQLVASVISAMANWPFAGIRSIGWGWTGVIWIFNIV 842
Query: 833 FYIPLDVMKFAIRYILSGKAWNNLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPE 892
Y+ LD +KF +RY LSGK+W+ +++ +TA T KK++G+EER A WA +RT HGL+ +
Sbjct: 843 TYMLLDPIKFLVRYALSGKSWDRMVEGRTALTGKKNFGQEERMAAWATEKRTQHGLETGQ 902
Query: 893 SSGIFNEKSSYRELSEIAEQAKRRAEVARL 922
E++S EL+ +AE+AKRRAE+AR+
Sbjct: 903 KP--VYERNSATELNNMAEEAKRRAEIARM 930
>At1g17260 H+-transporting ATPase AHA10
Length = 946
Score = 1318 bits (3411), Expect = 0.0
Identities = 661/912 (72%), Positives = 772/912 (84%), Gaps = 13/912 (1%)
Query: 17 VDLERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKILKFLGFMWNPLSW 76
+DL +P+EEVFE L+ + +GL S + RL+IFGPN+LEEK++++ +KFLGFMWNPLSW
Sbjct: 20 IDLGILPLEEVFEYLRTSPQGLLSGDAEERLKIFGPNRLEEKQENRFVKFLGFMWNPLSW 79
Query: 77 VMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNAAAALMAGLAPK 136
VMEAAALMAI LAN PDW+DF GI+CLL+IN+TISF EENNAGNAAAALMA LA K
Sbjct: 80 VMEAAALMAIALANSQSLGPDWEDFTGIVCLLLINATISFFEENNAGNAAAALMARLALK 139
Query: 137 TKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSALTGESLPVTRN 196
T+VLRDG+W EQ+A+ILVPGDIISIKLGDI+PADARLLEGDPLKIDQS LTGESLPVT+
Sbjct: 140 TRVLRDGQWQEQDASILVPGDIISIKLGDIIPADARLLEGDPLKIDQSVLTGESLPVTKK 199
Query: 197 PGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQVGHFQKVLTAIGNFCICSI 256
G++V+SGSTCKQGE+EAVVIATG TFFGK A LVDST+ GHFQ+VLT+IGNFCICSI
Sbjct: 200 KGEQVFSGSTCKQGEIEAVVIATGSTTFFGKTARLVDSTDVTGHFQQVLTSIGNFCICSI 259
Query: 257 AVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAIGSHRLSQQGAIT 316
AVGM+ EII+M+P+QHR YR GI+NLLVLLIGGIPIAMPTVLSVT+AIGSHRLSQQGAIT
Sbjct: 260 AVGMVLEIIIMFPVQHRSYRIGINNLLVLLIGGIPIAMPTVLSVTLAIGSHRLSQQGAIT 319
Query: 317 KRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLLAARASRVENQ 376
KRMTAIEEMAGMDVLC DKTGTLTLN L+VDKNLIEVF +DK+ ++LLA RASR+ENQ
Sbjct: 320 KRMTAIEEMAGMDVLCCDKTGTLTLNSLTVDKNLIEVFVDYMDKDTILLLAGRASRLENQ 379
Query: 377 DAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGNGNWHRASKGAPEQIMD 436
DAIDAAIV LADP+EARA +REIHFLPFNPVDKRTA+TYID +G W+RA+KGAPEQ+++
Sbjct: 380 DAIDAAIVSMLADPREARANIREIHFLPFNPVDKRTAITYIDSDGKWYRATKGAPEQVLN 439
Query: 437 LCKLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQFVGLLSLFDPPRH 496
LC+ + + + ++AIID+FAE+GLRSLAVA QE+PEK+ SPG PW+F GLL LFDPPRH
Sbjct: 440 LCQQKNEIAQRVYAIIDRFAEKGLRSLAVAYQEIPEKSNNSPGGPWRFCGLLPLFDPPRH 499
Query: 497 DSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLGQDKDANIAALPV 556
DS ETI RAL LGV VKMITGDQLAIAKETGRRLGMGTNMYPS++LLG + D + A+PV
Sbjct: 500 DSGETILRALSLGVCVKMITGDQLAIAKETGRRLGMGTNMYPSSSLLGHNNDEH-EAIPV 558
Query: 557 EELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRADIGIAVADATDA 616
+ELIE ADGFAGVFPEHKYEIV+ LQE KH+ GMTGDGVNDAPALK+ADIGIAVADATDA
Sbjct: 559 DELIEMADGFAGVFPEHKYEIVKILQEMKHVVGMTGDGVNDAPALKKADIGIAVADATDA 618
Query: 617 ARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFMFIALIWKFDF 676
AR ++DIVLT+PGLSVIISAVLTSRAIFQRM+NYT+YAVSITIRI+ GF +ALIW++DF
Sbjct: 619 ARSSADIVLTDPGLSVIISAVLTSRAIFQRMRNYTVYAVSITIRIL-GFTLLALIWEYDF 677
Query: 677 SPFMVLIIAILNDGTIMTISKDRVVPSPLPDSWKLKEIFATGIVLGGYLALMTVIFFWAM 736
PFMVLIIAILNDGTIMTISKDRV PSP P+SWKL +IFATGIV+G YLAL+TV+F+W +
Sbjct: 678 PPFMVLIIAILNDGTIMTISKDRVRPSPTPESWKLNQIFATGIVIGTYLALVTVLFYWII 737
Query: 737 KENDFFPDKFGVRKL--NHDEMMSALYLQVSIVSQALIFVTRSRGWSFLERPGALLVIAF 794
FF F V+ + N +++ SA+YLQVSI+SQALIFVTRSRGWSF ERPG LL+ AF
Sbjct: 738 VSTTFFEKHFHVKSIANNSEQVSSAMYLQVSIISQALIFVTRSRGWSFFERPGTLLIFAF 797
Query: 795 FIAQLIATIIAVYANWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKFAIRYILSGKAWN 854
+AQL AT+IAVYAN FAK+ GIGW WAGVIWLYS++FYIPLDV+KF Y LSG+AWN
Sbjct: 798 ILAQLAATLIAVYANISFAKITGIGWRWAGVIWLYSLIFYIPLDVIKFVFHYALSGEAWN 857
Query: 855 NLLDNKTAFTTKKDYGKEEREAQWAHAQRTLHGLQPPESSGIFNEKSSYRELSEIAEQAK 914
+LD KTAFT KKDYGK++ +QR S + S S IAEQ +
Sbjct: 858 LVLDRKTAFTYKKDYGKDDGSPNVTISQR---------SRSAEELRGSRSRASWIAEQTR 908
Query: 915 RRAEVARLIPFH 926
RRAE+ARL+ H
Sbjct: 909 RRAEIARLLEVH 920
>At4g11730 H+-transporting ATPase - like protein
Length = 813
Score = 607 bits (1564), Expect = e-173
Identities = 306/462 (66%), Positives = 382/462 (82%), Gaps = 10/462 (2%)
Query: 8 SLEQIKNETVD-LERIPVEEVFEQLKCTKEGLSSEEGANRLQIFGPNKLEEKKDSKI-LK 65
SLE IK E D LE+IP+EEVF++L+C++EGLS EG RL+IFGPNKLE KK I L+
Sbjct: 6 SLEDIKIEIDDDLEKIPIEEVFKKLRCSREGLSGAEGKERLKIFGPNKLENKKKEHITLR 65
Query: 66 FLGFMWNPLSWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNA 125
F M+ PLSWV++AAA+MA+ ANG+G+ Q F+GI+CLL++N+ I +++E++A N
Sbjct: 66 FFALMFKPLSWVIQAAAIMAMLFANGDGR----QLFLGIVCLLIVNTIICYLKEDDAANV 121
Query: 126 AAALMAGLAPKTKVLRDGKWSEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSA 185
A AGL+PKTKVLRDGKWSEQEA+ILVPGDI+SIK GDI+P DARLLEGD LK+DQSA
Sbjct: 122 VAMARAGLSPKTKVLRDGKWSEQEASILVPGDIVSIKPGDIIPCDARLLEGDTLKVDQSA 181
Query: 186 LTGESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDS-TNQVGHFQKV 244
LTGE P+T+ PG+EV+SG+TCKQGE+EAVVIATGVHTF G AHLVD+ TN+VGHF+KV
Sbjct: 182 LTGEFGPITKGPGEEVFSGTTCKQGEMEAVVIATGVHTFSGTTAHLVDNRTNKVGHFRKV 241
Query: 245 LTAIGNFCICSIAVGMLAEIIVMYPIQHRKYRDGIDNLLVLLIGGIPIAMPTVLSVTMAI 304
+T I N C+ SIA+G+ E+IVMY IQ R + D I+NLLVL+IGGIP+AMPTVL V M
Sbjct: 242 VTEIENLCVISIAIGISIEVIVMYWIQRRNFSDVINNLLVLVIGGIPLAMPTVLYVIMVT 301
Query: 305 GSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVM 364
GS RL + G IT+R+TAIE+MA +DVLCSDKTGTLTLNKLSVDKNLI+V+ K V+KE V+
Sbjct: 302 GSLRLYRTGTITQRITAIEDMAAIDVLCSDKTGTLTLNKLSVDKNLIKVYSKDVEKEQVL 361
Query: 365 LLAARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTYIDGNGNWH 424
LLAARASR+EN+D IDAA+VG+LADPKEARAG+RE+H FN VDKRTALTYIDGNG+WH
Sbjct: 362 LLAARASRIENRDGIDAAMVGSLADPKEARAGIREVH---FNLVDKRTALTYIDGNGDWH 418
Query: 425 RASKGAPEQIMDLCKLREDTKRNIHAIIDKFAERGLRSLAVA 466
R SKG PEQI+DLC R+D ++++H+ I +AERGL+S A++
Sbjct: 419 RVSKGTPEQILDLCNARDDLRKSVHSAIRNYAERGLKSFAIS 460
Score = 399 bits (1025), Expect = e-111
Identities = 203/291 (69%), Positives = 236/291 (80%), Gaps = 5/291 (1%)
Query: 572 EHKYEIVRKLQERKHICGMTGDGVNDAPALKRADIGIAVADATDAARGASDIVLTEPGLS 631
EHKY IV KLQER HICG+ GDGV+D P+LK+AD+GIAVA+AT+AAR ASDIVLTEPGLS
Sbjct: 480 EHKYHIVNKLQER-HICGLIGDGVDDVPSLKKADVGIAVANATEAARAASDIVLTEPGLS 538
Query: 632 VIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFMFIALIWKFDFSPFMVLIIAILNDGT 691
VII AVL SRAI Q+MK+YTIYAVSITIR+VFGFMFIALIWKFDFSPFMVL IA+LN+ T
Sbjct: 539 VIIDAVLASRAILQQMKHYTIYAVSITIRVVFGFMFIALIWKFDFSPFMVLAIALLNEET 598
Query: 692 IMTISKDRVV-PSPLPDSWKLKEIFATGIVLGGYLALMTVIFFWAMKENDFFPDKFGVRK 750
I+ D V PSP PDS KLKEIFATG+V G Y+AL+TV+FFWA D FP F VR
Sbjct: 599 TKAITMDNVTNPSPTPDSLKLKEIFATGVVYGSYMALITVVFFWAAYRTDIFPRTFHVRD 658
Query: 751 L--NHDEMMSALYLQVSIVSQALIFVTRSRGWSFLERPGALLVIAFFIAQLIATIIAVYA 808
L N EMM ALYLQVSI+SQAL FV +SR W F+ERPG LL ++F Q IAT +AVYA
Sbjct: 659 LRGNEAEMMCALYLQVSIMSQALFFVIQSRSWFFVERPGELLFLSFVTVQTIATTLAVYA 718
Query: 809 NWGFAKVQGIGWGWAGVIWLYSIVFYIPLDVMKFAIRYILSGKAWNNLLDN 859
+W A+++GIGW WAGVIWLY+I+F+ PLD+MKF IRYIL+GKA +L DN
Sbjct: 719 SWETARIEGIGWSWAGVIWLYNIIFFFPLDIMKFGIRYILTGKA-QSLFDN 768
>At3g22910 calmodulin-stimulated calcium-ATPase, putative
Length = 1017
