

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124970.5 - phase: 0
(952 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
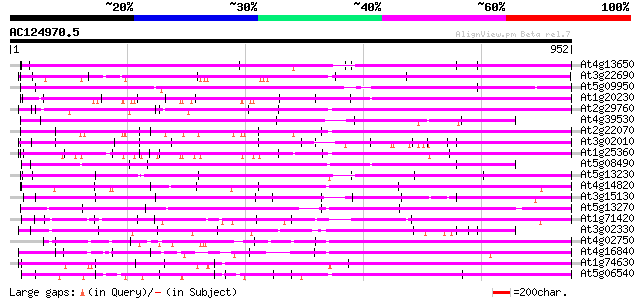
Score E
Sequences producing significant alignments: (bits) Value
At4g13650 unknown protein 556 e-158
At3g22690 hypothetical protein 521 e-148
At5g09950 selenium-binding protein-like 501 e-141
At1g20230 hypothetical protein 499 e-141
At2g29760 hypothetical protein 478 e-135
At4g39530 putative protein 477 e-134
At2g22070 hypothetical protein 473 e-133
At3g02010 hypothetical protein 449 e-126
At1g25360 449 e-126
At5g08490 selenium-binding protein-like 443 e-124
At5g13230 putative protein 437 e-122
At4g14820 hypothetical protein 433 e-121
At3g15130 hypothetical protein 432 e-121
At5g13270 putative protein 431 e-121
At1g71420 hypothetical protein 429 e-120
At3g02330 hypothetical protein 428 e-120
At4g02750 hypothetical protein 426 e-119
At4g16840 hypothetical protein 424 e-118
At1g74630 hypothetical protein 406 e-113
At5g06540 selenium-binding protein-like 405 e-113
>At4g13650 unknown protein
Length = 1024
Score = 556 bits (1433), Expect = e-158
Identities = 301/934 (32%), Positives = 492/934 (52%), Gaps = 36/934 (3%)
Query: 20 LPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKV-KQFHDDATRCGVMSDVSIG 78
L E ++ + + P++ F V +AC A V +Q H G+ +
Sbjct: 126 LIGEVFGLFVRMVSENVTPNEGTFSGVLEACRGGSVAFDVVEQIHARILYQGLRDSTVVC 185
Query: 79 NAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVK 138
N I Y + V+ ARRVFD L +D +W ++ + + + +F M + +
Sbjct: 186 NPLIDLYSRNGFVDLARRVFDGLRLKDHSSWVAMISGLSKNECEAEAIRLFCDMYVLGIM 245
Query: 139 ANPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTV 198
P SS+L C ++ L+ G+++HG V++ G D +V +A V+ Y + A+ +
Sbjct: 246 PTPYAFSSVLSACKKIESLEIGEQLHGLVLKLGFSSDTYVCNALVSLYFHLGNLISAEHI 305
Query: 199 FDLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQDL 258
F M RD VT+N+L + CG+ +K + +F+ M LDG++PD T++ ++ ACS L
Sbjct: 306 FSNMSQRDAVTYNTLINGLSQCGYGEKAMELFKRMHLDGLEPDSNTLASLVVACSADGTL 365
Query: 259 KSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASCY 318
G+ +H + K G N + AL+NLY C + A F NV+ WN + Y
Sbjct: 366 FRGQQLHAYTTKLGFASNNKIEGALLNLYAKCADIETALDYFLETEVENVVLWNVMLVAY 425
Query: 319 VNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDV 378
+ +FR+M + + P+ SIL C +L DL+ G+ IH +K +
Sbjct: 426 GLLDDLRNSFRIFRQMQIEEIVPNQYTYPSILKTCIRLGDLELGEQIHSQIIKTNFQLNA 485
Query: 379 FVCTALVNLYANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLN 438
+VC+ L+++YA + A + ++VV+W ++ + Y F K L FR+M+
Sbjct: 486 YVCSVLIDMYAKLGKLDTAWDILIRFAGKDVVSWTTMIAGYTQYNFDDKALTTFRQMLDR 545
Query: 439 GVKPDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREA 498
G++ D V + + + AC+ LQ LK G+ IH A G D+ NAL++LY++C + E+
Sbjct: 546 GIRSDEVGLTNAVSACAGLQALKEGQQIHAQACVSGFSSDLPFQNALVTLYSRCGKIEES 605
Query: 499 QVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGGCVKN 558
+ F+ + +WN +++ + + E+ L +F +MNR+ + + T+
Sbjct: 606 YLAFEQTEAGDNIAWNALVSGFQQSGNNEEALRVFVRMNREGIDNNNFTFG--------- 656
Query: 559 SRIEEAMEIFRKMQTMGFKPDETTIYSILRACSLSECLRMGKEIHCYVFRHWKDWDLART 618
S ++A S + ++ GK++H + + D +
Sbjct: 657 --------------------------SAVKAASETANMKQGKQVHAVITKTGYDSETEVC 690
Query: 619 NALVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKEALSLFEKMLLSMVK 678
NAL+ MYAKCG +S + F + K+ SWN +I A HG G EAL F++M+ S V+
Sbjct: 691 NALISMYAKCGSISDAEKQFLEVSTKNEVSWNAIINAYSKHGFGSEALDSFDQMIHSNVR 750
Query: 679 PDSATFTCVLSACSHSMLVEEGVQIFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYG 738
P+ T VLSACSH LV++G+ F SM+ ++ + P+ EHY CVVD+ +RAG L A
Sbjct: 751 PNHVTLVGVLSACSHIGLVDKGIAYFESMNSEYGLSPKPEHYVCVVDMLTRAGLLSRAKE 810
Query: 739 FIQRMPMEPTAIAWKAFLAGCRVYKNVELAKISAKKLFEIDPNGSANYVTLFNILVTAKL 798
FIQ MP++P A+ W+ L+ C V+KN+E+ + +A L E++P SA YV L N+ +K
Sbjct: 811 FIQEMPIKPDALVWRTLLSACVVHKNMEIGEFAAHHLLELEPEDSATYVLLSNLYAVSKK 870
Query: 799 WSEASKIRKLMKERGITKTPGCSWFHVGNRVHTFVAGDKSNMESDKIYNFLDELFAKIKA 858
W R+ MKE+G+ K PG SW V N +H+F GD+++ +D+I+ + +L +
Sbjct: 871 WDARDLTRQKMKEKGVKKEPGQSWIEVKNSIHSFYVGDQNHPLADEIHEYFQDLTKRASE 930
Query: 859 AGYKPDTDYVLHDIDQEEKAESLCNHSEKLAVAFGILNLNGQSTIRVFKNLRICGDCHNA 918
GY D +L+++ E+K + HSEKLA++FG+L+L I V KNLR+C DCH
Sbjct: 931 IGYVQDCFSLLNELQHEQKDPIIFIHSEKLAISFGLLSLPATVPINVMKNLRVCNDCHAW 990
Query: 919 IKYMSNVVGVTIVVRDSLRFHHFKNGNCSCKDFW 952
IK++S V I+VRD+ RFHHF+ G CSCKD+W
Sbjct: 991 IKFVSKVSNREIIVRDAYRFHHFEGGACSCKDYW 1024
Score = 263 bits (673), Expect = 3e-70
Identities = 185/762 (24%), Positives = 342/762 (44%), Gaps = 41/762 (5%)
Query: 34 RGIKPDKPVFMAVAKACAASRDAL-KVKQFHDDATRCGVMSDVSIGNAFIHAYGKCKCVE 92
RGI+P+ + + C + +L + ++ H + G+ S+ + Y +
Sbjct: 38 RGIRPNHQTLKWLLEGCLKTNGSLDEGRKLHSQILKLGLDSNGCLSEKLFDFYLFKGDLY 97
Query: 93 GARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKANPLTVSSILPGC- 151
GA +VFD++ R + TWN + + + +F +M V N T S +L C
Sbjct: 98 GAFKVFDEMPERTIFTWNKMIKELASRNLIGEVFGLFVRMVSENVTPNEGTFSGVLEACR 157
Query: 152 SDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTWN 211
++IH ++ G+ + V + ++ Y++ V A+ VFD + +D +W
Sbjct: 158 GGSVAFDVVEQIHARILYQGLRDSTVVCNPLIDLYSRNGFVDLARRVFDGLRLKDHSSWV 217
Query: 212 SLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQDLKSGKAIHGFALKH 271
++ S + + +F +M + G+ P P S +LSAC ++ L+ G+ +HG LK
Sbjct: 218 AMISGLSKNECEAEAIRLFCDMYVLGIMPTPYAFSSVLSACKKIESLEIGEQLHGLVLKL 277
Query: 272 GMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASCYVNCGFPQKGLNVF 331
G + +V NALV+LY + A+ +F M R+ +T+N+L + CG+ +K + +F
Sbjct: 278 GFSSDTYVCNALVSLYFHLGNLISAEHIFSNMSQRDAVTYNTLINGLSQCGYGEKAMELF 337
Query: 332 REMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANC 391
+ M L+G++PD ++S++ ACS L G+ +H + K G + + AL+NLYA C
