

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125475.5 + phase: 0 /pseudo
(574 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
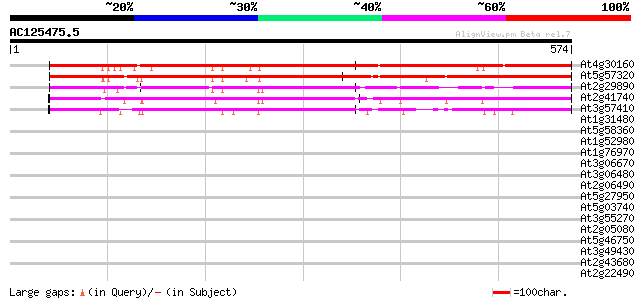
Score E
Sequences producing significant alignments: (bits) Value
At4g30160 putative villin 647 0.0
At5g57320 villin 619 e-177
At2g29890 villin 1 (VLN1) 349 3e-96
At2g41740 putative villin 2 protein 292 3e-79
At3g57410 villin 3 288 7e-78
At1g31480 hypothetical protein 39 0.010
At5g58360 similar to unknown protein (gb|AAB63094.1) 37 0.023
At1g52980 unknown protein 37 0.030
At1g76970 unknown protein 35 0.11
At3g06670 unknown protein 34 0.25
At3g06480 putative RNA helicase 34 0.25
At2g06490 putative PttA-like transposon protein 33 0.43
At5g27950 unknown protein 33 0.56
At5g03740 histone deacetylase -like protein 33 0.56
At3g55270 MAP kinase phosphatase (MKP1) 33 0.56
At2g05080 putative helicase 32 0.73
At5g46750 zinc finger protein Glo3-like 32 1.3
At3g49430 PRE-MRNA SPLICING FACTOR SF2-like protein 32 1.3
At2g43680 SF16 protein {Helianthus annuus} like protein 32 1.3
At2g22490 putative cyclin D 32 1.3
>At4g30160 putative villin
Length = 974
Score = 647 bits (1670), Expect = 0.0
Identities = 356/584 (60%), Positives = 422/584 (71%), Gaps = 55/584 (9%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVE------- 93
QVWRVNGQ K LL A D SKFYSGDCY+FQYSYPGE++EE LIGTW GK SVE
Sbjct: 395 QVWRVNGQAKTLLQAADHSKFYSGDCYVFQYSYPGEEKEEVLIGTWFGKQSVEEERGSAV 454
Query: 94 SFTSKQ---------NGRINE------VYSIYGSYL*RQ*NNSILFHP----PKLDCF*D 134
S SK RI E + I S++ + S + ++D D
Sbjct: 455 SMASKMVESMKFVPAQARIYEGKEPIQFFVIMQSFIVFKGGISSGYKKYIAEKEVD---D 511
Query: 135 ETYKEDGVALFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQ 194
+TY E+GVALFRIQGSGPE+MQAIQV+ A+SLNSSY YIL ++S VFTW GNL+ + DQ
Sbjct: 512 DTYNENGVALFRIQGSGPENMQAIQVDPVAASLNSSYYYILHNDSSVFTWAGNLSTATDQ 571
Query: 195 ELAERMLDLIKPDLQCRPQKEGAETEQFWELLGVKTEYSSQKIVREAENDPHLFSCNFSE 254
ELAER LDLIKP+ Q R QKEG+E+EQFWELLG K EYSSQK+ +E E DPHLFSC F++
Sbjct: 572 ELAERQLDLIKPNQQSRAQKEGSESEQFWELLGGKAEYSSQKLTKEPERDPHLFSCTFTK 631
Query: 255 ------EIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFLL 308
EI+NF+QDDLMTEDIFI+DCHS+IFVWVGQ+V PK ++ AL IGEKF+E+D LL
Sbjct: 632 EVLKVTEIYNFTQDDLMTEDIFIIDCHSEIFVWVGQEVVPKNKLLALTIGEKFIEKDSLL 691
Query: 309 ETISCSAPIYIVMEGSEPPFFTRFF-KWDSAKSAMLGNSYQRKLAIMKNGGTPPLVKPKR 367
E +S APIY++MEG EP FFTRFF WDS+KSAM GNS+QRKL I+KNGGTP KPKR
Sbjct: 692 EKLSPEAPIYVIMEGGEPSFFTRFFTSWDSSKSAMHGNSFQRKLKIVKNGGTPVADKPKR 751
Query: 368 RASVSYGGRSGGLPEKS-QRSRSMSVSPDRVRVRGRSPAFNALAATFENSNVRNLSTPPP 426
R SYGGR+ +P+KS QRSRSMS SPDRVRVRGRSPAFNALAATFE+ N RNLSTPPP
Sbjct: 752 RTPASYGGRA-SVPDKSQQRSRSMSFSPDRVRVRGRSPAFNALAATFESQNARNLSTPPP 810
Query: 427 MIRKLYPKSKTPDLATL--APKSSAISHLTSTFEPPSAREKLIPRSLKDTSKS----NPE 480
++RKLYP+S TPD + APKSSAI+ ++ FE +E IP+ +K + K+ PE
Sbjct: 811 VVRKLYPRSVTPDSSKFAPAPKSSAIASRSALFEKIPPQEPSIPKPVKASPKTPESPAPE 870
Query: 481 TNS----------DNENSTGSREESLTIQEDVNEGEPEDNEGLPVYPYESVKTDSTDPMP 530
+NS E S SR ESLTIQED EG ED E LP +PY+ +KT STDP+
Sbjct: 871 SNSKEQEEKKENDKEEGSMSSRIESLTIQEDAKEG-VEDEEDLPAHPYDRLKTTSTDPVS 929
Query: 531 DIDVTKREAYLSPEEFQERLGMTRSEFYKLPKWKQNKLKMAVQL 574
DIDVT+REAYLS EEF+E+ GMT+ FYKLPKWKQNK KMAVQL
Sbjct: 930 DIDVTRREAYLSSEEFKEKFGMTKEAFYKLPKWKQNKFKMAVQL 973
Score = 43.9 bits (102), Expect = 2e-04