Score = 199 bits (507), Expect = 5e-51
Identities = 179/704 (25%), Positives = 324/704 (45%), Gaps = 84/704 (11%)
Query: 34 TKEGLSSE--EGANRLQIFGPNKLEEKKDSKILKFLGFMWNPLS-WVMEAAALMAIGLA- 89
T+ G++ E E R FG N + + F+ + L+ ++ A +++G
Sbjct: 117 TRLGINEEGDEIQRRRSTFGSNTYTRQPSKGLFHFVVEAFKDLTILILLGCATLSLGFGI 176
Query: 90 NGNGKPPDWQD----FVGIICLLVINSTISFIEENNAGNAAAALMAGLAPKTKVLRDGKW 145
+G W D FV + L+V S +S +N + + + + + K V+R+G+
Sbjct: 177 KEHGLKEGWYDGGSIFVAVF-LVVAVSAVSNFRQNRQFDKLSKVSSNI--KIDVVRNGRR 233
Query: 146 SEQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSALTGES--LPVTRNPGDEVYS 203
E +V GDI+ + +GD VPAD +EG L +D+S++TGES + V+ ++S
Sbjct: 234 QEISIFDIVVGDIVCLNIGDQVPADGVFVEGHLLHVDESSMTGESDHVEVSLTGNTFLFS 293
Query: 204 GSTCKQGELEAVVIATGVHTFFGKA-AHLVDSTNQVGHFQ----KVLTAIGNFCICSIAV 258
G+ G + V + G++T +G+ +H+ TN+ Q K+ ++IG + +A
Sbjct: 294 GTKIADGFGKMAVTSVGMNTAWGQMMSHISRDTNEQTPLQSRLDKLTSSIGKVGLL-VAF 352
Query: 259 GMLAEIIVMY-------PIQHRKYR------DGIDNLLV--------LLIGGIPIAMPTV 297
+L +++ Y +R+Y D I N +V +++ IP +P
Sbjct: 353 LVLLVLLIRYFTGTTKDESGNREYNGKTTKSDEIVNAVVKMVAAAVTIIVVAIPEGLPLA 412
Query: 298 LSVTMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKL------------- 344
+++T+A R+ + A+ ++++A E M V+C+DKTGTLTLN++
Sbjct: 413 VTLTLAYSMKRMMKDNAMVRKLSACETMGSATVICTDKTGTLTLNQMKVTDFWFGLESGK 472
Query: 345 --SVDKNLIEVFEKGVDKE---HVMLLAARASRVENQDAIDAAIVGTLADPKEARAGVRE 399
SV + ++E+F +GV V A + + AI+ + E G+ +
Sbjct: 473 ASSVSQRVVELFHQGVAMNTTGSVFKAKAGTEYEFSGSPTEKAILSWAVE--ELEMGMEK 530
Query: 400 -------IHFLPFNPVDKRTALTYIDGNGNWHRAS---KGAPEQIMDLC----------- 438
+H FN KR+ + N KGA E+I+ +C
Sbjct: 531 VIEEHDVVHVEGFNSEKKRSGVLMKKKGVNTENNVVHWKGAAEKILAMCSTFCDGSGVVR 590
Query: 439 KLREDTKRNIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQFVGLLSLFDPPRHDS 498
+++ED K II A + LR +A A E E K+ +G++ + DP R
Sbjct: 591 EMKEDDKIQFEKIIQSMAAKSLRCIAFAYSEDNEDNKKLKEEKLSLLGIIGIKDPCRPGV 650
Query: 499 AETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGT--NMYPSATLLGQDKDANIAALPV 556
+ + GVN+KMITGD + A+ G+ T + S +L +K N
Sbjct: 651 KKAVEDCQFAGVNIKMITGDNIFTARAIAVECGILTPEDEMNSEAVLEGEKFRNYTQEER 710
Query: 557 EELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRADIGIAVA-DATD 615
E +E+ A P K +V+ L+E H+ +TGDG NDAPALK ADIG+++ T+
Sbjct: 711 LEKVERIKVMARSSPFDKLLMVKCLKELGHVVAVTGDGTNDAPALKEADIGLSMGIQGTE 770
Query: 616 AARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITI 659
A+ +SDIV+ + + + + + R ++ ++ + + +++ +
Sbjct: 771 VAKESSDIVILDDNFASVATVLKWGRCVYNNIQKFIQFQLTVNV 814
>At3g63380 Ca2+-transporting ATPase -like protein
Length = 1033
Score = 175 bits (444), Expect = 9e-44
Identities = 173/717 (24%), Positives = 316/717 (43%), Gaps = 100/717 (13%)
Query: 39 SSEEGANRLQIFGPNKLEEKKDSKILKFLGFMWNPLS-WVMEAAALMAIGLA-NGNGKPP 96
+ +E + R +FG N + +L F+ + L+ ++ A+ ++G +G
Sbjct: 129 NEQEVSRRRDLFGSNTYHKPPPKGLLFFVYEAFKDLTILILLVCAIFSLGFGIKEHGIKE 188
Query: 97 DWQD----FVGIICLLVINSTISFIEENNAGNAAAALMAGLAPKTKVLRDGKWSEQEAAI 152
W + FV + ++V+++ +F +E + K +VLRD +
Sbjct: 189 GWYEGGSIFVAVFLVIVVSALSNFRQERQFDKLSKI---SNNIKVEVLRDSRRQHISIFD 245
Query: 153 LVPGDIISIKLGDIVPADARLLEGDPLKIDQSALTGES--LPVTRNPGDEVYSGSTCKQG 210
+V GD++ +K+GD +PAD LEG L++D+S++TGES L V ++SG+ G
Sbjct: 246 VVVGDVVFLKIGDQIPADGLFLEGHSLQVDESSMTGESDHLEVDHKDNPFLFSGTKIVDG 305
Query: 211 ELEAVVIATGVHTFFGKAAHLV--DSTNQVG---HFQKVLTAIGNFCICSIAVGMLAEII 265
+ +V++ G+ T +G+ + DS+ + + + IG + ++A +L ++
Sbjct: 306 FAQMLVVSVGMSTTWGQTMSSINQDSSERTPLQVRLDTLTSTIGKIGL-TVAALVLVVLL 364
Query: 266 VMYPIQH------RKYR------DGIDNLLVLLIGG--------IPIAMPTVLSVTMAIG 305
