

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC125368.9 + phase: 0
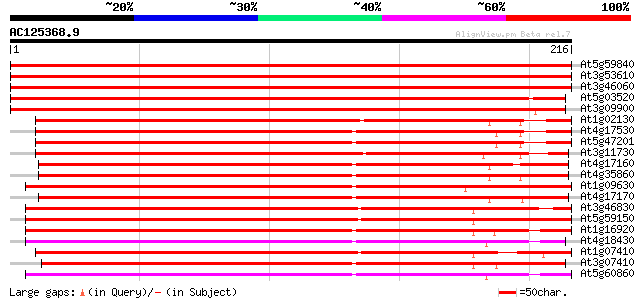
(216 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g59840 GTP-binding protein ara-3 409 e-115
At3g53610 GTPase AtRAB8 409 e-115
At3g46060 GTP-binding protein ara-3 408 e-114
At5g03520 GTP-binding protein - like 372 e-103
At3g09900 putative Ras-like GTP-binding protein 365 e-102
At1g02130 GTP-binding protein, ara-5 251 2e-67
At4g17530 ras-related small GTP-binding protein RAB1c 239 1e-63
At5g47201 ras-related small GTP-binding protein-like 237 4e-63
At3g11730 putative GTP-binding protein (ATFP8) 233 8e-62
At4g17160 GTP-binding RAB2A like protein 193 5e-50
At4g35860 GTP-binding protein GB2 192 1e-49
At1g09630 putative RAS-related protein, RAB11C 186 6e-48
At4g17170 GTP-binding RAB2A like protein 186 8e-48
At3g46830 GTP-binding protein Rab11 183 7e-47
At5g59150 GTP-binding protein rab11 - like 180 6e-46
At1g16920 unknown protein 178 2e-45
At4g18430 membrane-bound small GTP-binding - like protein 177 5e-45
At1g07410 small G protein, putative 177 5e-45
At3g07410 GTP-binding protein like 175 1e-44
At5g60860 GTP-binding protein - like 172 9e-44
>At5g59840 GTP-binding protein ara-3
Length = 216
Score = 409 bits (1052), Expect = e-115
Identities = 206/216 (95%), Positives = 210/216 (96%)
Query: 1 MAAPPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD 60
MAAPPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD
Sbjct: 1 MAAPPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD 60
Query: 61 GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDN 120
GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDN
Sbjct: 61 GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDN 120
Query: 121 VNKILVGNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQR 180
VNKILVGNKADMDESKRAVP SKGQALADEYGIKFFETSAKTNLNVEEVFFSIA+DIKQR
Sbjct: 121 VNKILVGNKADMDESKRAVPKSKGQALADEYGIKFFETSAKTNLNVEEVFFSIAKDIKQR 180
Query: 181 LADTDSKSEPQTIKINQPDQGAGAAQAAQKSACCGS 216
LADTDS++EP TIKI+Q DQ AGA QA QKSACCGS
Sbjct: 181 LADTDSRAEPATIKISQTDQAAGAGQATQKSACCGS 216
>At3g53610 GTPase AtRAB8
Length = 216
Score = 409 bits (1052), Expect = e-115
Identities = 204/216 (94%), Positives = 212/216 (97%)
Query: 1 MAAPPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD 60
MAAPPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD
Sbjct: 1 MAAPPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD 60
Query: 61 GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDN 120
GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASD+
Sbjct: 61 GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDS 120
Query: 121 VNKILVGNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQR 180
VNKILVGNKADMDESKRAVP SKGQALADEYG+KFFETSAKTNLNVEEVFFSIA+DIKQR
Sbjct: 121 VNKILVGNKADMDESKRAVPKSKGQALADEYGMKFFETSAKTNLNVEEVFFSIAKDIKQR 180