Sbjct: 338 KRMHLDGLEPDSNTLASLVVACSADGTLFRGQQLHAYTTKLGFASNNKIEGALLNLYAKC 397
Query: 392 LCVREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSIL 451
+ A F NVV WN + Y + +FR+M + + P+ T SIL
Sbjct: 398 ADIETALDYFLETEVENVVLWNVMLVAYGLLDDLRNSFRIFRQMQIEEIVPNQYTYPSIL 457
Query: 452 HACSDLQDLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREVA 511
C L DL+ G+ IH ++ + +VC+ L+ +YAK + A + ++V
Sbjct: 458 KTCIRLGDLELGEQIHSQIIKTNFQLNAYVCSVLIDMYAKLGKLDTAWDILIRFAGKDVV 517
Query: 512 SWNGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGGCVKNSRIEEAMEIFRKM 571
SW ++ Y +K L F QM +++DE+ + + C ++E +I +
Sbjct: 518 SWTTMIAGYTQYNFDDKALTTFRQMLDRGIRSDEVGLTNAVSACAGLQALKEGQQIHAQA 577
Query: 572 QTMGFKPDETTIYSILRACSLSECLRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGGL 631
GF DL NALV +Y++CG +
Sbjct: 578 CVSGFSS-----------------------------------DLPFQNALVTLYSRCGKI 602
Query: 632 SLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKEALSLFEKMLLSMVKPDSATFTCVLSAC 691
S F+ D +WN ++ GN +EAL +F +M + ++ TF + A
Sbjct: 603 EESYLAFEQTEAGDNIAWNALVSGFQQSGNNEEALRVFVRMNREGIDNNNFTFGSAVKAA 662
Query: 692 SHSMLVEEGVQIFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEPTAIA 751
S + +++G Q+ +++ + E E ++ +Y++ G + +A + + ++
Sbjct: 663 SETANMKQGKQVHAVITKTG-YDSETEVCNALISMYAKCGSISDAEKQFLEVSTK-NEVS 720
Query: 752 WKAFLAGCRVYKNVELAKISAKKLFEIDPNGSANYVTLFNIL 793
W A + + A S ++ I N N+VTL +L
Sbjct: 721 WNAIINAYSKHGFGSEALDSFDQM--IHSNVRPNHVTLVGVL 760
Score = 241 bits (615), Expect = 1e-63
Identities = 154/571 (26%), Positives = 275/571 (47%), Gaps = 19/571 (3%)
Query: 19 GLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSIG 78
G +A++++ G++PD ++ AC+A + +Q H T+ G S+ I
Sbjct: 328 GYGEKAMELFKRMHLDGLEPDSNTLASLVVACSADGTLFRGQQLHAYTTKLGFASNNKIE 387
Query: 79 NAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVK 138
A ++ Y KC +E A F + +VV WN + Y + +FR+M + ++
Sbjct: 388 GALLNLYAKCADIETALDYFLETEVENVVLWNVMLVAYGLLDDLRNSFRIFRQMQIEEIV 447
Query: 139 ANPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTV 198
N T SIL C L DL+ G++IH +++ + +V S ++ YAK + A +
Sbjct: 448 PNQYTYPSILKTCIRLGDLELGEQIHSQIIKTNFQLNAYVCSVLIDMYAKLGKLDTAWDI 507
Query: 199 FDLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQDL 258
+DVV+W ++ + Y F K L FR+M+ G++ D V ++ +SAC+ LQ L
Sbjct: 508 LIRFAGKDVVSWTTMIAGYTQYNFDDKALTTFRQMLDRGIRSDEVGLTNAVSACAGLQAL 567
Query: 259 KSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASCY 318
K G+ IH A G ++ NALV LY C + E+ F+ + I WN+L S +
Sbjct: 568 KEGQQIHAQACVSGFSSDLPFQNALVTLYSRCGKIEESYLAFEQTEAGDNIAWNALVSGF 627
Query: 319 VNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDV 378
G ++ L VF M G+ + S + A S+ ++K GK +H K G +
Sbjct: 628 QQSGNNEEALRVFVRMNREGIDNNNFTFGSAVKAASETANMKQGKQVHAVITKTGYDSET 687
Query: 379 FVCTALVNLYANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLN 438
VC AL+++YA C + +A+ F + +N V+WN++ + Y GF + L+ F +M+ +
Sbjct: 688 EVCNALISMYAKCGSISDAEKQFLEVSTKNEVSWNAIINAYSKHGFGSEALDSFDQMIHS 747
Query: 439 GVKPDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGMVEDV--------FVCNALLSLYA 490
V+P+ VT++ +L ACS + G V G A M + +VC ++ +
Sbjct: 748 NVRPNHVTLVGVLSACSHI-----GLVDKGIAYFESMNSEYGLSPKPEHYVC--VVDMLT 800
Query: 491 KCVCVREAQVVFDLIPHR-EVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWS 549
+ + A+ +P + + W +L+A +K E G F+ + E++ ++
Sbjct: 801 RAGLLSRAKEFIQEMPIKPDALVWRTLLSACVVHKNMEIG--EFAAHHLLELEPEDSATY 858
Query: 550 VVIGGCVKNSRIEEAMEIFR-KMQTMGFKPD 579
V++ S+ +A ++ R KM+ G K +
Sbjct: 859 VLLSNLYAVSKKWDARDLTRQKMKEKGVKKE 889
Score = 128 bits (321), Expect = 2e-29
Identities = 93/375 (24%), Positives = 163/375 (42%), Gaps = 40/375 (10%)
Query: 18 HGLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSI 77
+ ++A+ + RGI+ D+ ACA + + +Q H A G SD+
Sbjct: 529 YNFDDKALTTFRQMLDRGIRSDEVGLTNAVSACAGLQALKEGQQIHAQACVSGFSSDLPF 588
Query: 78 GNAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKV 137
NA + Y +C +E + F+ A D + WN+L + + G ++ L VF +M +
Sbjct: 589 QNALVTLYSRCGKIEESYLAFEQTEAGDNIAWNALVSGFQQSGNNEEALRVFVRMNREGI 648
Query: 138 KANPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQT 197
N T S + S+ ++K GK++H + + G + V +A ++ YAKC + +A+
Sbjct: 649 DNNNFTFGSAVKAASETANMKQGKQVHAVITKTGYDSETEVCNALISMYAKCGSISDAEK 708
Query: 198 VFDLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQD 257
F + ++ V+WN++ + Y GF + L+ F +M+ V+P+ VT+ +LSACS +
Sbjct: 709 QFLEVSTKNEVSWNAIINAYSKHGFGSEALDSFDQMIHSNVRPNHVTLVGVLSACSHIGL 768
Query: 258 LKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASC 317
+ G A E++ L E +CV
Sbjct: 769 VDKGIA---------YFESMNSEYGLSPKPEHYVCV---------------------VDM 798
Query: 318 YVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGM--- 374
G + +EM +KPD + ++L AC K+++ G+ FA H +
Sbjct: 799 LTRAGLLSRAKEFIQEM---PIKPDALVWRTLLSACVVHKNMEIGE----FAAHHLLELE 851
Query: 375 VEDVFVCTALVNLYA 389
ED L NLYA
Sbjct: 852 PEDSATYVLLSNLYA 866
Score = 65.1 bits (157), Expect = 2e-10
Identities = 46/189 (24%), Positives = 86/189 (45%), Gaps = 2/189 (1%)
Query: 571 MQTMGFKPDETTIYSILRAC-SLSECLRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCG 629
++ G +P+ T+ +L C + L G+++H + + D + + L D Y G
Sbjct: 35 VENRGIRPNHQTLKWLLEGCLKTNGSLDEGRKLHSQILKLGLDSNGCLSEKLFDFYLFKG 94
Query: 630 GLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKEALSLFEKMLLSMVKPDSATFTCVLS 689
L + VFD MP + +F+WN MI E LF +M+ V P+ TF+ VL
Sbjct: 95 DLYGAFKVFDEMPERTIFTWNKMIKELASRNLIGEVFGLFVRMVSENVTPNEGTFSGVLE 154
Query: 690 ACSHSMLVEEGVQIFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEPTA 749
AC + + V+ ++ + ++D+YSR G ++ A + ++ +
Sbjct: 155 ACRGGSVAFDVVEQIHARILYQGLRDSTVVCNPLIDLYSRNGFVDLARRVFDGLRLKDHS 214
Query: 750 IAWKAFLAG 758