V Y + R+Y D + N +V ++ IP +P +++T+A
Sbjct: 365 VRYFTGNTEKEGKREYNGSKTPVDTVVNSVVRIVAAAVTIVVVAIPEGLPLAVTLTLAYS 424
Query: 306 SHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDK------NLIEVFEKGVD 359
R+ A+ ++++A E M V+C+DKTGTLTLN++ V K ++ E K +
Sbjct: 425 MKRMMSDQAMVRKLSACETMGSATVICTDKTGTLTLNEMKVTKFWLGQESIHEDSTKMIS 484
Query: 360 KEHVMLL----------------AARASRVENQDAIDAAIVGTLADPKEARAGVREIHFL 403
+ + LL + A + T+ + V++ H +
Sbjct: 485 PDVLDLLYQGTGLNTTGSVCVSDSGSTPEFSGSPTEKALLSWTVLNLGMDMESVKQKHEV 544
Query: 404 ----PFNPVDKRT-ALTYIDGNGNWHRASKGAPEQIMDLCK-----------LREDTKRN 447
F+ KR+ L + H KGA E ++ +C + K
Sbjct: 545 LRVETFSSAKKRSGVLVRRKSDNTVHVHWKGAAEMVLAMCSHYYTSTGSVDLMDSTAKSR 604
Query: 448 IHAIIDKFAERGLRSLAVARQEVP-EKTKESPGAPWQFVGLLSLFDPPRHDSAETIRRAL 506
I AII A LR +A A + + E G +G++ L DP R ++ +
Sbjct: 605 IQAIIQGMAASSLRCIAFAHKIASNDSVLEEDGL--TLMGIVGLKDPCRPGVSKAVETCK 662
Query: 507 HLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLGQDKDANIAAL--------PVEE 558
GV +KMITGD + AK G+ L DKD A + EE
Sbjct: 663 LAGVTIKMITGDNVFTAKAIAFECGI---------LDHNDKDEEDAVVEGVQFRNYTDEE 713
Query: 559 LIEKADG---FAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRADIGIAVA-DAT 614
++K D A P K +V+ L+ + H+ +TGDG NDAPALK ADIG+++ T
Sbjct: 714 RMQKVDKIRVMARSSPSDKLLMVKCLRLKGHVVAVTGDGTNDAPALKEADIGLSMGIQGT 773
Query: 615 DAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITIRIVFGFMFIALI 671
+ A+ +SDIV+ + + + + + R ++ ++ + + +++ + + FIA I
Sbjct: 774 EVAKESSDIVILDDNFASVATVLKWGRCVYNNIQKFIQFQLTVNVAALV-INFIAAI 829
>At1g07670 endoplasmic reticulum-type calcium-transporting ATPase 4
(ECA4)
Length = 1061
Score = 141 bits (356), Expect = 1e-33
Identities = 117/431 (27%), Positives = 211/431 (48%), Gaps = 65/431 (15%)
Query: 8 SLEQIKNETVDLERIPVEEVFEQLKCTKE-GLSSEEGANRLQIFGPNKLEEKKDSKILKF 66
S E +K++T V E E+ ++E GLS++E R QI+G N+LE+ + + I K
Sbjct: 15 SSELVKSDTFPAWGKDVSECEEKFGVSREKGLSTDEVLKRHQIYGLNELEKPEGTSIFKL 74
Query: 67 LGFMWNP-LSWVMEAAALMAIGLANGNGKPPDWQDFVG------IICLLVINSTISFIEE 119
+ +N L ++ AAA+++ LA +G I +L++N+ + +E
Sbjct: 75 ILEQFNDTLVRILLAAAVISFVLAFFDGDEGGEMGITAFVEPLVIFLILIVNAIVGIWQE 134
Query: 120 NNAGNAAAALMAGLAPKTKVLRDG-KWSEQEAAILVPGDIISIKLGDIVPADARL--LEG 176
NA A AL + + V+RDG K S A LVPGDI+ +++GD VPAD R+ L
Sbjct: 135 TNAEKALEALKEIQSQQATVMRDGTKVSSLPAKELVPGDIVELRVGDKVPADMRVVALIS 194
Query: 177 DPLKIDQSALTGESLPVTRNPG--DE----------VYSGSTCKQGELEAVVIATGVHTF 224
L+++Q +LTGES V++ DE V++G+T G +V TG++T
Sbjct: 195 STLRVEQGSLTGESEAVSKTTKHVDENADIQGKKCMVFAGTTVVNGNCICLVTDTGMNTE 254
Query: 225 FGKAAHLVDSTNQVGH---FQKVLTAIGNFCICSIAVGMLAEIIVMYPIQHR---KYRDG 278
G+ + Q +K L G + ++ +G++ ++ + +++ +Y DG
Sbjct: 255 IGRVHSQIQEAAQHEEDTPLKKKLNEFGE--VLTMIIGLICALVWLINVKYFLSWEYVDG 312
Query: 279 ---------------IDNLLVLLIGGIPIAMPTVLSVTMAIGSHRLSQQGAITKRMTAIE 323
+ + L + IP +P V++ +A+G+ +++Q+ A+ +++ ++E
Sbjct: 313 WPRNFKFSFEKCTYYFEIAVALAVAAIPEGLPAVITTCLALGTRKMAQKNALVRKLPSVE 372
Query: 324 EMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLLAARASRVENQDAIDAAI 383
+ V+CSDKTGTLT N+++V K L A SR+ + + +
Sbjct: 373 TLGCTTVICSDKTGTLTTNQMAVSK-----------------LVAMGSRIGTLRSFN--V 413
Query: 384 VGTLADPKEAR 394
GT DP++ +
Sbjct: 414 EGTSFDPRDGK 424
Score = 114 bits (286), Expect = 2e-25
Identities = 89/271 (32%), Positives = 131/271 (47%), Gaps = 31/271 (11%)
Query: 484 FVGLLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLL 543
FVG + L DPPR + + I G+ V +ITGD + A+ R +G+
Sbjct: 621 FVGFVGLRDPPRKEVRQAIADCRTAGIRVMVITGDNKSTAEAICREIGV----------F 670