Query: 181 LADTDSKSEPQTIKINQPDQGAGAAQAAQKSACCGS 216
LADTD+++EPQTIKINQ DQGAG +QA QKSACCG+
Sbjct: 181 LADTDARAEPQTIKINQSDQGAGTSQATQKSACCGT 216
>At3g46060 GTP-binding protein ara-3
Length = 216
Score = 408 bits (1048), Expect = e-114
Identities = 204/216 (94%), Positives = 210/216 (96%)
Query: 1 MAAPPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD 60
MAAPPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD
Sbjct: 1 MAAPPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD 60
Query: 61 GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDN 120
GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDN
Sbjct: 61 GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDN 120
Query: 121 VNKILVGNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQR 180
VNKILVGNKADMDESKRAVPT+KGQALADEYGIKFFETSAKTNLNVEEVFFSI RDIKQR
Sbjct: 121 VNKILVGNKADMDESKRAVPTAKGQALADEYGIKFFETSAKTNLNVEEVFFSIGRDIKQR 180
Query: 181 LADTDSKSEPQTIKINQPDQGAGAAQAAQKSACCGS 216
L+DTDS++EP TIKI+Q DQ AGA QA QKSACCG+
Sbjct: 181 LSDTDSRAEPATIKISQTDQAAGAGQATQKSACCGT 216
>At5g03520 GTP-binding protein - like
Length = 216
Score = 372 bits (954), Expect = e-103
Identities = 186/214 (86%), Positives = 200/214 (92%), Gaps = 1/214 (0%)
Query: 1 MAAPPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD 60
MA PARAR+DYDYLIKLLLIGDSGVGKSCLLLRFSD +FTTSFITTIGIDFKIRT+ELD
Sbjct: 1 MAVAPARARSDYDYLIKLLLIGDSGVGKSCLLLRFSDDTFTTSFITTIGIDFKIRTVELD 60
Query: 61 GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDN 120
GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNW++NIEQHASDN
Sbjct: 61 GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWMKNIEQHASDN 120
Query: 121 VNKILVGNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQR 180
VNKILVGNKADMDESKRAVPT+KGQALADEYGIKFFETSAKTNLNVE VF SIA+DIKQR
Sbjct: 121 VNKILVGNKADMDESKRAVPTAKGQALADEYGIKFFETSAKTNLNVENVFMSIAKDIKQR 180
Query: 181 LADTDSKSEPQTIKINQPDQGAGAAQAAQKSACC 214
L +TD+K+EPQ IKI + D A ++ A+KSACC
Sbjct: 181 LTETDTKAEPQGIKITKQDT-AASSSTAEKSACC 213
>At3g09900 putative Ras-like GTP-binding protein
Length = 218
Score = 365 bits (938), Expect = e-102
Identities = 183/215 (85%), Positives = 199/215 (92%), Gaps = 1/215 (0%)
Query: 1 MAAPPARARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD 60
MA PARAR+DYDYLIKLLLIGDSGVGKSCLLLRFSD +FTTSFITTIGIDFKIRT+ELD
Sbjct: 1 MAVAPARARSDYDYLIKLLLIGDSGVGKSCLLLRFSDDTFTTSFITTIGIDFKIRTVELD 60
Query: 61 GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDN 120
GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNW++NIEQHASD+
Sbjct: 61 GKRIKLQIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWMKNIEQHASDS 120
Query: 121 VNKILVGNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQR 180
VNKILVGNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTN NVE+VF SIA+DIKQR
Sbjct: 121 VNKILVGNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNQNVEQVFLSIAKDIKQR 180
Query: 181 LADTDSKSEPQTIKINQPDQG-AGAAQAAQKSACC 214
L ++D+K+EPQ IKI + D A ++ +KSACC
Sbjct: 181 LTESDTKAEPQGIKITKQDANKASSSSTNEKSACC 215
>At1g02130 GTP-binding protein, ara-5
Length = 203
Score = 251 bits (642), Expect = 2e-67
Identities = 126/209 (60%), Positives = 160/209 (76%), Gaps = 12/209 (5%)
Query: 11 DYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWD 70
+YDYL KLLLIGDSGVGKSCLLLRFSD S+ S+I+TIG+DFKIRT+E DGK IKLQIWD
Sbjct: 4 EYDYLFKLLLIGDSGVGKSCLLLRFSDDSYVESYISTIGVDFKIRTVEQDGKTIKLQIWD 63
Query: 71 TAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKA 130
TAGQERFRTIT++YYRGA GI++VYDVTDE SFNN++ W+ I+++ASDNVNK+LVGNK+
Sbjct: 64 TAGQERFRTITSSYYRGAHGIIIVYDVTDEESFNNVKQWLSEIDRYASDNVNKLLVGNKS 123
Query: 131 DMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLAD--TDSKS 188
D+ E+ RA+P +A ADE GI F ETSAK NVE+ F +++ IK+R+A + +
Sbjct: 124 DLTEN-RAIPYETAKAFADEIGIPFMETSAKDATNVEQAFMAMSASIKERMASQPAGNNA 182
Query: 189 EPQTIKI-NQPDQGAGAAQAAQKSACCGS 216
P T++I QP AQK+ CC +
Sbjct: 183 RPPTVQIRGQP--------VAQKNGCCST 203
>At4g17530 ras-related small GTP-binding protein RAB1c
Length = 202
Score = 239 bits (609), Expect = 1e-63
Identities = 123/208 (59%), Positives = 154/208 (73%), Gaps = 11/208 (5%)
Query: 11 DYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWD 70
+YDYL KLLLIGDSGVGKSCLLLRF+D S+ S+I+TIG+DFKIRT+E DGK IKLQIWD
Sbjct: 4 EYDYLFKLLLIGDSGVGKSCLLLRFADDSYLDSYISTIGVDFKIRTVEQDGKTIKLQIWD 63
Query: 71 TAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKA 130
TAGQERFRTIT++YYRGA GI++ YDVTD SFNN++ W+ I+++AS+NVNK+LVGNK
Sbjct: 64 TAGQERFRTITSSYYRGAHGIIVTYDVTDLESFNNVKQWLNEIDRYASENVNKLLVGNKC 123
Query: 131 DMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDS-KSE 189
D+ S++ V T +A ADE GI F ETSAK NVEE F ++ IK R+A + S+
Sbjct: 124 DL-TSQKVVSTETAKAFADELGIPFLETSAKNATNVEEAFMAMTAAIKTRMASQPAGGSK 182
Query: 190 PQTIKI-NQPDQGAGAAQAAQKSACCGS 216
P T++I QP Q+S CC S
Sbjct: 183 PPTVQIRGQP--------VNQQSGCCSS 202
>At5g47201 ras-related small GTP-binding protein-like
Length = 202
Score = 237 bits (604), Expect = 4e-63
Identities = 122/208 (58%), Positives = 154/208 (73%), Gaps = 11/208 (5%)
Query: 11 DYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWD 70
+YDYL KLLLIGDSGVGKSCLLLRF+D S+ S+I+TIG+DFKIRT+E DGK IKLQIWD
Sbjct: 4 EYDYLFKLLLIGDSGVGKSCLLLRFADDSYLDSYISTIGVDFKIRTVEQDGKTIKLQIWD 63
Query: 71 TAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKA 130
TAGQERFRTIT++YYRGA GI++ YDVTD SFNN++ W+ I+++AS+NVNK+LVGNK
Sbjct: 64 TAGQERFRTITSSYYRGAHGIIVTYDVTDLESFNNVKQWLNEIDRYASENVNKLLVGNKN 123
Query: 131 DMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDS-KSE 189
D+ S++ V T +A ADE GI F ETSAK NVEE F ++ IK R+A + ++
Sbjct: 124 DL-TSQKVVSTETAKAFADELGIPFLETSAKNATNVEEAFMAMTAAIKTRMASQPAGGAK 182
Query: 190 PQTIKI-NQPDQGAGAAQAAQKSACCGS 216
P T++I QP Q+S CC S
Sbjct: 183 PPTVQIRGQP--------VNQQSGCCSS 202
>At3g11730 putative GTP-binding protein (ATFP8)