Identities = 75/357 (21%), Positives = 138/357 (38%), Gaps = 58/357 (16%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYI-FQYSYPGEDREEHLIGTWIGKNSVESFTSKQ 99
++WR+ + + KF++GD YI + + H I W+GK++ +
Sbjct: 22 EIWRIENFIPTPIPKSSIGKFFTGDSYIVLKTTALKTGALRHDIHYWLGKDTSQDEAGTA 81
Query: 100 NGRINEVYSIYGS----YL*RQ*NNSILFHPPKLDCF*DETYKEDGVA------------ 143
+ E+ + G Y Q + + F C + E GVA
Sbjct: 82 AVKTVELDAALGGRAVQYREVQGHETEKFLSYFKPCIIPQ---EGGVASGFKHVVAEEHI 138
Query: 144 --LFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERML 201
LF +G + +V A SSLN YIL ++S +F + G+ ++ ++ A ++
Sbjct: 139 TRLFVCRGK--HVVHVKEVPFARSSLNHDDIYILDTKSKIFQFNGSNSSIQERAKALEVV 196
Query: 202 DLIKP---DLQCRPQ--KEG-----AETEQFWELLGVKTEYSSQKIVREAENDP------ 245
IK D C ++G A++ +FW G + R+ ND
Sbjct: 197 QYIKDTYHDGTCEVATVEDGKLMADADSGEFWGFFG-----GFAPLPRKTANDEDKTYNS 251
Query: 246 ---HLFSCNFSE----EIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIG 298
LF + E ++ L T +ILDC ++FVW+G+ R A
Sbjct: 252 DITRLFCVEKGQANPVEGDTLKREMLDTNKCYILDCGIEVFVWMGRTTSLDDRKIASKAA 311
Query: 299 EKFLEQDFLLETISCSAPIYIVMEGSEP-PFFTRFFKWDSAKSAMLGNSYQRKLAIM 354
E+ + + + + ++EG E PF ++F W + + + ++A +
Sbjct: 312 EEMIR-----SSERPKSQMIRIIEGFETVPFRSKFESWTQETNTTVSEDGRGRVAAL 363
>At5g57320 villin
Length = 962
Score = 619 bits (1596), Expect = e-177
Identities = 329/570 (57%), Positives = 402/570 (69%), Gaps = 41/570 (7%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVE------- 93
QVWR+N +EK LL A +QSKFYSGDCYI QYSYPGEDREEHL+GTW GK SVE
Sbjct: 397 QVWRINCEEKILLEAAEQSKFYSGDCYILQYSYPGEDREEHLVGTWFGKQSVEEDRASAI 456
Query: 94 SFTSKQ---------NGRINEVYSIYGSYL*RQ*NNSILFHPPKLDCF*---------DE 135
S +K RINE ++ Q + I F D F D
Sbjct: 457 SLANKMVESMKFVPAQARINEGKEPIQFFVIMQ--SFITFKGGVSDAFKKYIAENDIPDT 514
Query: 136 TYKEDGVALFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQE 195
TY+ +GVALFR+QGSGPE+MQAIQ+ +A++ LNSS+CYIL +S VFTW GNLT+S+DQE
Sbjct: 515 TYEAEGVALFRVQGSGPENMQAIQIEAASAGLNSSHCYILHGDSTVFTWCGNLTSSEDQE 574
Query: 196 LAERMLDLIKPDLQCRPQKEGAETEQFWELLGVKTEYSSQKIVREAENDPHLFSCNFSE- 254
L ERMLDLIKP+ + QKEG+E+EQFWELLG K+EY SQKI R+ E+DPHLFSC ++
Sbjct: 575 LMERMLDLIKPNEPTKAQKEGSESEQFWELLGGKSEYPSQKIKRDGESDPHLFSCTYTNE 634
Query: 255 -----EIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFLLE 309
EI NF+QDDLMTEDIFILDCH+++FVWVGQQVDPK++ QAL IGE FL+ DFLLE
Sbjct: 635 SLKATEIFNFTQDDLMTEDIFILDCHTEVFVWVGQQVDPKKKPQALDIGENFLKHDFLLE 694
Query: 310 TISCSAPIYIVMEGSEPPFFTRFFKWDSAKSAMLGNSYQRKLAIMKNGGTPPLVKPKRRA 369
++ PIYIV EG+EPPFFTRFF WDS+KS M G+S+QRKLAI+ N G P L KPKRR
Sbjct: 695 NLASETPIYIVTEGNEPPFFTRFFTWDSSKSGMHGDSFQRKLAILTNKGKPLLDKPKRRV 754
Query: 370 SVSYGGRSGGLPEKSQ-RSRSMSVSPDRVRVRGRSPAFNALAATFENSNVRNLSTPP--- 425
+Y RS +P+KSQ RSRSM+ SPDR RVRGRSPAFNALAA FE N+RN STPP
Sbjct: 755 P-AYSSRS-TVPDKSQPRSRSMTFSPDRARVRGRSPAFNALAANFEKLNIRNQSTPPPMV 812
Query: 426 -PMIRKLYPKSKTPDLATLAPKSSAISHLTSTFEPPSAREKLIPRSLKDTSKSNPETNSD 484
PM+RKLYPKS PDL+ +APK SAI+ T+ FE P+ + P S + +N
Sbjct: 813 SPMVRKLYPKSHAPDLSKIAPK-SAIAARTALFEKPTPTSQEPPTSPSSSEATNQAEAPK 871
Query: 485 NENSTGSREESLTIQEDVNEGEPEDNEGLPVYPYESVKTDSTDPMPDIDVTKREAYLSPE 544
+ + T E +I ED E E E+ LP +PYE +KTDS DP+ D+D+T+REAYL+
Sbjct: 872 STSETNEEEAMSSINEDSKEEEAEEESSLPTFPYERLKTDSEDPVSDVDLTRREAYLTSV 931
Query: 545 EFQERLGMTRSEFYKLPKWKQNKLKMAVQL 574
EF+E+ MT++EFYKLPKWKQNKLKM+V L
Sbjct: 932 EFKEKFEMTKNEFYKLPKWKQNKLKMSVNL 961
Score = 54.7 bits (130), Expect = 1e-07
Identities = 73/339 (21%), Positives = 133/339 (38%), Gaps = 50/339 (14%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDRE-EHLIGTWIGKNSVES----- 94