Query: 544 GQDKDANIAALPVEELIEKADG-----------FAGVFPEHKYEIVRKLQERKHICGMTG 592
D+D + +L +E ++ D F+ P+HK EIVR L+E + MTG
Sbjct: 671 EADEDISSRSLTGKEFMDVKDQKNHLRQTGGLLFSRAEPKHKQEIVRLLKEDGEVVAMTG 730
Query: 593 DGVNDAPALKRADIGIAVA-DATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYT 651
DGVNDAPALK ADIG+A+ T+ A+ ASD+VL + S I++AV R+I+ MK +
Sbjct: 731 DGVNDAPALKLADIGVAMGISGTEVAKEASDLVLADDNFSTIVAAVGEGRSIYNNMKAFI 790
Query: 652 IYAVSITI-RIVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTI------SKD--RVVP 702
Y +S I + F+ AL P +L + ++ DG T KD + P
Sbjct: 791 RYMISSNIGEVASIFLTAALGIPEGMIPVQLLWVNLVTDGPPATALGFNPPDKDIMKKPP 850
Query: 703 SPLPDSWKLKEIFATGIVLGGYLALMTVIFF 733
DS I +V+G Y+ + TV F
Sbjct: 851 RRSDDSLITAWILFRYMVIGLYVGVATVGVF 881
>At1g07810 ER-type Ca2+-pump protein
Length = 1061
Score = 141 bits (355), Expect = 2e-33
Identities = 116/438 (26%), Positives = 213/438 (48%), Gaps = 65/438 (14%)
Query: 1 MAESKSISLEQIKNETVDLERIPVEEVFEQLKCTKE-GLSSEEGANRLQIFGPNKLEEKK 59
+ + +S++ + ++T V E E ++E GLSS+E R QI+G N+LE+ +
Sbjct: 8 LVKKESLNSTPVNSDTFPAWAKDVAECEEHFVVSREKGLSSDEVLKRHQIYGLNELEKPE 67
Query: 60 DSKILKFLGFMWNP-LSWVMEAAALMAIGLANGNGKPPDWQDFVG------IICLLVINS 112
+ I K + +N L ++ AAA+++ LA +G I +L++N+
Sbjct: 68 GTSIFKLILEQFNDTLVRILLAAAVISFVLAFFDGDEGGEMGITAFVEPLVIFLILIVNA 127
Query: 113 TISFIEENNAGNAAAALMAGLAPKTKVLRDG-KWSEQEAAILVPGDIISIKLGDIVPADA 171
+ +E NA A AL + + V+RDG K S A LVPGDI+ +++GD VPAD
Sbjct: 128 IVGIWQETNAEKALEALKEIQSQQATVMRDGTKVSSLPAKELVPGDIVELRVGDKVPADM 187
Query: 172 RL--LEGDPLKIDQSALTGESLPVTRNPG--DE----------VYSGSTCKQGELEAVVI 217
R+ L L+++Q +LTGES V++ DE V++G+T G +V
Sbjct: 188 RVVALISSTLRVEQGSLTGESEAVSKTTKHVDENADIQGKKCMVFAGTTVVNGNCICLVT 247
Query: 218 ATGVHTFFGKAAHLVDSTNQVGH---FQKVLTAIGNFCICSIAVGMLAEIIVMYPIQHR- 273
TG++T G+ + Q +K L G + ++ +G++ ++ + +++
Sbjct: 248 DTGMNTEIGRVHSQIQEAAQHEEDTPLKKKLNEFGE--VLTMIIGLICALVWLINVKYFL 305
Query: 274 --KYRDG---------------IDNLLVLLIGGIPIAMPTVLSVTMAIGSHRLSQQGAIT 316
+Y DG + + L + IP +P V++ +A+G+ +++Q+ A+
Sbjct: 306 SWEYVDGWPRNFKFSFEKCTYYFEIAVALAVAAIPEGLPAVITTCLALGTRKMAQKNALV 365
Query: 317 KRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLLAARASRVENQ 376
+++ ++E + V+CSDKTGTLT N+++V K L A SR+
Sbjct: 366 RKLPSVETLGCTTVICSDKTGTLTTNQMAVSK-----------------LVAMGSRIGTL 408
Query: 377 DAIDAAIVGTLADPKEAR 394
+ + + GT DP++ +
Sbjct: 409 RSFN--VEGTSFDPRDGK 424
Score = 112 bits (281), Expect = 7e-25
Identities = 89/271 (32%), Positives = 130/271 (47%), Gaps = 31/271 (11%)
Query: 484 FVGLLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLL 543
FVG + L DPPR + + I G+ V +ITGD + A+ R +G+
Sbjct: 621 FVGFVGLRDPPRKEVRQAIADCRTAGIRVMVITGDNKSTAEAICREIGV----------F 670
Query: 544 GQDKDANIAALPVEELIEKADG-----------FAGVFPEHKYEIVRKLQERKHICGMTG 592
D+D + +L E ++ D F+ P+HK EIVR L+E + MTG
Sbjct: 671 EADEDISSRSLTGIEFMDVQDQKNHLRQTGGLLFSRAEPKHKQEIVRLLKEDGEVVAMTG 730
Query: 593 DGVNDAPALKRADIGIAVA-DATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYT 651
DGVNDAPALK ADIG+A+ T+ A+ ASD+VL + S I++AV R+I+ MK +
Sbjct: 731 DGVNDAPALKLADIGVAMGISGTEVAKEASDMVLADDNFSTIVAAVGEGRSIYNNMKAFI 790
Query: 652 IYAVSITI-RIVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTI------SKD--RVVP 702
Y +S I + F+ AL P +L + ++ DG T KD + P
Sbjct: 791 RYMISSNIGEVASIFLTAALGIPEGMIPVQLLWVNLVTDGPPATALGFNPPDKDIMKKPP 850
Query: 703 SPLPDSWKLKEIFATGIVLGGYLALMTVIFF 733
DS I +V+G Y+ + TV F
Sbjct: 851 RRSDDSLITAWILFRYMVIGLYVGVATVGVF 881
>At1g10130 putative calcium ATPase
Length = 992
Score = 137 bits (345), Expect = 3e-32