Length = 205
Score = 233 bits (593), Expect = 8e-62
Identities = 119/209 (56%), Positives = 156/209 (73%), Gaps = 12/209 (5%)
Query: 11 DYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWD 70
+YDYL KLLLIGDS VGKSCLLLRF+D ++ S+I+TIG+DFKIRTIE DGK IKLQIWD
Sbjct: 4 EYDYLFKLLLIGDSSVGKSCLLLRFADDAYIDSYISTIGVDFKIRTIEQDGKTIKLQIWD 63
Query: 71 TAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKA 130
TAGQERFRTIT++YYRGA GI++VYD T+ SFNN++ W+ I+++A+++V K+L+GNK
Sbjct: 64 TAGQERFRTITSSYYRGAHGIIIVYDCTEMESFNNVKQWLSEIDRYANESVCKLLIGNKN 123
Query: 131 DMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRL---ADTDSK 187
DM ESK V T G+ALADE GI F ETSAK ++NVE+ F +IA +IK+++ + +
Sbjct: 124 DMVESK-VVSTETGRALADELGIPFLETSAKDSINVEQAFLTIAGEIKKKMGSQTNANKT 182
Query: 188 SEPQTIKI-NQPDQGAGAAQAAQKSACCG 215
S P T+++ QP Q CCG
Sbjct: 183 SGPGTVQMKGQPIQ-------QNNGGCCG 204
>At4g17160 GTP-binding RAB2A like protein
Length = 205
Score = 193 bits (491), Expect = 5e-50
Identities = 99/206 (48%), Positives = 132/206 (64%), Gaps = 5/206 (2%)
Query: 12 YDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDT 71
Y Y K ++IGD+GVGKSCLLL+F+D F TIG++F +TI +D K IKLQIWDT
Sbjct: 3 YAYRFKYIIIGDTGVGKSCLLLKFTDKRFQAVHDLTIGVEFGAKTITIDNKPIKLQIWDT 62
Query: 72 AGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKAD 131
AGQE FR++T +YYRG G LLVYD+T +FN++ +W+ QHAS+N+ +L+GNK D
Sbjct: 63 AGQESFRSVTRSYYRGRAGTLLVYDITRRETFNHLASWLEEARQHASENMTTMLIGNKCD 122
Query: 132 MDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLAD--TDSKSE 189
+ E KR V T +G+ A E+G+ F E SAKT NVEE F A I +R+ D D +E
Sbjct: 123 L-EDKRTVSTEEGEQFAREHGLIFMEASAKTAHNVEEAFVETAATIYKRIQDGVVDEANE 181
Query: 190 PQTIKINQPDQGAGAAQAAQKSACCG 215
P P G A+ + Q+ CCG
Sbjct: 182 PGITP--GPFGGKDASSSQQRRGCCG 205
>At4g35860 GTP-binding protein GB2
Length = 211
Score = 192 bits (487), Expect = 1e-49
Identities = 101/210 (48%), Positives = 133/210 (63%), Gaps = 7/210 (3%)
Query: 12 YDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDT 71
YDYL K ++IGD+GVGKSCLLL+F+D F TIG++F R + +DG+ IKLQIWDT
Sbjct: 3 YDYLFKYIIIGDTGVGKSCLLLQFTDKRFQPVHDLTIGVEFGARMVTVDGRPIKLQIWDT 62
Query: 72 AGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKAD 131
AGQE FR+IT +YYRGA G LLVYD+T +FN++ +W+ + QHA+ N++ +L+GNK D
Sbjct: 63 AGQESFRSITRSYYRGAAGALLVYDITRRETFNHLASWLEDARQHANPNMSIMLIGNKCD 122
Query: 132 MDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLAD--TDSKSE 189
+ KRAV +GQ A E+G+ F E SA+T NVEE F A I Q + D D +E
Sbjct: 123 L-AHKRAVSKEEGQQFAKEHGLLFLEASARTAQNVEEAFIETAAKILQNIQDGVFDVSNE 181
Query: 190 PQTIKI----NQPDQGAGAAQAAQKSACCG 215
IKI Q G +Q CCG
Sbjct: 182 SSGIKIGYGRTQGAAGGRDGTISQGGGCCG 211
>At1g09630 putative RAS-related protein, RAB11C
Length = 217
Score = 186 bits (473), Expect = 6e-48
Identities = 96/214 (44%), Positives = 136/214 (62%), Gaps = 5/214 (2%)
Query: 7 RARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKL 66