++WR+ + + KF++GD YI + H I W+GK+S +
Sbjct: 22 EIWRIENFKPVTVPQESHGKFFTGDSYIVLKTTASRSGSLHHDIHYWLGKDSSQDEAGAV 81
Query: 95 ------FTSKQNGRINEVYSIYGSYL*RQ*NNSILFHPPKL--------DCF*DETYKED 140
S GR + + G + + + P + F +E
Sbjct: 82 AVMTVELDSALGGRAVQYREVQG----HETEKFLSYFKPCIIPQEGGVASGFNHVKPEEH 137
Query: 141 GVALFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERM 200
L+ +G ++ +V S+LN +IL +ES +F + G+ ++ ++ A +
Sbjct: 138 QTRLYICKGK--HVVRVKEVPFVRSTLNHEDVFILDTESKIFQFSGSKSSIQERAKALEV 195
Query: 201 LDLIKP---DLQC--RPQKEG-----AETEQFWELLGVKTEYSSQKIVRE----AENDPH 246
+ IK D +C ++G AE +FW L G + V + A +
Sbjct: 196 VQYIKDTYHDGKCDIAAVEDGRMMADAEAGEFWGLFGGFAPLPKKPAVNDDETAASDGIK 255
Query: 247 LFSCNFSE----EIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFL 302
LFS + E +++ L T +ILDC ++FVW G+ +R A E+F
Sbjct: 256 LFSVEKGQTDAVEAECLTKELLDTNKCYILDCGLELFVWKGRSTSIDQRKSATEAAEEFF 315
Query: 303 EQDFLLETISCSAPIYIVMEGSEPPFF-TRFFKWDSAKS 340
+ + + VMEG E F ++F W ++ +
Sbjct: 316 R-----SSEPPKSNLVSVMEGYETVMFRSKFDSWPASST 349
>At2g29890 villin 1 (VLN1)
Length = 909
Score = 349 bits (895), Expect = 3e-96
Identities = 215/571 (37%), Positives = 307/571 (53%), Gaps = 91/571 (15%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHLIGTWIGKNSVESF----- 95
+VWRV+G + +LL+ DQ+K ++GDCY+ QY Y ++R EHL+ WIG S++
Sbjct: 392 KVWRVDGDDVSLLSIPDQTKLFTGDCYLVQYKYTYKERTEHLLYVWIGCESIQQDRADAI 451
Query: 96 --------TSKQNGRINEVYS---------------IYGSYL*RQ*NNSILFHPPKLDCF 132
T+K + +Y ++ L R+ +L K+
Sbjct: 452 TNASAIVGTTKGESVLCHIYQGNEPSRFFPMFQSLVVFKGGLSRR-YKVLLAEKEKIG-- 508
Query: 133 *DETYKEDGVALFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSD 192
E Y E+ +LFR+ G+ P +MQAIQVN A+SLNSSY YILQ + FTW G L++
Sbjct: 509 --EEYNENKASLFRVVGTSPRNMQAIQVNLVATSLNSSYSYILQYGASAFTWIGKLSSDS 566
Query: 193 DQELAERMLDLIKPDLQCRPQ--KEGAETEQFWELLGVKTEYSSQKIVREAENDPHLFSC 250
D E+ +RML + D C+P +EG ET+ FW LLG K+EY +K +R+ +PHLF+C
Sbjct: 567 DHEVLDRMLYFL--DTSCQPIYIREGNETDTFWNLLGGKSEYPKEKEMRKQIEEPHLFTC 624
Query: 251 NFS------EEIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQ 304
+ S +EI+NF QDDL TED+F+LDC S+++VW+G + K + +AL +G KFLE
Sbjct: 625 SCSSDVLKVKEIYNFVQDDLTTEDVFLLDCQSEVYVWIGSNSNIKSKEEALTLGLKFLEM 684
Query: 305 DFLLETISCSAPIYIVMEGSEPPFFTRFFKWDSAKSAMLGNSYQRKLAIMKNGGTPPLVK 364
D L E ++ P+Y+V EG EPPFFTRFF+W K+ M GNS++RKLA +K T
Sbjct: 685 DILEEGLTMRTPVYVVTEGHEPPFFTRFFEWVPEKANMHGNSFERKLASLKGKKT----S 740
Query: 365 PKRRASVSYGGRSGGLPEKSQRSRSMSVSPDRVRVRGRSP-AFNALAATFENSNVRNLST 423
KR + Y +S + +SRS+S + RG SP + L + ++ N S
Sbjct: 741 TKRSSGSQYRSQSKDNASRDLQSRSVSSNGSE---RGVSPCSSEKLLSLSSAEDMTNSSN 797
Query: 424 PPPMIRKLYPKSKTPDLATLAPKSSAISHLTSTFEPPSAREKLIPRSLKDTSKSNPETNS 483
P+++KL+ +S D P + S D SK P
Sbjct: 798 STPVVKKLFSESLLVD-------------------PNDGVARQESSSKSDISKQKPRVGI 838
Query: 484 DNENSTGSREESLTIQEDVNEGEPEDNEGLPVYPYESVKTDSTDPMPDIDVTKREAYLSP 543
NS S ESL Y YE ++ DS P+ DID T+REAYL+
Sbjct: 839 ---NSDLSSLESL------------------AYSYEQLRVDSQKPVTDIDATRREAYLTE 877
Query: 544 EEFQERLGMTRSEFYKLPKWKQNKLKMAVQL 574
+EF+ER GM +SEFY LPKWKQNKLK+++ L
Sbjct: 878 KEFEERFGMAKSEFYALPKWKQNKLKISLHL 908
Score = 64.3 bits (155), Expect = 2e-10
Identities = 62/240 (25%), Positives = 107/240 (43%), Gaps = 29/240 (12%)
Query: 135 ETYKEDGVALFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQ 194
ETY+ V L R +G ++ +V SSLN +IL + S VF + G +++ ++