+W A ++G
Sbjct: 215 -SWVAMISG 222
>At3g22690 hypothetical protein
Length = 978
Score = 521 bits (1343), Expect = e-148
Identities = 292/825 (35%), Positives = 461/825 (55%), Gaps = 16/825 (1%)
Query: 134 LNKVKANPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVR 193
LN+ K T SS L C + +LK H + + G+ DV + V + L R
Sbjct: 26 LNQSKCTKATPSS-LKNCKTIDELKM---FHRSLTKQGLDNDVSTITKLVARSCE-LGTR 80
Query: 194 E----AQTVFDLMP-HRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCI 248
E A+ VF+ + +NSL Y + G + + +F M+ G+ PD T
Sbjct: 81 ESLSFAKEVFENSESYGTCFMYNSLIRGYASSGLCNEAILLFLRMMNSGISPDKYTFPFG 140
Query: 249 LSACSDLQDLKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNV 308
LSAC+ + +G IHG +K G +++FV N+LV+ Y C + A+ VFD M RNV
Sbjct: 141 LSACAKSRAKGNGIQIHGLIVKMGYAKDLFVQNSLVHFYAECGELDSARKVFDEMSERNV 200
Query: 309 ITWNSLASCYVNCGFPQKGLNVFREMGLNG-VKPDPMAMSSILPACSQLKDLKSGKTIHG 367
++W S+ Y F + +++F M + V P+ + M ++ AC++L+DL++G+ ++
Sbjct: 201 VSWTSMICGYARRDFAKDAVDLFFRMVRDEEVTPNSVTMVCVISACAKLEDLETGEKVYA 260
Query: 368 FAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQK 427
F G+ + + +ALV++Y C + A+ +FD N+ N+++S YV G ++
Sbjct: 261 FIRNSGIEVNDLMVSALVDMYMKCNAIDVAKRLFDEYGASNLDLCNAMASNYVRQGLTRE 320
Query: 428 GLNVFREMVLNGVKPDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGMVEDVFVCNALLS 487
L VF M+ +GV+PD ++MLS + +CS L+++ GK HG+ +R+G +CNAL+
Sbjct: 321 ALGVFNLMMDSGVRPDRISMLSAISSCSQLRNILWGKSCHGYVLRNGFESWDNICNALID 380
Query: 488 LYAKCVCVREAQVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKADEIT 547
+Y KC A +FD + ++ V +WN I+ Y N E + F M + ++
Sbjct: 381 MYMKCHRQDTAFRIFDRMSNKTVVTWNSIVAGYVENGEVDAAWETFETMPEKNI----VS 436
Query: 548 WSVVIGGCVKNSRIEEAMEIFRKMQTM-GFKPDETTIYSILRACSLSECLRMGKEIHCYV 606
W+ +I G V+ S EEA+E+F MQ+ G D T+ SI AC L + K I+ Y+
Sbjct: 437 WNTIISGLVQGSLFEEAIEVFCSMQSQEGVNADGVTMMSIASACGHLGALDLAKWIYYYI 496
Query: 607 FRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKEAL 666
++ D+ LVDM+++CG + ++F+ + +DV +W I A M GN + A+
Sbjct: 497 EKNGIQLDVRLGTTLVDMFSRCGDPESAMSIFNSLTNRDVSAWTAAIGAMAMAGNAERAI 556
Query: 667 SLFEKMLLSMVKPDSATFTCVLSACSHSMLVEEGVQIFNSMSRDHLVEPEAEHYTCVVDI 726
LF+ M+ +KPD F L+ACSH LV++G +IF SM + H V PE HY C+VD+
Sbjct: 557 ELFDDMIEQGLKPDGVAFVGALTACSHGGLVQQGKEIFYSMLKLHGVSPEDVHYGCMVDL 616
Query: 727 YSRAGCLEEAYGFIQRMPMEPTAIAWKAFLAGCRVYKNVELAKISAKKLFEIDPNGSANY 786
RAG LEEA I+ MPMEP + W + LA CRV NVE+A +A+K+ + P + +Y
Sbjct: 617 LGRAGLLEEAVQLIEDMPMEPNDVIWNSLLAACRVQGNVEMAAYAAEKIQVLAPERTGSY 676
Query: 787 VTLFNILVTAKLWSEASKIRKLMKERGITKTPGCSWFHVGNRVHTFVAGDKSNMESDKIY 846
V L N+ +A W++ +K+R MKE+G+ K PG S + + H F +GD+S+ E I
Sbjct: 677 VLLSNVYASAGRWNDMAKVRLSMKEKGLRKPPGTSSIQIRGKTHEFTSGDESHPEMPNIE 736
Query: 847 NFLDELFAKIKAAGYKPDTDYVLHDIDQEEKAESLCNHSEKLAVAFGILNLNGQSTIRVF 906
LDE+ + G+ PD VL D+D++EK L HSEKLA+A+G+++ N +TIR+
Sbjct: 737 AMLDEVSQRASHLGHVPDLSNVLMDVDEKEKIFMLSRHSEKLAMAYGLISSNKGTTIRIV 796
Query: 907 KNLRICGDCHNAIKYMSNVVGVTIVVRDSLRFHHFKNGNCSCKDF 951
KNLR+C DCH+ K+ S V I++RD+ RFH+ + G CSC DF
Sbjct: 797 KNLRVCSDCHSFAKFASKVYNREIILRDNNRFHYIRQGKCSCGDF 841
Score = 258 bits (660), Expect = 9e-69
Identities = 184/679 (27%), Positives = 331/679 (48%), Gaps = 81/679 (11%)
Query: 40 KPVFMAVAKACAASRDALK-------VKQFHDDATRCGVMSDVSIGNAFIHA---YGKCK 89
KP + +K A+ +LK +K FH T+ G+ +DVS + G +
Sbjct: 22 KPSLLNQSKCTKATPSSLKNCKTIDELKMFHRSLTKQGLDNDVSTITKLVARSCELGTRE 81
Query: 90 CVEGARRVFDDLVARDVV-TWNSLSACYVNCGFPQQGLNVFRKMGLNKVKANPLTVSSIL 148
+ A+ VF++ + +NSL Y + G + + +F +M + + + T L
Sbjct: 82 SLSFAKEVFENSESYGTCFMYNSLIRGYASSGLCNEAILLFLRMMNSGISPDKYTFPFGL 141
Query: 149 PGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVV 208
C+ + +G +IHG +V+ G +D+FV ++ V+FYA+C + A+ VFD M R+VV
Sbjct: 142 SACAKSRAKGNGIQIHGLIVKMGYAKDLFVQNSLVHFYAECGELDSARKVFDEMSERNVV 201
Query: 209 TWNSLSSCYVNCGFPQKGLNVFREMVLD-GVKPDPVTVSCILSACSDLQDLKSGKAIHGF 267
+W S+ Y F + +++F MV D V P+ VT+ C++SAC+ L+DL++G+ ++ F
Sbjct: 202 SWTSMICGYARRDFAKDAVDLFFRMVRDEEVTPNSVTMVCVISACAKLEDLETGEKVYAF 261
Query: 268 ALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASCYVNCGFPQKG 327
G+ N + +ALV++Y C + A+ +FD N+ N++AS YV G ++
Sbjct: 262 IRNSGIEVNDLMVSALVDMYMKCNAIDVAKRLFDEYGASNLDLCNAMASNYVRQGLTREA 321
Query: 328 LNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNL 387
L VF M +GV+PD ++M S + +CSQL+++ GK+ HG+ +++G +C AL+++
Sbjct: 322 LGVFNLMMDSGVRPDRISMLSAISSCSQLRNILWGKSCHGYVLRNGFESWDNICNALIDM 381
Query: 388 YANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCG-----------FPQKGL------- 429
Y C A +FD M ++ VVTWNS+ + YV G P+K +
Sbjct: 382 YMKCHRQDTAFRIFDRMSNKTVVTWNSIVAGYVENGEVDAAWETFETMPEKNIVSWNTII 441
Query: 430 ------NVFREMV--------LNGVKPDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGM 475
++F E + GV D VTM+SI AC L L K I+ + ++G+
Sbjct: 442 SGLVQGSLFEEAIEVFCSMQSQEGVNADGVTMMSIASACGHLGALDLAKWIYYYIEKNGI 501
Query: 476 VEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQ 535
DV + L+ ++++C A +F+ + +R+V++W + A E+ + +F
Sbjct: 502 QLDVRLGTTLVDMFSRCGDPESAMSIFNSLTNRDVSAWTAAIGAMAMAGNAERAIELFDD 561
Query: 536 MNRDEVKADEITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKPDETTIYSILRACSLSEC 595
M +K D + + + C +++ EIF YS+L+ +S
Sbjct: 562 MIEQGLKPDGVAFVGALTACSHGGLVQQGKEIF---------------YSMLKLHGVSP- 605
Query: 596 LRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIK--DVFSWNTMI 653
+++H +VD+ + G L + + + MP++ DV WN+++
Sbjct: 606 ----EDVH--------------YGCMVDLLGRAGLLEEAVQLIEDMPMEPNDVI-WNSLL 646
Query: 654 FANGMHGNGKEALSLFEKM 672
A + GN + A EK+
Sbjct: 647 AACRVQGNVEMAAYAAEKI 665
Score = 248 bits (633), Expect = 1e-65
Identities = 160/569 (28%), Positives = 281/569 (49%), Gaps = 38/569 (6%)