R +YDYL K++LIGDSGVGKS LL RF+ F +TIG++F RT++++G+ +K
Sbjct: 4 RPDEEYDYLFKVVLIGDSGVGKSNLLSRFTRNEFCLESKSTIGVEFATRTLQVEGRTVKA 63
Query: 67 QIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILV 126
QIWDTAGQER+R IT+AYYRGA+G LLVYDVT ++F N+ W++ + HA N+ +L+
Sbjct: 64 QIWDTAGQERYRAITSAYYRGALGALLVYDVTKPTTFENVSRWLKELRDHADSNIVIMLI 123
Query: 127 GNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIA----RDIKQRLA 182
GNK D+ + RAV T Q+ A++ G+ F ETSA LNVE+ F +I R I ++
Sbjct: 124 GNKTDL-KHLRAVATEDAQSYAEKEGLSFIETSALEALNVEKAFQTILSEVYRIISKKSI 182
Query: 183 DTDSKSEPQTIKINQPDQGAGAAQAAQKSACCGS 216
+D + IK Q A +++ K CC S
Sbjct: 183 SSDQTTANANIKEGQTIDVAATSESNAKKPCCSS 216
>At4g17170 GTP-binding RAB2A like protein
Length = 211
Score = 186 bits (472), Expect = 8e-48
Identities = 98/210 (46%), Positives = 131/210 (61%), Gaps = 7/210 (3%)
Query: 12 YDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDT 71
Y YL K ++IGD+GVGKSCLLL+F+D F TIG++F R I +D K IKLQIWDT
Sbjct: 3 YAYLFKYIIIGDTGVGKSCLLLQFTDKRFQPVHDLTIGVEFGARMITIDNKPIKLQIWDT 62
Query: 72 AGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKAD 131
AGQE FR+IT +YYRGA G LLVYD+T +FN++ +W+ + QHA+ N+ +L+GNK D
Sbjct: 63 AGQESFRSITRSYYRGAAGALLVYDITRRETFNHLASWLEDARQHANANMTIMLIGNKCD 122
Query: 132 MDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLAD--TDSKSE 189
+ +RAV T +G+ A E+G+ F E SAKT NVEE F A I +++ D D +E
Sbjct: 123 L-AHRRAVSTEEGEQFAKEHGLIFMEASAKTAQNVEEAFIKTAATIYKKIQDGVFDVSNE 181
Query: 190 PQTIKIN----QPDQGAGAAQAAQKSACCG 215
IK+ G +Q CCG
Sbjct: 182 SYGIKVGYGGIPGPSGGRDGSTSQGGGCCG 211
>At3g46830 GTP-binding protein Rab11
Length = 217
Score = 183 bits (464), Expect = 7e-47
Identities = 99/220 (45%), Positives = 134/220 (60%), Gaps = 16/220 (7%)
Query: 7 RARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKL 66
R +YDYL K++LIGDSGVGKS +L RF+ F +TIG++F RT +++GK IK
Sbjct: 4 RVDQEYDYLFKIVLIGDSGVGKSNILSRFTRNEFCLESKSTIGVEFATRTTQVEGKTIKA 63
Query: 67 QIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILV 126
QIWDTAGQER+R IT+AYYRGA+G LLVYD+T +F+N+ W+R + HA N+ ++
Sbjct: 64 QIWDTAGQERYRAITSAYYRGAVGALLVYDITKRQTFDNVLRWLRELRDHADSNIVIMMA 123
Query: 127 GNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDI--------- 177
GNK+D++ R+V GQ+LA++ G+ F ETSA NVE+ F +I +I
Sbjct: 124 GNKSDLNH-LRSVAEEDGQSLAEKEGLSFLETSALEATNVEKAFQTILGEIYHIISKKAL 182
Query: 178 -KQRLADTDSKSEPQTIKINQPDQGAGAAQAAQKSACCGS 216
Q A +S Q IN D GA K ACC S
Sbjct: 183 AAQEAAAANSAIPGQGTTINVDDTSGGA-----KRACCSS 217
>At5g59150 GTP-binding protein rab11 - like
Length = 217
Score = 180 bits (456), Expect = 6e-46
Identities = 92/215 (42%), Positives = 131/215 (60%), Gaps = 6/215 (2%)
Query: 7 RARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKL 66
R DYDYL K++LIGDSGVGK+ +L RF+ F +TIG++F RT++++GK +K
Sbjct: 4 RVEQDYDYLFKIVLIGDSGVGKTNILSRFTRNEFCLESKSTIGVEFATRTLQVEGKTVKA 63