Sbjct: 134 ETYQ---VTLLRCKGD--HVVRVKEVPFLRSSLNHDDVFILDTASKVFLFAGCNSSTQEK 188
Query: 195 ELAERMLDLIKP---DLQCRPQ--KEG-----AETEQFWELLG--VKTEYSSQKIVREAE 242
A +++ IK D +C ++G ++ +FW G S +E
Sbjct: 189 AKAMEVVEYIKDNKHDGRCEVATIEDGKFSGDSDAGEFWSFFGGYAPIPKLSSSTTQEQT 248
Query: 243 NDP--HLFSCNFSEEIH-----NFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQAL 295
P LF + +H + +D L ++LDCHS++FVW+G+ R ++
Sbjct: 249 QTPCAELFWIDTKGNLHPTGTSSLDKDMLEKNKCYMLDCHSEVFVWMGRNTSLTERKTSI 308
Query: 296 PIGEKFLEQDFLLETISCSAPIYIVMEGSEPPFFTRFF-KWDSAKSAMLGNSYQRKLAIM 354
E+FL + E S + + ++ EG E F FF KW + L N + K+A +
Sbjct: 309 SSSEEFLRK----EGRSTTTSLVLLTEGLENARFRSFFNKWPQTVESSLYNEGREKVAAL 364
>At2g41740 putative villin 2 protein
Length = 976
Score = 292 bits (748), Expect = 3e-79
Identities = 191/593 (32%), Positives = 294/593 (49%), Gaps = 69/593 (11%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSY-PGEDREEHLIGTWIGKNSVESFTSKQ 99
+VW VNG+ K L D K YSGDCY+ Y+Y GE ++E+ + W GK S+ Q
Sbjct: 393 EVWYVNGKVKTPLPKEDIGKLYSGDCYLVLYTYHSGERKDEYFLSCWFGKKSIPE---DQ 449
Query: 100 NGRINEVYSIYGSYL*RQ*NNSILF--HPPKLDCF*------------------------ 133
+ I ++ S R I PP+
Sbjct: 450 DTAIRLANTMSNSLKGRPVQGRIYEGKEPPQFVALFQPMVVLKGGLSSGYKSSMGESEST 509
Query: 134 DETYKEDGVALFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDD 193
DETY + +AL ++ G+G + +A+QV + A+SLNS C++LQS + +F W+GN + +
Sbjct: 510 DETYTPESIALVQVSGTGVHNNKAVQVETVATSLNSYECFLLQSGTSMFLWHGNQSTHEQ 569
Query: 194 QELAERMLDLIKPDLQCRPQKEGAETEQFWELLGVKTEYSSQKIVREAENDPHLFSCNFS 253
ELA ++ + +KP + + KEG E+ FW LG K ++S+K E DPHLFS F+
Sbjct: 570 LELATKVAEFLKPGITLKHAKEGTESSTFWFALGGKQNFTSKKASSETIRDPHLFSFAFN 629
Query: 254 ------EEIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDFL 307
EEI+NF+QDDL+TEDI+ LD H+++FVWVGQ V+PK + IG+K+++
Sbjct: 630 RGKFQVEEIYNFAQDDLLTEDIYFLDTHAEVFVWVGQCVEPKEKQTVFEIGQKYIDLAGS 689
Query: 308 LETISCSAPIYIVMEGSEPPFFTRFFKWDSAKSAMLGNSYQRKLAIMKNGGTPPLVKPKR 367
LE + PIY + EG+EP FFT +F WD+ K+ + GNS+Q+K +++ GT +V+ K
Sbjct: 690 LEGLHPKVPIYKINEGNEPCFFTTYFSWDATKAIVQGNSFQKKASLL--FGTHHVVEDK- 746
Query: 368 RASVSYGGRSG-----------GLPEKSQRSRSMSVSPDRVR-----VRGRSPAFNALAA 411
S GG G S +R S DR+ R R+ A AL++
Sbjct: 747 ----SNGGNQGLRQRAEALAALNSAFNSSSNRPAYSSQDRLNESHDGPRQRAEALAALSS 802
Query: 412 TFENSNVRNLSTPPPM-IRKLYPKSKTPDLATLAP------KSSAISHLTSTFEPPSARE 464
F +S+ S PPP + + +A L+ K S + T + +
Sbjct: 803 AFNSSSSSTKSPPPPRPVGTSQASQRAAAVAALSQVLVAENKKSPDTSPTRRSTSSNPAD 862
Query: 465 KLIPRSLKDTSKSNPETN---SDNENSTGSREESLTIQEDVNEGEPEDNEGLPVYPYESV 521
+ KD +++ + E + + +E+ QE +G+ E + YE +
Sbjct: 863 DIPLTEAKDEEEASEVAGLEAKEEEEVSPAADETEAKQETEEQGDSEIQPSGATFTYEQL 922
Query: 522 KTDSTDPMPDIDVTKREAYLSPEEFQERLGMTRSEFYKLPKWKQNKLKMAVQL 574
+ S +P+ ID +REAYLS EEFQ G+ + F LP+WKQ+ LK L
Sbjct: 923 RAKSENPVTGIDFKRREAYLSEEEFQSVFGIEKEAFNNLPRWKQDLLKKKFDL 975
Score = 50.4 bits (119), Expect = 3e-06
Identities = 78/358 (21%), Positives = 138/358 (37%), Gaps = 49/358 (13%)
Query: 40 SQVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHL-IGTWIGKNSVESFTSK 98
+++WR+ E + ++ KFY GD YI + + I WIGK++ +
Sbjct: 19 TEIWRIENFEAVPVPKSEHGKFYMGDTYIVLQTTQNKGGAYLFDIHFWIGKDTSQDEAGT 78
Query: 99 QNGRINEVYSIYGSYL*R----Q*NNSILFHPPKLDCF*D---------ETYKEDGVALF 145
+ E+ ++ G + Q + S F C +T +E+
Sbjct: 79 AAVKTVELDAVLGGRAVQHREIQGHESDKFLSYFKPCIIPLEGGVASGFKTVEEEVFETR 138
Query: 146 RIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLDLIK 205
G +++ QV A SSLN +IL +E ++ + G +N ++ A ++ +K