Query: 19 GLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSIG 78
GL NEAI ++ GI PDK F ACA SR Q H + G D+ +
Sbjct: 113 GLCNEAILLFLRMMNSGISPDKYTFPFGLSACAKSRAKGNGIQIHGLIVKMGYAKDLFVQ 172
Query: 79 NAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNK-V 137
N+ +H Y +C ++ AR+VFD++ R+VV+W S+ Y F + +++F +M ++ V
Sbjct: 173 NSLVHFYAECGELDSARKVFDEMSERNVVSWTSMICGYARRDFAKDAVDLFFRMVRDEEV 232
Query: 138 KANPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQT 197
N +T+ ++ C+ L+DL++G++++ F+ G+ + + SA V+ Y KC + A+
Sbjct: 233 TPNSVTMVCVISACAKLEDLETGEKVYAFIRNSGIEVNDLMVSALVDMYMKCNAIDVAKR 292
Query: 198 VFDLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQD 257
+FD ++ N+++S YV G ++ L VF M+ GV+PD +++ +S+CS L++
Sbjct: 293 LFDEYGASNLDLCNAMASNYVRQGLTREALGVFNLMMDSGVRPDRISMLSAISSCSQLRN 352
Query: 258 LKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASC 317
+ GK+ HG+ L++G + NAL+++Y C A +FD M ++ V+TWNS+ +
Sbjct: 353 ILWGKSCHGYVLRNGFESWDNICNALIDMYMKCHRQDTAFRIFDRMSNKTVVTWNSIVAG 412
Query: 318 YVNCG-----------FPQKGL-------------NVFRE--------MGLNGVKPDPMA 345
YV G P+K + ++F E GV D +
Sbjct: 413 YVENGEVDAAWETFETMPEKNIVSWNTIISGLVQGSLFEEAIEVFCSMQSQEGVNADGVT 472
Query: 346 MSSILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMP 405
M SI AC L L K I+ + K+G+ DV + T LV++++ C A ++F+ +
Sbjct: 473 MMSIASACGHLGALDLAKWIYYYIEKNGIQLDVRLGTTLVDMFSRCGDPESAMSIFNSLT 532
Query: 406 HRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHACSDLQDLKSGK- 464
+R+V W + G ++ + +F +M+ G+KPD V + L ACS ++ GK
Sbjct: 533 NRDVSAWTAAIGAMAMAGNAERAIELFDDMIEQGLKPDGVAFVGALTACSHGGLVQQGKE 592
Query: 465 VIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREA-QVVFDLIPHREVASWNGILTAYFTN 523
+ + HG+ + ++ L + + EA Q++ D+ WN +L A
Sbjct: 593 IFYSMLKLHGVSPEDVHYGCMVDLLGRAGLLEEAVQLIEDMPMEPNDVIWNSLLAACRVQ 652
Query: 524 KEYEKGLYMFSQMNRDEVKADEITWSVVI 552
E Y ++ +V A E T S V+
Sbjct: 653 GNVEMAAYAAEKI---QVLAPERTGSYVL 678
Score = 122 bits (306), Expect = 1e-27
Identities = 93/339 (27%), Positives = 154/339 (44%), Gaps = 35/339 (10%)
Query: 16 IPHGLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDV 75
+ GL EA+ ++ G++PD+ ++ +C+ R+ L K H R G S
Sbjct: 313 VRQGLTREALGVFNLMMDSGVRPDRISMLSAISSCSQLRNILWGKSCHGYVLRNGFESWD 372
Query: 76 SIGNAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCG--------------- 120
+I NA I Y KC + A R+FD + + VVTWNS+ A YV G
Sbjct: 373 NICNALIDMYMKCHRQDTAFRIFDRMSNKTVVTWNSIVAGYVENGEVDAAWETFETMPEK 432
Query: 121 ----------------FPQQGLNVFRKMGLNK-VKANPLTVSSILPGCSDLQDLKSGKEI 163
++ + VF M + V A+ +T+ SI C L L K I
Sbjct: 433 NIVSWNTIISGLVQGSLFEEAIEVFCSMQSQEGVNADGVTMMSIASACGHLGALDLAKWI 492
Query: 164 HGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTWNSLSSCYVNCGFP 223
+ ++ ++G+ DV + + V+ +++C A ++F+ + +RDV W + G
Sbjct: 493 YYYIEKNGIQLDVRLGTTLVDMFSRCGDPESAMSIFNSLTNRDVSAWTAAIGAMAMAGNA 552
Query: 224 QKGLNVFREMVLDGVKPDPVTVSCILSACSDLQDLKSGKAIHGFALK-HGMVENVFVSNA 282
++ + +F +M+ G+KPD V L+ACS ++ GK I LK HG+
Sbjct: 553 ERAIELFDDMIEQGLKPDGVAFVGALTACSHGGLVQQGKEIFYSMLKLHGVSPEDVHYGC 612
Query: 283 LVNLYESCLCVREA-QAVFDLMPHRNVITWNS-LASCYV 319
+V+L + EA Q + D+ N + WNS LA+C V
Sbjct: 613 MVDLLGRAGLLEEAVQLIEDMPMEPNDVIWNSLLAACRV 651
>At5g09950 selenium-binding protein-like
Length = 995
Score = 501 bits (1289), Expect = e-141
Identities = 314/956 (32%), Positives = 494/956 (50%), Gaps = 61/956 (6%)
Query: 18 HGLPNEAIKIYTSSRARGIKPDKPVFMAVAKACA--ASRDALKVKQFHDDATRCGVMSDV 75
+G EA+ GI ++ F++V +AC S L +Q H + D
Sbjct: 80 NGEHKEALVFLRDMVKEGIFSNQYAFVSVLRACQEIGSVGILFGRQIHGLMFKLSYAVDA 139
Query: 76 SIGNAFIHAYGKCKCVEG-ARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGL 134
+ N I Y KC G A F D+ ++ V+WNS+ + Y G + +F M
Sbjct: 140 VVSNVLISMYWKCIGSVGYALCAFGDIEVKNSVSWNSIISVYSQAGDQRSAFRIFSSMQY 199
Query: 135 NKVKANPLTVSSILPGCSDLQ--DLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCV 192
+ + T S++ L D++ ++I + + G++ D+FV S V+ +AK +
Sbjct: 200 DGSRPTEYTFGSLVTTACSLTEPDVRLLEQIMCTIQKSGLLTDLFVGSGLVSAFAKSGSL 259
Query: 193 REAQTVFDLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREM--VLDGVKPDPVTVSCILS 250
A+ VF+ M R+ VT N L V + ++ +F +M ++D V P+ + +LS
Sbjct: 260 SYARKVFNQMETRNAVTLNGLMVGLVRQKWGEEATKLFMDMNSMID-VSPESYVI--LLS 316
Query: 251 ACSDLQ-----DLKSGKAIHGFALKHGMVE-NVFVSNALVNLYESCLCVREAQAVFDLMP 304
+ + LK G+ +HG + G+V+ V + N LVN+Y C + +A+ VF M
Sbjct: 317 SFPEYSLAEEVGLKKGREVHGHVITTGLVDFMVGIGNGLVNMYAKCGSIADARRVFYFMT 376
Query: 305 HRNVITWNSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKT 364
++ ++WNS+ + G + + ++ M + + P + S L +C+ LK K G+
Sbjct: 377 DKDSVSWNSMITGLDQNGCFIEAVERYKSMRRHDILPGSFTLISSLSSCASLKWAKLGQQ 436
Query: 365 IHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNC-- 422
IHG ++K G+ +V V AL+ LYA + E + +F MP + V+WNS+
Sbjct: 437 IHGESLKLGIDLNVSVSNALMTLYAETGYLNECRKIFSSMPEHDQVSWNSIIGALARSER 496
Query: 423 GFPQKGLNVFREMVLNGVKPDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGMVEDVFVC 482
P+ + F G K + +T S+L A S L + GK IHG A+++ + ++
Sbjct: 497 SLPE-AVVCFLNAQRAGQKLNRITFSSVLSAVSSLSFGELGKQIHGLALKNNIADEATTE 555
Query: 483 NALLSLYAKCVCVREAQVVFD-LIPHREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEV 541
NAL++ Y KC + + +F + R+ +WN +++ Y N+ K L + M +
Sbjct: 556 NALIACYGKCGEMDGCEKIFSRMAERRDNVTWNSMISGYIHNELLAKALDLVWFMLQTGQ 615
Query: 542 KADEITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKPDETTIYSILRACSLSECLRMGKE 601
+ D ++ V+ + +E ME+ ACS+ CL
Sbjct: 616 RLDSFMYATVLSAFASVATLERGMEV--------------------HACSVRACL----- 650
Query: 602 IHCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGN 661
+ D+ +ALVDMY+KCG L + F+ MP+++ +SWN+MI HG
Sbjct: 651 ----------ESDVVVGSALVDMYSKCGRLDYALRFFNTMPVRNSYSWNSMISGYARHGQ 700