Identities = 106/362 (29%), Positives = 167/362 (45%), Gaps = 40/362 (11%)
Query: 427 SKGAPEQIMDLCK------------LREDTKRNIHAIIDKFAERGLRSLAVARQEVP--- 471
SKGAPE I+ C L + + + F + LR LA+A + VP
Sbjct: 504 SKGAPESIIARCNKILCNGDGSVVPLTAAGRAELESRFYSFGDETLRCLALAFKTVPHGQ 563
Query: 472 EKTKESPGAPWQFVGLLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLG 531
+ F+GL+ + DPPR + + + + G+ V ++TGD + A+ R++G
Sbjct: 564 QTISYDNENDLTFIGLVGMLDPPREEVRDAMLACMTAGIRVIVVTGDNKSTAESLCRKIG 623
Query: 532 MGTNMYP-SATLLGQDKDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGM 590
N+ S + + A+ + + F+ V P HK +V LQ++ + M
Sbjct: 624 AFDNLVDFSGMSYTASEFERLPAVQQTLALRRMTLFSRVEPSHKRMLVEALQKQNEVVAM 683
Query: 591 TGDGVNDAPALKRADIGIAVADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNY 650
TGDGVNDAPALK+ADIGIA+ T A+ ASD+VL + + I++AV RAI+ K +
Sbjct: 684 TGDGVNDAPALKKADIGIAMGSGTAVAKSASDMVLADDNFASIVAAVAEGRAIYNNTKQF 743
Query: 651 TIYAVSITI-RIVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTI------------SK 697
Y +S I +V F+ L +P +L + ++ DG T +K
Sbjct: 744 IRYMISSNIGEVVCIFVAAVLGIPDTLAPVQLLWVNLVTDGLPATAIGFNKQDSDVMKAK 803
Query: 698 DRVVPSPLPDSWKLKEIFATGIVLGGYLALMTVI-FFWAMKENDFFPDKFGVRKLNHDEM 756
R V + W +F +V+G Y+ L TV F W +D G KL + E+
Sbjct: 804 PRKVGEAVVTGW----LFFRYLVIGVYVGLATVAGFIWWFVYSD------GGPKLTYSEL 853
Query: 757 MS 758
M+
Sbjct: 854 MN 855
Score = 118 bits (295), Expect = 2e-26
Identities = 98/345 (28%), Positives = 167/345 (48%), Gaps = 43/345 (12%)
Query: 36 EGLSSEEGANRLQIFGPN-----KLEEKK-DSKILKFLGFMWNPLSWVMEAAALMAIGLA 89
+GLS + + +++G N KL K+ D ++K L ++ A + LA
Sbjct: 22 KGLSDSQVVHHSRLYGRNGTPFWKLVLKQFDDLLVKIL---------IVAAIVSFVLALA 72
Query: 90 NGN-GKPPDWQDFVGIICLLVINSTISFIEENNAGNAAAALMAGLAPKTKVLRDGKWSEQ 148
NG G + FV I+ +L N+ + I E NA A L A A VLR+G +S
Sbjct: 73 NGETGLTAFLEPFV-ILLILAANAAVGVITETNAEKALEELRAYQANIATVLRNGCFSIL 131
Query: 149 EAAILVPGDIISIKLGDIVPADARLLE--GDPLKIDQSALTGESLPVTRNPG-------- 198
A LVPGDI+ + +G +PAD R++E + ++DQ+ LTGES V ++
Sbjct: 132 PATELVPGDIVEVTVGCKIPADLRMIEMSSNTFRVDQAILTGESCSVEKDVDCTLTTNAV 191
Query: 199 -----DEVYSGSTCKQGELEAVVIATGVHTFFGKAAH-LVDSTNQVGHFQKVLTAIGNFC 252
+ ++SG+ G AVVI G +T G ++ + ++ +K L G+F
Sbjct: 192 YQDKKNILFSGTDVVAGRGRAVVIGVGSNTAMGSIHDSMLQTDDEATPLKKKLDEFGSF- 250
Query: 253 ICSIAVGMLAEIIVMY------PIQHRKYRDGIDNLLV---LLIGGIPIAMPTVLSVTMA 303
+ + G+ + V+ P ++ I + L + IP +P V++ +A
Sbjct: 251 LAKVIAGICVLVWVVNIGHFSDPSHGGFFKGAIHYFKIAVALAVAAIPEGLPAVVTTCLA 310
Query: 304 IGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDK 348
+G+ ++++ AI + + ++E + V+CSDKTGTLT N +SV K
Sbjct: 311 LGTKKMARLNAIVRSLPSVETLGCTTVICSDKTGTLTTNMMSVSK 355
>At4g00900 Ca2+-transporting ATPase - like protein
Length = 1054
Score = 127 bits (318), Expect = 4e-29
Identities = 103/369 (27%), Positives = 180/369 (47%), Gaps = 50/369 (13%)
Query: 24 VEEVFEQLKCTKE-GLSSEEGANRLQIFGPNKLEEKKDSKILKFLGFMWNP-LSWVMEAA 81
VE+ ++ K + GL+SE+ R Q +G N+L ++K + + ++ L ++ A
Sbjct: 14 VEQCLKEYKTRLDKGLTSEDVQIRRQKYGFNELAKEKGKPLWHLVLEQFDDTLVKILLGA 73
Query: 82 ALMAIGLA-------NGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNAAAALMAGLA 134
A ++ LA +G+G + FV I+ +L++N+ + +E+NA A AL
Sbjct: 74 AFISFVLAFLGEEHGSGSGFEAFVEPFV-IVLILILNAVVGVWQESNAEKALEALKEMQC 132
Query: 135 PKTKVLRDGK-WSEQEAAILVPGDIISIKLGDIVPADARL--LEGDPLKIDQSALTGESL 191
KVLRDG A LVPGDI+ + +GD VPAD R+ L+ L+++QS+LTGE++
Sbjct: 133 ESAKVLRDGNVLPNLPARELVPGDIVELNVGDKVPADMRVSGLKTSTLRVEQSSLTGEAM 192
Query: 192 PVTRNPG-------------DEVYSGSTCKQGELEAVVIATGVHTFFGKAA---HLVDST 235
PV + + V++G+T G +V + G+ T GK H