Sbjct: 139 LYTCKGKRAIRLKQVPFARSSLNHDDVFILDTEEKIYQFNGANSNIQERAKALEVVQYLK 198
Query: 206 PDLQ--------CRPQKEGAETEQ--FWELLGVKTEYSSQKIVREAENDPHLFSCNFSEE 255
K E++ FW L G I R+ ND + + +
Sbjct: 199 DKYHEGTCDVAIVDDGKLDTESDSGAFWVLFG-----GFAPIGRKVANDDDIVPESTPPK 253
Query: 256 IH------------NFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLE 303
++ + S+ L ++LDC ++I++WVG+ R A E+FL
Sbjct: 254 LYCITDGKMEPIDGDLSKSMLENTKCYLLDCGAEIYIWVGRVTQVDERKAASQSAEEFLA 313
Query: 304 QDFLLETISCSAPIYIVMEGSEP-PFFTRFFKWDSAKSAMLGNSYQR--KLAIMKNGG 358
E + + V++G E F + F W S SA GN R A++K G
Sbjct: 314 S----ENRPKATHVTRVIQGYESHSFKSNFDSWPSG-SATPGNEEGRGKVAALLKQQG 366
>At3g57410 villin 3
Length = 965
Score = 288 bits (736), Expect = 7e-78
Identities = 194/614 (31%), Positives = 292/614 (46%), Gaps = 124/614 (20%)
Query: 41 QVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSY-PGEDREEHLIGTWIGKNS-------- 91
+VW ++ K +L+ K YSGDCY+ Y+Y GE +E++ + W GKNS
Sbjct: 395 EVWYIDANSKTVLSKDHVGKLYSGDCYLVLYTYHSGERKEDYFLCCWFGKNSNQEDQETA 454
Query: 92 ---VESFTSKQNGRINEVYSIYGSYL*RQ*NNSILFHPPKLDCF*D-------------- 134
+ T+ GR + G PP+
Sbjct: 455 VRLASTMTNSLKGRPVQARIFEGK------------EPPQFVALFQHMVVLKGGLSSGYK 502
Query: 135 ----------ETYKEDGVALFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTW 184
ETY + +AL ++ G+G + +A+QV + A+SLNS C++LQS + +F W
Sbjct: 503 NSMTEKGSSGETYTPESIALIQVSGTGVHNNKALQVEAVATSLNSYDCFLLQSGTSMFLW 562
Query: 185 YGNLTNSDDQELAERMLDLIKPDLQCRPQKEGAETEQFWELLGVKTEYSSQKIVREAEND 244
GN + + QELA ++ + +KP + KEG E+ FW LG K ++S+K+ E D
Sbjct: 563 VGNHSTHEQQELAAKVAEFLKPGTTIKHAKEGTESSSFWFALGGKQNFTSKKVSSETVRD 622
Query: 245 PHLFSCNFS------EEIHNFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIG 298
PHLFS +F+ EEIHNF QDDL+TE++ +LD H+++FVWVGQ VDPK + A IG
Sbjct: 623 PHLFSFSFNRGKFQVEEIHNFDQDDLLTEEMHLLDTHAEVFVWVGQCVDPKEKQTAFEIG 682
Query: 299 EKFLEQDFLLETISCSAPIYIVMEGSEPPFFTRFFKWDSAKSAMLGNSYQRKLAIMKNGG 358
++++ LE +S P+Y + EG+EP FFT +F WDS K+ + GNSYQ+K A++ G
Sbjct: 683 QRYINLAGSLEGLSPKVPLYKITEGNEPCFFTTYFSWDSTKATVQGNSYQKKAALLL--G 740
Query: 359 TPPLVK---------PKRRASVSYGGRSGGLPEKSQRSRSMSVSPDRVRVRG-------R 402
T +V+ P++RA+ L S + SP R R G R
Sbjct: 741 THHVVEDQSSSGNQGPRQRAAA-----LAALTSAFNSSSGRTSSPSRDRSNGSQGGPRQR 795
Query: 403 SPAFNALAATFENSNVRNLSTPPPMIRKLYPKSKTPDLATLAPKSSAISHLTSTFEPPSA 462
+ A AL + F +S P SK+P P+ S LTS +A
Sbjct: 796 AEALAALTSAFNSS----------------PSSKSP------PRRSG---LTSQASQRAA 830
Query: 463 REKLIPRSLKDTSKSNPETNSD--------------NENSTGSREE---SLTIQEDVNEG 505
+ + L K +P+T+ E +T ++EE S + E
Sbjct: 831 AVAALSQVLTAEKKKSPDTSPSAEAKDEKAFSEVEATEEATEAKEEEEVSPAAEASAEEA 890
Query: 506 EPEDNEGL-----PVYPYESVKTDSTDPMPDIDVTKREAYLSPEEFQERLGMTRSEFYKL 560
+P+ ++ + YE ++ S P+ ID +REAYLS EF+ GM + FYKL
Sbjct: 891 KPKQDDSEVETTGVTFTYERLQAKSEKPVTGIDFKRREAYLSEVEFKTVFGMEKESFYKL 950
Query: 561 PKWKQNKLKMAVQL 574
P WKQ+ LK L
Sbjct: 951 PGWKQDLLKKKFNL 964
Score = 57.4 bits (137), Expect = 2e-08
Identities = 76/349 (21%), Positives = 137/349 (38%), Gaps = 40/349 (11%)
Query: 40 SQVWRVNGQEKNLLAATDQSKFYSGDCYIFQYSYPGEDREEHL-IGTWIGKNSVESFTSK 98
+++WR+ E + ++ KFY GD YI + + I WIGK++ +
Sbjct: 21 TEIWRIENFEPVPVPKSEHGKFYMGDTYIVLQTTQNKGGAYLFDIHFWIGKDTSQDEAGT 80
Query: 99 QNGRINEVYSIYGS----YL*RQ*NNSILFHPPKLDC-----------F*DETYKEDGVA 143
+ E+ + G Y Q + S F C F +E
Sbjct: 81 AAVKTVELDAALGGRAVQYREIQGHESDKFLSYFKPCIIPLEGGVASGFKKPEEEEFETR 140
Query: 144 LFRIQGSGPESMQAIQVNSAASSLNSSYCYILQSESVVFTWYGNLTNSDDQELAERMLDL 203