Query: 662 GKEALSLFEKMLL-SMVKPDSATFTCVLSACSHSMLVEEGVQIFNSMSRDHLVEPEAEHY 720
G+EAL LFE M L PD TF VLSACSH+ L+EEG + F SMS + + P EH+
Sbjct: 701 GEEALKLFETMKLDGQTPPDHVTFVGVLSACSHAGLLEEGFKHFESMSDSYGLAPRIEHF 760
Query: 721 TCVVDIYSRAGCLEEAYGFIQRMPMEPTAIAWKAFLAGC--RVYKNVELAKISAKKLFEI 778
+C+ D+ RAG L++ FI++MPM+P + W+ L C + EL K +A+ LF++
Sbjct: 761 SCMADVLGRAGELDKLEDFIEKMPMKPNVLIWRTVLGACCRANGRKAELGKKAAEMLFQL 820
Query: 779 DPNGSANYVTLFNILVTAKLWSEASKIRKLMKERGITKTPGCSWFHVGNRVHTFVAGDKS 838
+P + NYV L N+ W + K RK MK+ + K G SW + + VH FVAGDKS
Sbjct: 821 EPENAVNYVLLGNMYAAGGRWEDLVKARKKMKDADVKKEAGYSWVTMKDGVHMFVAGDKS 880
Query: 839 NMESDKIYNFLDELFAKIKAAGYKPDTDYVLHDIDQEEKAESLCNHSEKLAVAFGILNLN 898
+ ++D IY L EL K++ AGY P T + L+D++QE K E L HSEKLAVAF +L
Sbjct: 881 HPDADVIYKKLKELNRKMRDAGYVPQTGFALYDLEQENKEEILSYHSEKLAVAF-VLAAQ 939
Query: 899 GQST--IRVFKNLRICGDCHNAIKYMSNVVGVTIVVRDSLRFHHFKNGNCSCKDFW 952
ST IR+ KNLR+CGDCH+A KY+S + G I++RDS RFHHF++G CSC DFW
Sbjct: 940 RSSTLPIRIMKNLRVCGDCHSAFKYISKIEGRQIILRDSNRFHHFQDGACSCSDFW 995
Score = 246 bits (629), Expect = 3e-65
Identities = 203/765 (26%), Positives = 359/765 (46%), Gaps = 58/765 (7%)
Query: 44 MAVAKACAASRDALKVKQFHDDATRCGVMSDVSIGNAFIHAYGKCKCVEGARRVFDDLVA 103
++ ++C R A + FH + + DV + N I+AY + AR+VFD++
Sbjct: 7 LSFVQSCVGHRGAARF--FHSRLYKNRLDKDVYLCNNLINAYLETGDSVSARKVFDEMPL 64
Query: 104 RDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKANPLTVSSILPGCSDLQD--LKSGK 161
R+ V+W + + Y G ++ L R M + +N S+L C ++ + G+
Sbjct: 65 RNCVSWACIVSGYSRNGEHKEALVFLRDMVKEGIFSNQYAFVSVLRACQEIGSVGILFGR 124
Query: 162 EIHGFVVRHGMVEDVFVSSAFVNFYAKCL-CVREAQTVFDLMPHRDVVTWNSLSSCYVNC 220
+IHG + + D VS+ ++ Y KC+ V A F + ++ V+WNS+ S Y
Sbjct: 125 QIHGLMFKLSYAVDAVVSNVLISMYWKCIGSVGYALCAFGDIEVKNSVSWNSIISVYSQA 184
Query: 221 GFPQKGLNVFREMVLDGVKPDPVTV-SCILSACSDLQ-DLKSGKAIHGFALKHGMVENVF 278
G + +F M DG +P T S + +ACS + D++ + I K G++ ++F
Sbjct: 185 GDQRSAFRIFSSMQYDGSRPTEYTFGSLVTTACSLTEPDVRLLEQIMCTIQKSGLLTDLF 244
Query: 279 VSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASCYVNCGFPQKGLNVFREM-GLN 337
V + LV+ + + A+ VF+ M RN +T N L V + ++ +F +M +
Sbjct: 245 VGSGLVSAFAKSGSLSYARKVFNQMETRNAVTLNGLMVGLVRQKWGEEATKLFMDMNSMI 304
Query: 338 GVKPDP-MAMSSILPACSQLKD--LKSGKTIHGFAVKHGMVE-DVFVCTALVNLYANCLC 393
V P+ + + S P S ++ LK G+ +HG + G+V+ V + LVN+YA C
Sbjct: 305 DVSPESYVILLSSFPEYSLAEEVGLKKGREVHGHVITTGLVDFMVGIGNGLVNMYAKCGS 364
Query: 394 VREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHA 453
+ +A+ VF M ++ V+WNS+ + G + + ++ M + + P T++S L +
Sbjct: 365 IADARRVFYFMTDKDSVSWNSMITGLDQNGCFIEAVERYKSMRRHDILPGSFTLISSLSS 424
Query: 454 CSDLQDLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREVASW 513
C+ L+ K G+ IHG +++ G+ +V V NAL++LYA+ + E + +F +P + SW
Sbjct: 425 CASLKWAKLGQQIHGESLKLGIDLNVSVSNALMTLYAETGYLNECRKIFSSMPEHDQVSW 484
Query: 514 NGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGGCVKNSR-IEEAMEIFRKMQ 572
N I IG ++ R + EA+ F Q
Sbjct: 485 NSI-----------------------------------IGALARSERSLPEAVVCFLNAQ 509
Query: 573 TMGFKPDETTIYSILRACSLSECLRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGGLS 632
G K + T S+L A S +GK+IH ++ + NAL+ Y KCG +
Sbjct: 510 RAGQKLNRITFSSVLSAVSSLSFGELGKQIHGLALKNNIADEATTENALIACYGKCGEMD 569
Query: 633 LSRNVFD-MMPIKDVFSWNTMIFANGMHGNGKEALSLFEKMLLSMVKPDSATFTCVLSAC 691
+F M +D +WN+MI + +AL L ML + + DS + VLSA
Sbjct: 570 GCEKIFSRMAERRDNVTWNSMISGYIHNELLAKALDLVWFMLQTGQRLDSFMYATVLSAF 629
Query: 692 SHSMLVEEGVQIFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEPTAIA 751
+ +E G+++ ++ S +E + + +VD+YS+ G L+ A F MP+ + +
Sbjct: 630 ASVATLERGMEV-HACSVRACLESDVVVGSALVDMYSKCGRLDYALRFFNTMPVR-NSYS 687
Query: 752 WKAFLAGCRVYKNVELAKISAKKLFE---IDPNGSANYVTLFNIL 793
W + ++G + E A KLFE +D ++VT +L
Sbjct: 688 WNSMISGYARHGQGE----EALKLFETMKLDGQTPPDHVTFVGVL 728
>At1g20230 hypothetical protein
Length = 760
Score = 499 bits (1284), Expect = e-141
Identities = 257/724 (35%), Positives = 410/724 (56%), Gaps = 37/724 (5%)
Query: 265 HGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASCYVNCGFP 324
H LK G + ++S L+ Y + C +A V +P + +++SL
Sbjct: 38 HARILKSGAQNDGYISAKLIASYSNYNCFNDADLVLQSIPDPTIYSFSSLIYALTKAKLF 97
Query: 325 QKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTAL 384
+ + VF M +G+ PD + ++ C++L K GK IH + G+ D FV ++
Sbjct: 98 TQSIGVFSRMFSHGLIPDSHVLPNLFKVCAELSAFKVGKQIHCVSCVSGLDMDAFVQGSM 157
Query: 385 VNLYANCLCVREAQTVFDLMPHRNVVT--------------------------------- 411
++Y C + +A+ VFD M ++VVT
Sbjct: 158 FHMYMRCGRMGDARKVFDRMSDKDVVTCSALLCAYARKGCLEEVVRILSEMESSGIEANI 217
Query: 412 --WNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHACSDLQDLKSGKVIHGF 469
WN + S + G+ ++ + +F+++ G PD VT+ S+L + D + L G++IHG+
Sbjct: 218 VSWNGILSGFNRSGYHKEAVVMFQKIHHLGFCPDQVTVSSVLPSVGDSEMLNMGRLIHGY 277
Query: 470 AVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREVASWNGILTAYFTNKEYEKG 529
++ G+++D V +A++ +Y K V +F+ E N +T N +K
Sbjct: 278 VIKQGLLKDKCVISAMIDMYGKSGHVYGIISLFNQFEMMEAGVCNAYITGLSRNGLVDKA 337
Query: 530 LYMFSQMNRDEVKADEITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKPDETTIYSILRA 589
L MF ++ + ++W+ +I GC +N + EA+E+FR+MQ G KP+ TI S+L A
Sbjct: 338 LEMFELFKEQTMELNVVSWTSIIAGCAQNGKDIEALELFREMQVAGVKPNHVTIPSMLPA 397
Query: 590 CSLSECLRMGKEIHCYVFR-HWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIKDVFS 648
C L G+ H + R H D ++ +AL+DMYAKCG ++LS+ VF+MMP K++
Sbjct: 398 CGNIAALGHGRSTHGFAVRVHLLD-NVHVGSALIDMYAKCGRINLSQIVFNMMPTKNLVC 456
Query: 649 WNTMIFANGMHGNGKEALSLFEKMLLSMVKPDSATFTCVLSACSHSMLVEEGVQIFNSMS 708
WN+++ MHG KE +S+FE ++ + +KPD +FT +LSAC L +EG + F MS
Sbjct: 457 WNSLMNGFSMHGKAKEVMSIFESLMRTRLKPDFISFTSLLSACGQVGLTDEGWKYFKMMS 516
Query: 709 RDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEPTAIAWKAFLAGCRVYKNVELA 768