Query: 67 QIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILV 126
QIWDTAGQER+R IT+AYYRGA+G LLVYD+T +F+N+ W+R + HA N+ ++
Sbjct: 64 QIWDTAGQERYRAITSAYYRGAVGALLVYDITKRQTFDNVLRWLRELRDHADSNIVIMMA 123
Query: 127 GNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDI-----KQRL 181
GNKAD++ R+V GQ LA+ G+ F ETSA NVE+ F ++ +I K+ L
Sbjct: 124 GNKADLNH-LRSVAEEDGQTLAETEGLSFLETSALEATNVEKAFQTVLAEIYHIISKKAL 182
Query: 182 ADTDSKSEPQTIKINQPDQGAGAAQAAQKSACCGS 216
A ++ + I A K CC +
Sbjct: 183 AAQEAAAANSAIPGQGTTINVEDTSGAGKRGCCST 217
>At1g16920 unknown protein
Length = 216
Score = 178 bits (451), Expect = 2e-45
Identities = 98/217 (45%), Positives = 134/217 (61%), Gaps = 12/217 (5%)
Query: 7 RARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKL 66
R DYDYL K++LIGDSGVGKS LL RF+ F +TIG++F RT+++DGK +K
Sbjct: 5 RVEDDYDYLFKVVLIGDSGVGKSNLLSRFTKNEFNLESKSTIGVEFATRTLKVDGKVVKA 64
Query: 67 QIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILV 126
QIWDTAGQER+R IT+AYYRGA+G LLVYDVT ++F N+ W++ ++ H N+ +LV
Sbjct: 65 QIWDTAGQERYRAITSAYYRGAVGALLVYDVTRRATFENVDRWLKELKNHTDPNIVVMLV 124
Query: 127 GNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDI------KQR 180
GNK+D+ AVPT G++ A++ + F ETSA NVE+ F + I KQ
Sbjct: 125 GNKSDL-RHLLAVPTEDGKSYAEQESLCFMETSALEATNVEDAFAEVLTQIYRITSKKQV 183
Query: 181 LADTD-SKSEPQTIKINQPDQGAGAAQAAQKSACCGS 216
A D + S P+ KI + A +K CC +
Sbjct: 184 EAGEDGNASVPKGEKIEVKND----VSALKKLGCCSN 216
>At4g18430 membrane-bound small GTP-binding - like protein
Length = 217
Score = 177 bits (448), Expect = 5e-45
Identities = 97/216 (44%), Positives = 131/216 (59%), Gaps = 13/216 (6%)
Query: 7 RARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKL 66
RA DYDYL KL+LIGDSGVGKS LL RF+ F+ +TIG++F R++ +D K IK
Sbjct: 5 RADDDYDYLFKLVLIGDSGVGKSNLLSRFTRNEFSIESKSTIGVEFATRSVHVDEKIIKA 64
Query: 67 QIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILV 126
Q+WDTAGQER+R IT+AYYRGA+G LLVYD+T +F N+ W++ + H NV +LV
Sbjct: 65 QLWDTAGQERYRAITSAYYRGAVGALLVYDITRHITFENVERWLKELRDHTDANVVIMLV 124
Query: 127 GNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLA---- 182
GNKAD+ RAVPT + ++ ++ + F ETSA NVE+ F + I + ++
Sbjct: 125 GNKADL-RHLRAVPTEEARSFSERENMFFMETSALDATNVEQAFTHVLTQIYRVMSRKAL 183
Query: 183 ----DTDSKSEPQTIKINQPDQGAGAAQAAQKSACC 214
D S + QTI I D A + S CC
Sbjct: 184 DGTGDPMSLPKGQTIDIGNKDD----VTAVKSSGCC 215
>At1g07410 small G protein, putative
Length = 214
Score = 177 bits (448), Expect = 5e-45
Identities = 91/215 (42%), Positives = 134/215 (62%), Gaps = 17/215 (7%)
Query: 11 DYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWD 70
+YDYL K++LIGDSGVGKS +L RF+ F +TIG++F RT++++GK +K QIWD
Sbjct: 8 EYDYLFKIVLIGDSGVGKSNILSRFTRNEFCLESKSTIGVEFATRTLQVEGKTVKAQIWD 67
Query: 71 TAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKA 130
TAGQER+R IT+AYYRGA+G LLVYD+T +F N+ W+R + HA N+ ++ GNK+
Sbjct: 68 TAGQERYRAITSAYYRGAVGALLVYDITKRQTFENVLRWLRELRDHADSNIVIMMAGNKS 127