L+ +G ++ QV A SSLN +IL ++ ++ + G +N ++ A ++
Sbjct: 141 LYTCKGK--RAVHLKQVPFARSSLNHDDVFILDTKEKIYQFNGANSNIQERAKALVVIQY 198
Query: 204 IKPDLQCRPQKEG----------AETEQFWELLG----VKTEYSSQKIVREAENDPHLFS 249
+K +++ +FW L G + + +S+ + P L+S
Sbjct: 199 LKDKFHEGTSDVAIVDDGKLDTESDSGEFWVLFGGFAPIARKVASEDEIIPETTPPKLYS 258
Query: 250 CNFS--EEIH-NFSQDDLMTEDIFILDCHSQIFVWVGQQVDPKRRVQALPIGEKFLEQDF 306
E I + S+ L ++LDC S+IF+WVG+ + R A+ E F+
Sbjct: 259 IADGQVESIDGDLSKSMLENNKCYLLDCGSEIFIWVGRVTQVEERKTAIQAAEDFVAS-- 316
Query: 307 LLETISCSAPIYIVMEGSEP-PFFTRFFKWDSAKSAMLGNSYQRKLAIM 354
E + I V++G EP F + F W S + + K+A +
Sbjct: 317 --ENRPKATRITRVIQGYEPHSFKSNFDSWPSGSATPANEEGRGKVAAL 363
Score = 36.2 bits (82), Expect = 0.051
Identities = 41/194 (21%), Positives = 74/194 (38%), Gaps = 42/194 (21%)
Query: 147 IQGSGPESMQAIQVNSAA-------SSLNSSYCYIL---------QSESVVFTWYGNLTN 190
++G G + I NS L S CY++ + + + W+G +N
Sbjct: 388 LEGGGKLEVWYIDANSKTVLSKDHVGKLYSGDCYLVLYTYHSGERKEDYFLCCWFGKNSN 447
Query: 191 SDDQELAERMLDLIKPDLQCRPQK----EGAETEQFWELL--------GVKTEYSSQKIV 238
+DQE A R+ + L+ RP + EG E QF L G+ + Y +
Sbjct: 448 QEDQETAVRLASTMTNSLKGRPVQARIFEGKEPPQFVALFQHMVVLKGGLSSGYKNSMTE 507
Query: 239 REAENDPH------LFSCNFSEEIHNFS-------QDDLMTEDIFILDCHSQIFVWVGQQ 285
+ + + + L + +HN L + D F+L + +F+WVG
Sbjct: 508 KGSSGETYTPESIALIQVS-GTGVHNNKALQVEAVATSLNSYDCFLLQSGTSMFLWVGNH 566
Query: 286 VDPKRRVQALPIGE 299
+++ A + E
Sbjct: 567 STHEQQELAAKVAE 580
>At1g31480 hypothetical protein
Length = 869
Score = 38.5 bits (88), Expect = 0.010
Identities = 27/91 (29%), Positives = 44/91 (47%), Gaps = 15/91 (16%)
Query: 420 NLSTPPPM---IRKLYPKSKTPDLATLAPKSSAISHLTSTFEPPSAREKLIPRSLKDTSK 476
NLS+P PM +K +P ++P A K + SH +S FEP K
Sbjct: 459 NLSSPFPMDSVYKKFFPDEESPPTPAKADKPCS-SHPSSNFEPE-----------KSDQL 506
Query: 477 SNPETNSDNENSTGSREESLTIQEDVNEGEP 507
+NPE + +N+T ++E ++ DV + +P
Sbjct: 507 NNPEKITGQDNNTMAKEPTVLEHHDVIQEDP 537
>At5g58360 similar to unknown protein (gb|AAB63094.1)
Length = 296
Score = 37.4 bits (85), Expect = 0.023
Identities = 44/165 (26%), Positives = 72/165 (42%), Gaps = 14/165 (8%)
Query: 416 SNVRNLSTPPPMIRKLYPKSKTPDLATLAPKSSAISHLTS--TFEPPSAREKL-IPRSLK 472
S+ R L P +R+L + P + PKSS+ T PS+R KL SL
Sbjct: 48 SSSRKLRDP---LRRLSSTAHHPQASNSPPKSSSFKRKIKRKTVYKPSSRLKLSTSSSLN 104
Query: 473 DTSKSNPETNSDNENSTGSREESLTIQEDVN-EGEPEDNEGLPVYPYESV-KTDSTDPMP 530
SKS+ N+ ++++ GS + ++ D N +PE + + + SV K D P
Sbjct: 105 HRSKSSSSANAISDSAVGSFLDRVSSPSDQNFVHDPEPHSSIDIKDELSVRKLDDVPEDP 164
Query: 531 DIDVTKREAYLSPEEFQE-RLGMTRSEFYKLPKWKQNKLKMAVQL 574
+ LSPE +E M + K PK + +K+ ++
Sbjct: 165 SVSPN-----LSPETAKEPPFEMMTQQKLKKPKAHSSGIKIPTKI 204
>At1g52980 unknown protein
Length = 576
Score = 37.0 bits (84), Expect = 0.030
Identities = 33/131 (25%), Positives = 55/131 (41%), Gaps = 9/131 (6%)
Query: 400 RGRSPAFNALAATFENSNVRNLSTPPPMIRKLYPKSKTPDLATLAPKSSAISHLTSTFEP 459
RGR P F + N+++ +I K D + A AI+ + ST +
Sbjct: 452 RGRIPFF------VPPPKLDNVASESEVIVPGIDKEAIADNSQAAAALKAIAGIMSTQQQ 505
Query: 460 ---PSAREKLIPRSLKDTSKSNPETNSDNENSTGSREESLTIQEDVNEGEPEDNEGLPVY 516
P R+ + LKD K+ T +D EN T + E+ + ED E + + +E
Sbjct: 506 KDVPVQRDFYDEKDLKDDKKAKESTETDAENGTDAEEDEDAVSEDGVESDSDADEDAVSE 565
Query: 517 PYESVKTDSTD 527
E ++DS +
Sbjct: 566 NDEEDESDSAE 576
>At1g76970 unknown protein
Length = 446
Score = 35.0 bits (79), Expect = 0.11
Identities = 25/95 (26%), Positives = 39/95 (40%), Gaps = 3/95 (3%)