++ ++P EHY+C+V++ RAG L+EAY I+ MP EP + W A L CR+ NV+LA
Sbjct: 517 EEYGIKPRLEHYSCMVNLLGRAGKLQEAYDLIKEMPFEPDSCVWGALLNSCRLQNNVDLA 576
Query: 769 KISAKKLFEIDPNGSANYVTLFNILVTAKLWSEASKIRKLMKERGITKTPGCSWFHVGNR 828
+I+A+KLF ++P YV L NI +W+E IR M+ G+ K PGCSW V NR
Sbjct: 577 EIAAEKLFHLEPENPGTYVLLSNIYAAKGMWTEVDSIRNKMESLGLKKNPGCSWIQVKNR 636
Query: 829 VHTFVAGDKSNMESDKIYNFLDELFAKIKAAGYKPDTDYVLHDIDQEEKAESLCNHSEKL 888
V+T +AGDKS+ + D+I +DE+ +++ +G++P+ D+ LHD++++E+ + L HSEKL
Sbjct: 637 VYTLLAGDKSHPQIDQITEKMDEISKEMRKSGHRPNLDFALHDVEEQEQEQMLWGHSEKL 696
Query: 889 AVAFGILNLNGQSTIRVFKNLRICGDCHNAIKYMSNVVGVTIVVRDSLRFHHFKNGNCSC 948
AV FG+LN + ++V KNLRICGDCH IK++S+ G I +RD+ RFHHFK+G CSC
Sbjct: 697 AVVFGLLNTPDGTPLQVIKNLRICGDCHAVIKFISSYAGREIFIRDTNRFHHFKDGICSC 756
Query: 949 KDFW 952
DFW
Sbjct: 757 GDFW 760
Score = 191 bits (485), Expect = 2e-48
Identities = 119/493 (24%), Positives = 234/493 (47%), Gaps = 71/493 (14%)
Query: 157 LKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTWNSLSSC 216
L + H +++ G D ++S+ + Y+ C +A V +P + +++SL
Sbjct: 31 LSKTTQAHARILKSGAQNDGYISAKLIASYSNYNCFNDADLVLQSIPDPTIYSFSSLIYA 90
Query: 217 YVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQDLKSGKAIHGFALKHGMVEN 276
+ + VF M G+ PD + + C++L K GK IH + G+ +
Sbjct: 91 LTKAKLFTQSIGVFSRMFSHGLIPDSHVLPNLFKVCAELSAFKVGKQIHCVSCVSGLDMD 150
Query: 277 VFVSNALVNLYESCLCVREAQAVFDLMPHR------------------------------ 306
FV ++ ++Y C + +A+ VFD M +
Sbjct: 151 AFVQGSMFHMYMRCGRMGDARKVFDRMSDKDVVTCSALLCAYARKGCLEEVVRILSEMES 210
Query: 307 -----NVITWNSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKS 361
N+++WN + S + G+ ++ + +F+++ G PD + +SS+LP+ + L
Sbjct: 211 SGIEANIVSWNGILSGFNRSGYHKEAVVMFQKIHHLGFCPDQVTVSSVLPSVGDSEMLNM 270
Query: 362 GKTIHGFAVKHGMVEDVFVCTALVNLYANC-------------------LC--------- 393
G+ IHG+ +K G+++D V +A++++Y +C
Sbjct: 271 GRLIHGYVIKQGLLKDKCVISAMIDMYGKSGHVYGIISLFNQFEMMEAGVCNAYITGLSR 330
Query: 394 ---VREAQTVFDLMPHR----NVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVT 446
V +A +F+L + NVV+W S+ + G + L +FREM + GVKP+ VT
Sbjct: 331 NGLVDKALEMFELFKEQTMELNVVSWTSIIAGCAQNGKDIEALELFREMQVAGVKPNHVT 390
Query: 447 MLSILHACSDLQDLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIP 506
+ S+L AC ++ L G+ HGFAVR ++++V V +AL+ +YAKC + +Q+VF+++P
Sbjct: 391 IPSMLPACGNIAALGHGRSTHGFAVRVHLLDNVHVGSALIDMYAKCGRINLSQIVFNMMP 450
Query: 507 HREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGGCVKNSRIEEAME 566
+ + WN ++ + + + ++ + +F + R +K D I+++ ++ C + +E +
Sbjct: 451 TKNLVCWNSLMNGFSMHGKAKEVMSIFESLMRTRLKPDFISFTSLLSACGQVGLTDEGWK 510
Query: 567 IFRKM-QTMGFKP 578
F+ M + G KP
Sbjct: 511 YFKMMSEEYGIKP 523
Score = 188 bits (478), Expect = 1e-47
Identities = 131/534 (24%), Positives = 248/534 (45%), Gaps = 72/534 (13%)
Query: 58 KVKQFHDDATRCGVMSDVSIGNAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYV 117
K Q H + G +D I I +Y C A V + + +++SL
Sbjct: 33 KTTQAHARILKSGAQNDGYISAKLIASYSNYNCFNDADLVLQSIPDPTIYSFSSLIYALT 92
Query: 118 NCGFPQQGLNVFRKMGLNKVKANPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVF 177
Q + VF +M + + + + ++ C++L K GK+IH G+ D F
Sbjct: 93 KAKLFTQSIGVFSRMFSHGLIPDSHVLPNLFKVCAELSAFKVGKQIHCVSCVSGLDMDAF 152
Query: 178 VSSAFVNFYAKCLCVREAQTVFDLMPHRDVVT---------------------------- 209
V + + Y +C + +A+ VFD M +DVVT
Sbjct: 153 VQGSMFHMYMRCGRMGDARKVFDRMSDKDVVTCSALLCAYARKGCLEEVVRILSEMESSG 212
Query: 210 -------WNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQDLKSGK 262
WN + S + G+ ++ + +F+++ G PD VTVS +L + D + L G+
Sbjct: 213 IEANIVSWNGILSGFNRSGYHKEAVVMFQKIHHLGFCPDQVTVSSVLPSVGDSEMLNMGR 272
Query: 263 AIHGFALKHGMVENVFVSNALVNLY-------------------ESCLC----------- 292
IHG+ +K G++++ V +A++++Y E+ +C
Sbjct: 273 LIHGYVIKQGLLKDKCVISAMIDMYGKSGHVYGIISLFNQFEMMEAGVCNAYITGLSRNG 332
Query: 293 -VREAQAVFDLMPHR----NVITWNSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMS 347
V +A +F+L + NV++W S+ + G + L +FREM + GVKP+ + +
Sbjct: 333 LVDKALEMFELFKEQTMELNVVSWTSIIAGCAQNGKDIEALELFREMQVAGVKPNHVTIP 392
Query: 348 SILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPHR 407
S+LPAC + L G++ HGFAV+ ++++V V +AL+++YA C + +Q VF++MP +
Sbjct: 393 SMLPACGNIAALGHGRSTHGFAVRVHLLDNVHVGSALIDMYAKCGRINLSQIVFNMMPTK 452
Query: 408 NVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHACSDLQDLKSG-KVI 466
N+V WNSL + + G ++ +++F ++ +KPD ++ S+L AC + G K
Sbjct: 453 NLVCWNSLMNGFSMHGKAKEVMSIFESLMRTRLKPDFISFTSLLSACGQVGLTDEGWKYF 512
Query: 467 HGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHR-EVASWNGILTA 519
+ +G+ + + +++L + ++EA + +P + W +L +
Sbjct: 513 KMMSEEYGIKPRLEHYSCMVNLLGRAGKLQEAYDLIKEMPFEPDSCVWGALLNS 566
Score = 172 bits (437), Expect = 6e-43
Identities = 130/509 (25%), Positives = 238/509 (46%), Gaps = 75/509 (14%)
Query: 23 EAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKV-KQFHDDATRCGVMSDVSIGNAF 81
++I +++ + G+ PD V + K C A A KV KQ H + G+ D + +
Sbjct: 99 QSIGVFSRMFSHGLIPDSHVLPNLFKVC-AELSAFKVGKQIHCVSCVSGLDMDAFVQGSM 157
Query: 82 IHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKAN- 140
H Y +C + AR+VFD + +DVVT ++L Y G ++ + + +M + ++AN
Sbjct: 158 FHMYMRCGRMGDARKVFDRMSDKDVVTCSALLCAYARKGCLEEVVRILSEMESSGIEANI 217
Query: 141 ----------------------------------PLTVSSILPGCSDLQDLKSGKEIHGF 166
+TVSS+LP D + L G+ IHG+
Sbjct: 218 VSWNGILSGFNRSGYHKEAVVMFQKIHHLGFCPDQVTVSSVLPSVGDSEMLNMGRLIHGY 277
Query: 167 VVRHGMVEDVFVSSAFVNFYAKC-------------------LC------------VREA 195
V++ G+++D V SA ++ Y K +C V +A
Sbjct: 278 VIKQGLLKDKCVISAMIDMYGKSGHVYGIISLFNQFEMMEAGVCNAYITGLSRNGLVDKA 337
Query: 196 QTVFDLMPHR----DVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSA 251
+F+L + +VV+W S+ + G + L +FREM + GVKP+ VT+ +L A