Sbjct: 193 PVLKGANLVVMDDCELQGKENMVFAGTTVVNGSCVCIVTSIGMDTEIGKIQRQIHEASLE 252
Query: 236 NQVGHFQKVLTAIGN-----FCICSIAVGML--------------AEIIVMYPIQHRKYR 276
+K L G+ CI + V M+ + + + + Y
Sbjct: 253 ESETPLKKKLDEFGSRLTTAICIVCVLVWMINYKNFVSWDVVDGYKPVNIKFSFEKCTYY 312
Query: 277 DGIDNLLVLLIGGIPIAMPTVLSVTMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKT 336
I + L + IP +P V++ +A+G+ +++Q+ AI +++ ++E + V+CSDKT
Sbjct: 313 FKI--AVALAVAAIPEGLPAVITTCLALGTRKMAQKNAIVRKLPSVETLGCTTVICSDKT 370
Query: 337 GTLTLNKLS 345
GTLT N++S
Sbjct: 371 GTLTTNQMS 379
Score = 125 bits (314), Expect = 1e-28
Identities = 114/387 (29%), Positives = 168/387 (42%), Gaps = 59/387 (15%)
Query: 398 REIHFLPFNPVDKRTALTYIDGNGNWHRASKGAPEQIMD-----------LCKLREDTKR 446
+++ L F+ V K ++ + NG KGA E I++ L L E ++
Sbjct: 499 KKVATLEFDRVRKSMSVIVSEPNGQNRLLVKGAAESILERSSFAQLADGSLVALDESSRE 558
Query: 447 NIHAIIDKFAERGLRSLAVA------------RQEVPEKTKESPGAPWQ-------FVGL 487
I + +GLR L +A +E P K + + FVG+
Sbjct: 559 VILKKHSEMTSKGLRCLGLAYKDELGEFSDYSSEEHPSHKKLLDPSSYSNIETNLIFVGV 618
Query: 488 LSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLGQDK 547
+ L DPPR + I G+ V +ITGD K T + ++ L Q
Sbjct: 619 VGLRDPPREEVGRAIEDCRDAGIRVMVITGDN----KSTAEAICCEIRLFSENEDLSQSS 674
Query: 548 --DANIAALPVE---ELIEKADG--FAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPA 600
+LP E++ K+ G F+ P HK EIVR L+E I MTGDGVNDAPA
Sbjct: 675 FTGKEFMSLPASRRSEILSKSGGKVFSRAEPRHKQEIVRMLKEMGEIVAMTGDGVNDAPA 734
Query: 601 LKRADIGIAVA-DATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVSITI 659
LK ADIGIA+ T+ A+ ASD+VL + S I+SAV R+I+ MK + Y +S +
Sbjct: 735 LKLADIGIAMGITGTEVAKEASDMVLADDNFSTIVSAVAEGRSIYNNMKAFIRYMISSNV 794
Query: 660 -RIVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTI------------SKDRVVPSPLP 706
++ F+ AL P +L + ++ DG T R L
Sbjct: 795 GEVISIFLTAALGIPECMIPVQLLWVNLVTDGPPATALGFNPADIDIMKKPPRKSDDCLI 854
Query: 707 DSWKLKEIFATGIVLGGYLALMTVIFF 733
DSW + +V+G Y+ + TV F
Sbjct: 855 DSW----VLIRYLVIGSYVGVATVGIF 877
>At5g44790 COPPER-TRANSPORTING ATPASE RAN1 (RESPONSIVE-TO-ANTAGONIST
1)
Length = 1001
Score = 121 bits (304), Expect = 2e-27
Identities = 148/641 (23%), Positives = 253/641 (39%), Gaps = 109/641 (17%)
Query: 75 SWVMEAAALMAIGLANGNGKPPDWQDFVGIICLLVINSTISFIEENNAGNAAAALMAGLA 134
S+ AL+ G G P + +I +++ + + + +A L+ L
Sbjct: 381 SYFYSVGALL-YGAVTGFWSPTYFDASAMLITFVLLGKYLESLAKGKTSDAMKKLVQ-LT 438
Query: 135 PKTKVL-RDGKWS------EQEAAILVPGDIISIKLGDIVPADARLLEGDPLKIDQSALT 187
P T +L +GK E +A ++ PGD + + G +PAD ++ G +++S +T
Sbjct: 439 PATAILLTEGKGGKLVGEREIDALLIQPGDTLKVHPGAKIPADGVVVWGSSY-VNESMVT 497
Query: 188 GESLPVTRNPGDEVYSGSTCKQGELEAVVIATGVHTFFGKAAHLVDSTNQ---------- 237
GES+PV++ V G+ G L G + LV++
Sbjct: 498 GESVPVSKEVDSPVIGGTINMHGALHMKATKVGSDAVLSQIISLVETAQMSKAPIQKFAD 557
Query: 238 --VGHFQKVLTAIGNFCICSIAVGMLAEIIVMYPIQHRKYRD---------GIDNLLVLL 286
F V+ + F + ++G + YP + I +++
Sbjct: 558 YVASIFVPVVITLALFTLVGWSIG---GAVGAYPDEWLPENGTHFVFSLMFSISVVVIAC 614
Query: 287 IGGIPIAMPTVLSVTMAIGSHRLSQQGAITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSV 346
+ +A PT + V +G + G + K A+E+ + + DKTGTLT K +V
Sbjct: 615 PCALGLATPTAVMVATGVG----ATNGVLIKGGDALEKAHKVKYVIFDKTGTLTQGKATV 670
Query: 347 DKNLIEVFEKGVDKEHVMLLAARASRVENQDAIDAAIVGTLADPKEARAGVREIHFLPFN 406
+VF + +D+ + L A A ++ + AIV A R HF +
Sbjct: 671 --TTTKVFSE-MDRGEFLTLVASA-EASSEHPLAKAIV----------AYARHFHFFDES 716
Query: 407 PVDKRTALTYIDGNGNW------HRASKGAPEQ--------IMDLCKLREDTKRNIHAII 452
D T + +G W A G Q ++ KL + NI +
Sbjct: 717 TEDGETNNKDLQNSG-WLLDTSDFSALPGKGIQCLVNEKMILVGNRKLMSENAINIPDHV 775