Query: 415 NSNVRNLSTPPPMIRKLYPKSKTPDLATLAPKSSAISHLTSTFEPPSAREKLIPRSLKDT 474
NS+ + + PPP S +P +P+ S S + PP +R + +
Sbjct: 331 NSSSQGVKKPPPPPPHTSSSSSSPVFDDASPQQSKSSEVIRNLPPPPSRHNQRQQFFEHH 390
Query: 475 SKSNPETNSDNENSTGSREESLTIQEDVNEGEPED 509
S + SD+ +R SLT E E +PED
Sbjct: 391 HSS---SGSDSSYEGQTRNLSLTSSEPQKEEKPED 422
>At3g06670 unknown protein
Length = 863
Score = 33.9 bits (76), Expect = 0.25
Identities = 45/201 (22%), Positives = 71/201 (34%), Gaps = 25/201 (12%)
Query: 336 DSAKSAMLGNSYQRKLA--IMKNGGTPPLVKPKRRASVSYGGRSGGLPEKSQRSRSMSVS 393
D SA N+ + K A I K P L + S RSGGL +
Sbjct: 664 DEEDSASASNTQKEKPASNIQKEQPKPHLSNGVAASPTSSSPRSGGLVDYEDDEDDEDYK 723
Query: 394 PDRVRVRGRSPAFNALAATFENSNVRNLSTPPPMIRKLYPKSKTPDLATLAPKSSAISHL 453
P P A+ E + L ++ + SK P L + + + + L
Sbjct: 724 PP--------PRKQPEASEDEEGELLRLKRKSALVEREQEPSKKPRLGKSSKRENVFAVL 775
Query: 454 TSTFEPP------------SAREKLIPRSLKDTSKSNPETNSDNENSTGSREE--SLTIQ 499
ST SA ++ + +D SKS+ E NS + + +++ S +
Sbjct: 776 CSTLSHAVLTGKKSPGPAGSAARSIVAKGAED-SKSSEENNSSSSDDENHKDDGVSSSEH 834
Query: 500 EDVNEGEPEDNEGLPVYPYES 520
E + G+ E L V P S
Sbjct: 835 ETSDNGKLNGEESLVVAPKSS 855
>At3g06480 putative RNA helicase
Length = 1088
Score = 33.9 bits (76), Expect = 0.25
Identities = 34/118 (28%), Positives = 49/118 (40%), Gaps = 14/118 (11%)
Query: 359 TPPLVKPKRRASVSYG---GRSGGLPEKSQRSRSMSVSPDRVRVRGRSPAFNALAATFEN 415
+P LV+P+RR+S SY RSG RSRS S R R RSP +
Sbjct: 958 SPDLVRPRRRSS-SYSRSRSRSGSYSRSRSRSRSWS------RSRSRSPRHSRDRGGHNR 1010
Query: 416 SNVRNLSTPPPMIRKLYPKSKTPDLATLAPKSSAISHLTSTFEPPSAREKLIPRSLKD 473
S + S P R + + P ++ K S + +A E ++P SL +
Sbjct: 1011 SRSYSRSPSPVYER----RDRAPRVSGFDIKPPVESVVNLDMNAAAAIENVVPTSLSE 1064
>At2g06490 putative PttA-like transposon protein
Length = 491
Score = 33.1 bits (74), Expect = 0.43
Identities = 27/135 (20%), Positives = 52/135 (38%)
Query: 375 GRSGGLPEKSQRSRSMSVSPDRVRVRGRSPAFNALAATFENSNVRNLSTPPPMIRKLYPK 434
GR G + S + V + V G S + ++ NS ++ + P
Sbjct: 14 GRGRGRGRVRESSTGVQVFQGKNPVLGSSGHRASSHSSNPNSATQSHRSRPSQYPPATVP 73
Query: 435 SKTPDLATLAPKSSAISHLTSTFEPPSAREKLIPRSLKDTSKSNPETNSDNENSTGSREE 494
TP TL P +S L+ + +PP L P + S + + + + + +
Sbjct: 74 QTTPPQTTLTPTNSRQPLLSGSQQPPPVHRNLPPTRQRQAPTSQQQPPTCQQKAPSRQHQ 133
Query: 495 SLTIQEDVNEGEPED 509
SL +Q+ + E ++
Sbjct: 134 SLGLQQQPRQTEQQN 148
>At5g27950 unknown protein
Length = 625
Score = 32.7 bits (73), Expect = 0.56
Identities = 22/65 (33%), Positives = 29/65 (43%), Gaps = 1/65 (1%)
Query: 336 DSAKSAMLGNSYQRKLAIMKNGGTPPLVKPKRRASVSYGGRSGGLPEKSQRSRSMSVSPD 395
DS A+L SY + L N GTPP PKR + R S RS+ ++ S
Sbjct: 550 DSRSKALLRRSYTKPLQAAANSGTPP-ETPKRHIKDNSLQRKNMNDTSSPRSKMVTSSDP 608
Query: 396 RVRVR 400
VR +
Sbjct: 609 NVRAK 613
>At5g03740 histone deacetylase -like protein
Length = 294
Score = 32.7 bits (73), Expect = 0.56
Identities = 21/68 (30%), Positives = 32/68 (46%), Gaps = 3/68 (4%)
Query: 444 APKSSAISHLTSTFEPPSAREKLIPRSLKDTSKSNPETNSDNENSTGSREESLTIQEDVN 503
APKS+A F+ P+ K D S+ + + D+ENS EE +T + D
Sbjct: 117 APKSAAKQ---VNFQLPNEDVKAKQDDDADGSEEDSSDDDDSENSGDEEEEKVTAESDSE 173
Query: 504 EGEPEDNE 511
E + D+E
Sbjct: 174 EDDSSDDE 181
>At3g55270 MAP kinase phosphatase (MKP1)
Length = 732
Score = 32.7 bits (73), Expect = 0.56
Identities = 27/85 (31%), Positives = 37/85 (42%), Gaps = 25/85 (29%)
Query: 270 FILDCHSQIFVWVGQQV------DPKRRVQALPIGEKFLEQDFLLETISCSAPIYIVMEG 323
FI+ S I++WVG+Q D K V + EK API +V EG