Sbjct: 338 LEMFELFKEQTMELNVVSWTSIIAGCAQNGKDIEALELFREMQVAGVKPNHVTIPSMLPA 397
Query: 252 CSDLQDLKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITW 311
C ++ L G++ HGFA++ +++NV V +AL+++Y C + +Q VF++MP +N++ W
Sbjct: 398 CGNIAALGHGRSTHGFAVRVHLLDNVHVGSALIDMYAKCGRINLSQIVFNMMPTKNLVCW 457
Query: 312 NSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSG-KTIHGFAV 370
NSL + + G ++ +++F + +KPD ++ +S+L AC Q+ G K +
Sbjct: 458 NSLMNGFSMHGKAKEVMSIFESLMRTRLKPDFISFTSLLSACGQVGLTDEGWKYFKMMSE 517
Query: 371 KHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPHR-NVVTWNS-LSSCYVNCGFPQKG 428
++G+ + + +VNL ++EA + MP + W + L+SC +
Sbjct: 518 EYGIKPRLEHYSCMVNLLGRAGKLQEAYDLIKEMPFEPDSCVWGALLNSCRLQNNVDLAE 577
Query: 429 LNVFREMVLNGVKPDLVTMLSILHACSDL 457
+ + L P +LS ++A +
Sbjct: 578 IAAEKLFHLEPENPGTYVLLSNIYAAKGM 606
Score = 93.6 bits (231), Expect = 5e-19
Identities = 75/333 (22%), Positives = 141/333 (41%), Gaps = 37/333 (11%)
Query: 19 GLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSIG 78
G EA+ ++ G PD+ +V + S + H + G++ D +
Sbjct: 231 GYHKEAVVMFQKIHHLGFCPDQVTVSSVLPSVGDSEMLNMGRLIHGYVIKQGLLKDKCVI 290
Query: 79 NAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVK 138
+A I YGK V G +F+ + N+ G + L +F ++
Sbjct: 291 SAMIDMYGKSGHVYGIISLFNQFEMMEAGVCNAYITGLSRNGLVDKALEMFELFKEQTME 350
Query: 139 ANPLTVSSI-----------------------------------LPGCSDLQDLKSGKEI 163
N ++ +SI LP C ++ L G+
Sbjct: 351 LNVVSWTSIIAGCAQNGKDIEALELFREMQVAGVKPNHVTIPSMLPACGNIAALGHGRST 410
Query: 164 HGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTWNSLSSCYVNCGFP 223
HGF VR ++++V V SA ++ YAKC + +Q VF++MP +++V WNSL + + G
Sbjct: 411 HGFAVRVHLLDNVHVGSALIDMYAKCGRINLSQIVFNMMPTKNLVCWNSLMNGFSMHGKA 470
Query: 224 QKGLNVFREMVLDGVKPDPVTVSCILSACSDLQDLKSG-KAIHGFALKHGMVENVFVSNA 282
++ +++F ++ +KPD ++ + +LSAC + G K + ++G+ + +
Sbjct: 471 KEVMSIFESLMRTRLKPDFISFTSLLSACGQVGLTDEGWKYFKMMSEEYGIKPRLEHYSC 530
Query: 283 LVNLYESCLCVREAQAVFDLMPHR-NVITWNSL 314
+VNL ++EA + MP + W +L
Sbjct: 531 MVNLLGRAGKLQEAYDLIKEMPFEPDSCVWGAL 563
Score = 64.7 bits (156), Expect = 2e-10
Identities = 46/202 (22%), Positives = 94/202 (45%), Gaps = 3/202 (1%)
Query: 18 HGLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSI 77
+G EA++++ + G+KP+ ++ AC + H A R ++ +V +
Sbjct: 366 NGKDIEALELFREMQVAGVKPNHVTIPSMLPACGNIAALGHGRSTHGFAVRVHLLDNVHV 425
Query: 78 GNAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKV 137
G+A I Y KC + ++ VF+ + +++V WNSL + G ++ +++F + ++
Sbjct: 426 GSALIDMYAKCGRINLSQIVFNMMPTKNLVCWNSLMNGFSMHGKAKEVMSIFESLMRTRL 485
Query: 138 KANPLTVSSILPGCSDLQDLKSG-KEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQ 196
K + ++ +S+L C + G K +G+ + S VN + ++EA
Sbjct: 486 KPDFISFTSLLSACGQVGLTDEGWKYFKMMSEEYGIKPRLEHYSCMVNLLGRAGKLQEAY 545
Query: 197 TVFDLMPHR-DVVTWNS-LSSC 216
+ MP D W + L+SC
Sbjct: 546 DLIKEMPFEPDSCVWGALLNSC 567
Score = 34.3 bits (77), Expect = 0.34
Identities = 37/199 (18%), Positives = 73/199 (36%), Gaps = 33/199 (16%)
Query: 593 SECLRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTM 652
S L + H + + D + L+ Y+ + + V +P ++S++++
Sbjct: 28 SSSLSKTTQAHARILKSGAQNDGYISAKLIASYSNYNCFNDADLVLQSIPDPTIYSFSSL 87
Query: 653 IFANGMHGNGKEALSLFEKMLLSMVKPDSATFTCVLSACSHSMLVEEGVQIF-----NSM 707
I+A +++ +F +M + PDS + C+ + G QI + +
Sbjct: 88 IYALTKAKLFTQSIGVFSRMFSHGLIPDSHVLPNLFKVCAELSAFKVGKQIHCVSCVSGL 147
Query: 708 SRDHLVEPEAEHY-----------------------TC--VVDIYSRAGCLEEAYGFIQR 742
D V+ H TC ++ Y+R GCLEE +
Sbjct: 148 DMDAFVQGSMFHMYMRCGRMGDARKVFDRMSDKDVVTCSALLCAYARKGCLEEVVRILSE 207
Query: 743 MP---MEPTAIAWKAFLAG 758
M +E ++W L+G
Sbjct: 208 MESSGIEANIVSWNGILSG 226
>At2g29760 hypothetical protein
Length = 738
Score = 478 bits (1230), Expect = e-135
Identities = 243/710 (34%), Positives = 406/710 (56%), Gaps = 12/710 (1%)
Query: 248 ILSACSDLQDLKSGKAIHGFALKHGMVENVFVSNALVNL--YESCLCVREAQAVFDLMPH 305
++ C L+ LK HG ++ G + + ++ L + S + A+ VFD +P
Sbjct: 36 LIERCVSLRQLKQ---THGHMIRTGTFSDPYSASKLFAMAALSSFASLEYARKVFDEIPK 92
Query: 306 RNVITWNSLASCYVNCGFPQKGLNVFREM-GLNGVKPDPMAMSSILPACSQLKDLKSGKT 364
N WN+L Y + P + F +M + P+ ++ A +++ L G++
Sbjct: 93 PNSFAWNTLIRAYASGPDPVLSIWAFLDMVSESQCYPNKYTFPFLIKAAAEVSSLSLGQS 152
Query: 365 IHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCGF 424
+HG AVK + DVFV +L++ Y +C + A VF + ++VV+WNS+ + +V G
Sbjct: 153 LHGMAVKSAVGSDVFVANSLIHCYFSCGDLDSACKVFTTIKEKDVVSWNSMINGFVQKGS 212
Query: 425 PQKGLNVFREMVLNGVKPDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGMVEDVFVCNA 484
P K L +F++M VK VTM+ +L AC+ +++L+ G+ + + + + ++ + NA
Sbjct: 213 PDKALELFKKMESEDVKASHVTMVGVLSACAKIRNLEFGRQVCSYIEENRVNVNLTLANA 272
Query: 485 LLSLYAKCVCVREAQVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKAD 544
+L +Y KC + +A+ +FD + ++ +W +L Y +++YE + + M + ++
Sbjct: 273 MLDMYTKCGSIEDAKRLFDAMEEKDNVTWTTMLDGYAISEDYEAAREVLNSMPQKDI--- 329
Query: 545 EITWSVVIGGCVKNSRIEEAMEIFRKMQTM-GFKPDETTIYSILRACSLSECLRMGKEIH 603
+ W+ +I +N + EA+ +F ++Q K ++ T+ S L AC+ L +G+ IH
Sbjct: 330 -VAWNALISAYEQNGKPNEALIVFHELQLQKNMKLNQITLVSTLSACAQVGALELGRWIH 388
Query: 604 CYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGK 663
Y+ +H + T+AL+ MY+KCG L SR VF+ + +DVF W+ MI MHG G
Sbjct: 389 SYIKKHGIRMNFHVTSALIHMYSKCGDLEKSREVFNSVEKRDVFVWSAMIGGLAMHGCGN 448
Query: 664 EALSLFEKMLLSMVKPDSATFTCVLSACSHSMLVEEGVQIFNSMSRDHLVEPEAEHYTCV 723
EA+ +F KM + VKP+ TFT V ACSH+ LV+E +F+ M ++ + PE +HY C+
Sbjct: 449 EAVDMFYKMQEANVKPNGVTFTNVFCACSHTGLVDEAESLFHQMESNYGIVPEEKHYACI 508
Query: 724 VDIYSRAGCLEEAYGFIQRMPMEPTAIAWKAFLAGCRVYKNVELAKISAKKLFEIDPNGS 783
VD+ R+G LE+A FI+ MP+ P+ W A L C+++ N+ LA+++ +L E++P
Sbjct: 509 VDVLGRSGYLEKAVKFIEAMPIPPSTSVWGALLGACKIHANLNLAEMACTRLLELEPRND 568
Query: 784 ANYVTLFNILVTAKLWSEASKIRKLMKERGITKTPGCSWFHVGNRVHTFVAGDKSNMESD 843