Query: 453 DKFAERGLRSLAVARQEVPEKTKESPGAPWQFVGLLSLFDPPRHDSAETIRRALHLGVNV 512
+KF E E KT + VG++ + DP + ++A + L +GV
Sbjct: 776 EKFVE---------DLEESGKTGVIVAYNGKLVGVMGIADPLKREAALVVEGLLRMGVRP 826
Query: 513 KMITGDQLAIAKETGRRLGMGTNMYPSATLLGQDKDANIAALPVEELIEKADGFAGVFPE 572
M+TGD A+ + +G+ D A V P
Sbjct: 827 IMVTGDNWRTARAVAKEVGI------------------------------EDVRAEVMPA 856
Query: 573 HKYEIVRKLQERKHICGMTGDGVNDAPALKRADIGIAVADATDAARGASDIVLTEPGLSV 632
K +++R LQ+ M GDG+ND+PAL AD+G+A+ TD A A+D VL L
Sbjct: 857 GKADVIRSLQKDGSTVAMVGDGINDSPALAAADVGMAIGAGTDVAIEAADYVLMRNNLED 916
Query: 633 IISAVLTSRAIFQRMKNYTIYAVS---ITIRIVFGFMFIAL 670
+I+A+ SR R++ ++A++ ++I I G F L
Sbjct: 917 VITAIDLSRKTLTRIRLNYVFAMAYNVVSIPIAAGVFFPVL 957
>At5g21930 metal-transporting ATPase - like protein
Length = 856
Score = 111 bits (277), Expect = 2e-24
Identities = 136/586 (23%), Positives = 240/586 (40%), Gaps = 106/586 (18%)
Query: 156 GDIISIKLGDIVPADARLLEGDPLKIDQSALTGESLPVTRNPGDEVYSGSTCKQGELEAV 215
GD + + G+ P D +L G + +D+S LTGESLPV + G V +G+ G L
Sbjct: 327 GDSLLVLPGETFPVDGSVLAGRSV-VDESMLTGESLPVFKEEGCSVSAGTINWDGPLRIK 385
Query: 216 VIATGVHTFFGKAAHLV-DSTNQVGHFQKVLTAIGNFCICSIAVGMLAEIIVMYPIQHRK 274
+TG ++ K +V D+ Q++ AI + +I Y +
Sbjct: 386 ASSTGSNSTISKIVRMVEDAQGNAAPVQRLADAIAGPFVYTIMSLSAMTFAFWYYVGSHI 445
Query: 275 YRD--------------------GIDNLLVLLIGGIPIAMPTVLSVTMAIGSHRLSQQGA 314
+ D +D L+V + +A PT + + ++G+ R G
Sbjct: 446 FPDVLLNDIAGPDGDALALSLKLAVDVLVVSCPCALGLATPTAILIGTSLGAKR----GY 501
Query: 315 ITKRMTAIEEMAGMDVLCSDKTGTLTLNKLSVDKNLIEVFEKGVDKEHVMLLAARASRVE 374
+ + +E +A +D + DKTGTLT + V V G +++ V+ +AA
Sbjct: 502 LIRGGDVLERLASIDCVALDKTGTLTEGRPVVSG----VASLGYEEQEVLKMAA------ 551
Query: 375 NQDAIDAAIVGTLADPKEARAGVREIHFLPFNPVDKRTALTY--------IDGNGNWHRA 426
A+ T P A+A V E L + R LT IDG
Sbjct: 552 -------AVEKTATHPI-AKAIVNEAESLNLKTPETRGQLTEPGFGTLAEIDGRF----V 599
Query: 427 SKGAPEQIMDLCKLREDTKR--NIHAIIDKFAERGLRSLAVARQEVPEKTKESPGAPWQF 484
+ G+ E + D + D+ + +++D S + + V +E G
Sbjct: 600 AVGSLEWVSDRFLKKNDSSDMVKLESLLDHKLSN-TSSTSRYSKTVVYVGREGEG----I 654
Query: 485 VGLLSLFDPPRHDSAETIRRALHLGVNVKMITGDQLAIAKETGRRLGMGTNMYPSATLLG 544
+G +++ D R D+ T+ R G+ +++GD+ + +G+
Sbjct: 655 IGAIAISDCLRQDAEFTVARLQEKGIKTVLLSGDREGAVATVAKNVGI------------ 702
Query: 545 QDKDANIAALPVEELIEKADGFAGVFPEHKYEIVRKLQERKHICGMTGDGVNDAPALKRA 604
+ + N + + PE K+E + LQ H M GDG+NDAP+L +A
Sbjct: 703 KSESTNYS----------------LSPEKKFEFISNLQSSGHRVAMVGDGINDAPSLAQA 746
Query: 605 DIGIA--VADATDAARGASDIVLTEPGLSVIISAVLTSRAIFQRMKNYTIYAVS---ITI 659
D+GIA + +AA A+ ++L LS ++ A+ ++A ++ +A++ I+I
Sbjct: 747 DVGIALKIEAQENAASNAASVILVRNKLSHVVDALSLAQATMSKVYQNLAWAIAYNVISI 806
Query: 660 RIVFGFMFIALIWKFDFSPFMVLIIAILNDGTIMTISKDRVVPSPL 705
I G L+ ++DF+ L G +M +S VV + L
Sbjct: 807 PIAAG----VLLPQYDFAMTPSL------SGGLMALSSIFVVSNSL 842
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.138 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,961,784
Number of Sequences: 26719
Number of extensions: 856417
Number of successful extensions: 2194
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 49
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 1979
Number of HSP's gapped (non-prelim): 115
length of query: 932
length of database: 11,318,596
effective HSP length: 109
effective length of query: 823
effective length of database: 8,406,225
effective search space: 6918323175
effective search space used: 6918323175
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 65 (29.6 bits)
Medicago: description of AC125475.9