Query: 131 DMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDI-----KQRLADTD 185
D++ R+V G++LA++ G+ F ETSA N+E+ F +I +I K+ LA +
Sbjct: 128 DLNH-LRSVADEDGRSLAEKEGLSFLETSALEATNIEKAFQTILSEIYHIISKKALAAQE 186
Query: 186 SKSEPQTIKINQPDQGAGA----AQAAQKSACCGS 216
+ N P QG + A + CC +
Sbjct: 187 AAG-------NLPGQGTAINISDSSATNRKGCCST 214
>At3g07410 GTP-binding protein like
Length = 217
Score = 175 bits (444), Expect = 1e-44
Identities = 89/207 (42%), Positives = 137/207 (65%), Gaps = 6/207 (2%)
Query: 13 DYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTA 72
+YL K++LIGDS VGKS LL RFS F T+ TIG++F+ + +E++GK +K QIWDTA
Sbjct: 10 EYLFKIVLIGDSAVGKSNLLSRFSRDEFDTNSKATIGVEFQTQLVEIEGKEVKAQIWDTA 69
Query: 73 GQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKADM 132
GQERFR +T+AYYRGA G L+VYD+T +F +++ W++ + H V ++LVGNK D+
Sbjct: 70 GQERFRAVTSAYYRGAFGALIVYDITRGDTFESVKRWLQELNTHCDTAVAQMLVGNKCDL 129
Query: 133 DESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDI----KQRLADTDS-K 187
E RAV +G+ALA+E G+ F ETSA NV++ F + R+I ++L ++D+ K
Sbjct: 130 -EDIRAVSVEEGKALAEEEGLFFMETSALDATNVDKAFEIVIREIFNNVSRKLLNSDAYK 188
Query: 188 SEPQTIKINQPDQGAGAAQAAQKSACC 214
+E +++ + G+ + + +CC
Sbjct: 189 AELSVNRVSLVNNQDGSESSWRNPSCC 215
>At5g60860 GTP-binding protein - like
Length = 217
Score = 172 bits (437), Expect = 9e-44
Identities = 94/218 (43%), Positives = 129/218 (59%), Gaps = 13/218 (5%)
Query: 7 RARADYDYLIKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKL 66
RA +YDYL K++LIGDSGVGKS LL RF+ F+ +TIG++F R+I +D K +K
Sbjct: 5 RADDEYDYLFKVVLIGDSGVGKSNLLSRFTRNEFSLESKSTIGVEFATRSIHVDDKIVKA 64
Query: 67 QIWDTAGQERFRTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILV 126
QIWDTAGQER+R IT+AYYRGA+G LLVYDVT +F N+ W++ + H N+ + V
Sbjct: 65 QIWDTAGQERYRAITSAYYRGAVGALLVYDVTRHVTFENVERWLKELRDHTDANIVIMFV 124
Query: 127 GNKADMDESKRAVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLA---- 182
GNKAD+ RAV T +A A+ F ETSA ++NVE F + I + ++
Sbjct: 125 GNKADL-RHLRAVSTEDAKAFAERENTFFMETSALESMNVENAFTEVLSQIYRVVSRKAL 183
Query: 183 ----DTDSKSEPQTIKINQPDQGAGAAQAAQKSACCGS 216
D + + QTI + D A +K CC +
Sbjct: 184 DIGDDPAALPKGQTINVGSKDD----VSAVKKVGCCSN 217
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.134 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,371,622
Number of Sequences: 26719
Number of extensions: 164847
Number of successful extensions: 731
Number of sequences better than 10.0: 115
Number of HSP's better than 10.0 without gapping: 97
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 511
Number of HSP's gapped (non-prelim): 119
length of query: 216
length of database: 11,318,596
effective HSP length: 95
effective length of query: 121
effective length of database: 8,780,291
effective search space: 1062415211
effective search space used: 1062415211
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC125368.9