Sbjct: 283 FIIQLPSAIYIWVGRQCETIMEKDAKAAVCQIARYEK------------VEAPIMVVREG 330
Query: 324 SEPPFFTRFFKWDSAKS--AMLGNS 346
EP ++ WD+ S M+G S
Sbjct: 331 DEPVYY-----WDAFASILPMIGGS 350
>At2g05080 putative helicase
Length = 1219
Score = 32.3 bits (72), Expect = 0.73
Identities = 23/63 (36%), Positives = 28/63 (43%), Gaps = 6/63 (9%)
Query: 493 EESLTIQEDVNEGEPEDNEGLPVYPYESVKTDSTDPMPDI------DVTKREAYLSPEEF 546
EE + D GEP D E L P E + T + DP+ I D+TK P F
Sbjct: 937 EEWILAVGDGRIGEPNDGEALIDIPSEFLITKAKDPIQAICTEIYGDITKIHEQKDPVFF 996
Query: 547 QER 549
QER
Sbjct: 997 QER 999
>At5g46750 zinc finger protein Glo3-like
Length = 402
Score = 31.6 bits (70), Expect = 1.3
Identities = 33/154 (21%), Positives = 63/154 (40%), Gaps = 15/154 (9%)
Query: 373 YGGRSGGLPEKSQRSRSMSVSPDRVRVRGRSPAFNALAATFENSNVRNLSTPPPMIRKLY 432
+G GG E SR+ D R A+A ++ +++T P+
Sbjct: 93 HGWNDGGKIEAKYTSRAA----DMYRQTLAKEVAKAMAEETVLPSLSSVATSQPV----- 143
Query: 433 PKSKTPDLATLAPKSSAISHLTSTFEPPSAREKLIPRSLKDTSKSNPETNSDNENSTGSR 492
+S + +PK S++ + P A +K++ + K K S G+R
Sbjct: 144 -ESSENGFTSESPKESSLKQEAAVVSSPKASQKVVASTFK---KPLVSRKSGKTGGLGAR 199
Query: 493 EESLTIQEDVNEGEPEDNEGLPVYPYESVKTDST 526
+ + ++++ E +PE E +PV P S D++
Sbjct: 200 KLTTKSKDNLYEQKPE--EPVPVIPAASPTNDTS 231
>At3g49430 PRE-MRNA SPLICING FACTOR SF2-like protein
Length = 300
Score = 31.6 bits (70), Expect = 1.3
Identities = 32/116 (27%), Positives = 55/116 (46%), Gaps = 12/116 (10%)
Query: 382 EKSQRSRSMSVSPDRVRVRGRSPAFNALAATFENSNVRNLSTPPPMIRKLYPKSKTPDLA 441
+K + SRS S SP R R R RS + + +S R+LS KS DL+
Sbjct: 196 KKYESSRSRSRSPSRSRSRSRSRS-RSRGRGRSHSRSRSLSR---------SKSPRKDLS 245
Query: 442 TLAPKSSAISHLTSTFEPPSAREKLIPRSL-KDTSKSNPETNSDNENSTGSREESL 496
+P+ S ++ + P ++K PR++ + S+S + S +++ RE S+
Sbjct: 246 K-SPRRSLSRSISKSRSPSPDKKKSPPRAMSRSKSRSRSRSRSPSKSPPKVREGSV 300
>At2g43680 SF16 protein {Helianthus annuus} like protein
Length = 668
Score = 31.6 bits (70), Expect = 1.3
Identities = 20/64 (31%), Positives = 31/64 (48%), Gaps = 3/64 (4%)
Query: 419 RNLSTPPPMIRKLYPKSKTPDLATLAPKSSAISHLTSTFEPPSAR---EKLIPRSLKDTS 475
R+LS PP R P+S +P + SSA ++ T P S R +++ P S+
Sbjct: 139 RSLSPKPPSPRADLPRSLSPKPFDRSKPSSASANAPPTLRPASTRVPSQRITPHSVPSPR 198
Query: 476 KSNP 479
S+P
Sbjct: 199 PSSP 202
>At2g22490 putative cyclin D
Length = 361
Score = 31.6 bits (70), Expect = 1.3
Identities = 31/135 (22%), Positives = 55/135 (39%), Gaps = 16/135 (11%)
Query: 360 PPLVKPKRRASVSYGGRSGGLPEKSQRSRSMSVSPDRVRVRGRSPAFNALAATFENSNVR 419
P + SVS G + + E+ S + V +RV+ N + + NVR
Sbjct: 242 PSEIAAAAAVSVSISGETECIDEEKALSSLIYVKQERVK-----RCLNLMRSLTGEENVR 296
Query: 420 NLSTPPPMIRKLYPKSKTPDLATLAPKSSAISHLTSTFEPPSAREKLIPRSLKDTSKSNP 479
S R +A A +S + L +T + E+ + S ++S+S+P
Sbjct: 297 GTSLSQEQAR----------VAVRAVPASPVGVLEATCLSYRSEERTV-ESCTNSSQSSP 345
Query: 480 ETNSDNENSTGSREE 494
+ N++N NS R +
Sbjct: 346 DNNNNNNNSNKRRRK 360
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,527,429
Number of Sequences: 26719
Number of extensions: 610558
Number of successful extensions: 2329
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 47
Number of HSP's that attempted gapping in prelim test: 2244
Number of HSP's gapped (non-prelim): 95
length of query: 574
length of database: 11,318,596
effective HSP length: 105
effective length of query: 469
effective length of database: 8,513,101
effective search space: 3992644369
effective search space used: 3992644369
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC125475.5