+V L NI W S++RK M+ G+ K PGCS + +H F++GD ++ S+
Sbjct: 569 GAHVLLSNIYAKLGKWENVSELRKHMRVTGLKKEPGCSSIEIDGMIHEFLSGDNAHPMSE 628
Query: 844 KIYNFLDELFAKIKAAGYKPDTDYVLHDIDQEE-KAESLCNHSEKLAVAFGILNLNGQST 902
K+Y L E+ K+K+ GY+P+ VL I++EE K +SL HSEKLA+ +G+++
Sbjct: 629 KVYGKLHEVMEKLKSNGYEPEISQVLQIIEEEEMKEQSLNLHSEKLAICYGLISTEAPKV 688
Query: 903 IRVFKNLRICGDCHNAIKYMSNVVGVTIVVRDSLRFHHFKNGNCSCKDFW 952
IRV KNLR+CGDCH+ K +S + I+VRD RFHHF+NG CSC DFW
Sbjct: 689 IRVIKNLRVCGDCHSVAKLISQLYDREIIVRDRYRFHHFRNGQCSCNDFW 738
Score = 192 bits (489), Expect = 6e-49
Identities = 120/447 (26%), Positives = 228/447 (50%), Gaps = 38/447 (8%)
Query: 44 MAVAKACAASRDALKVKQFHDDATRCGVMSD-VSIGNAF-IHAYGKCKCVEGARRVFDDL 101
+++ + C + R ++KQ H R G SD S F + A +E AR+VFD++
Sbjct: 34 ISLIERCVSLR---QLKQTHGHMIRTGTFSDPYSASKLFAMAALSSFASLEYARKVFDEI 90
Query: 102 VARDVVTWNSLSACYVNCGFPQQGLNVFRKM-GLNKVKANPLTVSSILPGCSDLQDLKSG 160
+ WN+L Y + P + F M ++ N T ++ +++ L G
Sbjct: 91 PKPNSFAWNTLIRAYASGPDPVLSIWAFLDMVSESQCYPNKYTFPFLIKAAAEVSSLSLG 150
Query: 161 KEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTWNSLSSCYVNC 220
+ +HG V+ + DVFV+++ ++ Y C + A VF + +DVV+WNS+ + +V
Sbjct: 151 QSLHGMAVKSAVGSDVFVANSLIHCYFSCGDLDSACKVFTTIKEKDVVSWNSMINGFVQK 210
Query: 221 GFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQDLKSGKAIHGFALKHGMVENVFVS 280
G P K L +F++M + VK VT+ +LSAC+ +++L+ G+ + + ++ + N+ ++
Sbjct: 211 GSPDKALELFKKMESEDVKASHVTMVGVLSACAKIRNLEFGRQVCSYIEENRVNVNLTLA 270
Query: 281 NALVNLYESCLCVREAQAVFDL-------------------------------MPHRNVI 309
NA++++Y C + +A+ +FD MP ++++
Sbjct: 271 NAMLDMYTKCGSIEDAKRLFDAMEEKDNVTWTTMLDGYAISEDYEAAREVLNSMPQKDIV 330
Query: 310 TWNSLASCYVNCGFPQKGLNVFREMGL-NGVKPDPMAMSSILPACSQLKDLKSGKTIHGF 368
WN+L S Y G P + L VF E+ L +K + + + S L AC+Q+ L+ G+ IH +
Sbjct: 331 AWNALISAYEQNGKPNEALIVFHELQLQKNMKLNQITLVSTLSACAQVGALELGRWIHSY 390
Query: 369 AVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKG 428
KHG+ + V +AL+++Y+ C + +++ VF+ + R+V W+++ G +
Sbjct: 391 IKKHGIRMNFHVTSALIHMYSKCGDLEKSREVFNSVEKRDVFVWSAMIGGLAMHGCGNEA 450
Query: 429 LNVFREMVLNGVKPDLVTMLSILHACS 455
+++F +M VKP+ VT ++ ACS
Sbjct: 451 VDMFYKMQEANVKPNGVTFTNVFCACS 477
Score = 174 bits (442), Expect = 2e-43
Identities = 104/401 (25%), Positives = 201/401 (49%), Gaps = 33/401 (8%)
Query: 38 PDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSIGNAFIHAYGKCKCVEGARRV 97
P+K F + KA A + H A + V SDV + N+ IH Y C ++ A +V
Sbjct: 129 PNKYTFPFLIKAAAEVSSLSLGQSLHGMAVKSAVGSDVFVANSLIHCYFSCGDLDSACKV 188
Query: 98 FDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKANPLTVSSILPGCSDLQDL 157
F + +DVV+WNS+ +V G P + L +F+KM VKA+ +T+ +L C+ +++L
Sbjct: 189 FTTIKEKDVVSWNSMINGFVQKGSPDKALELFKKMESEDVKASHVTMVGVLSACAKIRNL 248
Query: 158 KSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDL---------------- 201
+ G+++ ++ + + ++ +++A ++ Y KC + +A+ +FD
Sbjct: 249 EFGRQVCSYIEENRVNVNLTLANAMLDMYTKCGSIEDAKRLFDAMEEKDNVTWTTMLDGY 308
Query: 202 ---------------MPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLD-GVKPDPVTV 245
MP +D+V WN+L S Y G P + L VF E+ L +K + +T+
Sbjct: 309 AISEDYEAAREVLNSMPQKDIVAWNALISAYEQNGKPNEALIVFHELQLQKNMKLNQITL 368
Query: 246 SCILSACSDLQDLKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPH 305
LSAC+ + L+ G+ IH + KHG+ N V++AL+++Y C + +++ VF+ +
Sbjct: 369 VSTLSACAQVGALELGRWIHSYIKKHGIRMNFHVTSALIHMYSKCGDLEKSREVFNSVEK 428
Query: 306 RNVITWNSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTI 365
R+V W+++ G + +++F +M VKP+ + +++ ACS + +++
Sbjct: 429 RDVFVWSAMIGGLAMHGCGNEAVDMFYKMQEANVKPNGVTFTNVFCACSHTGLVDEAESL 488
Query: 366 -HGFAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMP 405
H +G+V + +V++ + +A + MP
Sbjct: 489 FHQMESNYGIVPEEKHYACIVDVLGRSGYLEKAVKFIEAMP 529
Score = 115 bits (287), Expect = 2e-25
Identities = 69/270 (25%), Positives = 130/270 (47%), Gaps = 32/270 (11%)
Query: 16 IPHGLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDV 75
+ G P++A++++ + +K + V ACA R+ +Q V ++
Sbjct: 208 VQKGSPDKALELFKKMESEDVKASHVTMVGVLSACAKIRNLEFGRQVCSYIEENRVNVNL 267
Query: 76 SIGNAFIHAYGKCKCVEGARRVFD-------------------------------DLVAR 104
++ NA + Y KC +E A+R+FD + +
Sbjct: 268 TLANAMLDMYTKCGSIEDAKRLFDAMEEKDNVTWTTMLDGYAISEDYEAAREVLNSMPQK 327
Query: 105 DVVTWNSLSACYVNCGFPQQGLNVFRKMGLNK-VKANPLTVSSILPGCSDLQDLKSGKEI 163
D+V WN+L + Y G P + L VF ++ L K +K N +T+ S L C+ + L+ G+ I
Sbjct: 328 DIVAWNALISAYEQNGKPNEALIVFHELQLQKNMKLNQITLVSTLSACAQVGALELGRWI 387
Query: 164 HGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTWNSLSSCYVNCGFP 223
H ++ +HG+ + V+SA ++ Y+KC + +++ VF+ + RDV W+++ G
Sbjct: 388 HSYIKKHGIRMNFHVTSALIHMYSKCGDLEKSREVFNSVEKRDVFVWSAMIGGLAMHGCG 447
Query: 224 QKGLNVFREMVLDGVKPDPVTVSCILSACS 253
+ +++F +M VKP+ VT + + ACS
Sbjct: 448 NEAVDMFYKMQEANVKPNGVTFTNVFCACS 477
>At4g39530 putative protein
Length = 834
Score = 477 bits (1227), Expect = e-134
Identities = 265/813 (32%), Positives = 438/813 (53%), Gaps = 43/813 (5%)
Query: 52 ASRDALKVKQF-HDDATRCGVMSDVSIGNAFIHAYGKCKCVEGARRVFDDLVARDVVTWN 110
AS D L + H G+ D + N I+ Y + + AR+VF+ + R++V+W+
Sbjct: 55 ASDDLLHYQNVVHGQIIVWGLELDTYLSNILINLYSRAGGMVYARKVFEKMPERNLVSWS 114
Query: 111 SLSACYVNCGFPQQGLNVFRKMG-LNKVKANPLTVSSILPGCSDLQDLKSGK--EIHGFV 167
++ + + G ++ L VF + K N +SS + CS L ++ F+
Sbjct: 115 TMVSACNHHGIYEESLVVFLEFWRTRKDSPNEYILSSFIQACSGLDGRGRWMVFQLQSFL 174
Query: 168 VRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTWNSLSSCYVNCGFPQKGL 227
V+ G DV+V + ++FY K + A+ VFD +P + VTW ++ S V G L
Sbjct: 175 VKSGFDRDVYVGTLLIDFYLKDGNIDYARLVFDALPEKSTVTWTTMISGCVKMGRSYVSL 234
Query: 228 NVFREMVLDGVKPDPVTVSCILSACSDLQDLKSGKAIHGFALKHGMVENVFVSNALVNLY 287
+F +++ D V PD +S +LSACS L L+ GK IH L++G+ + + N L++ Y
Sbjct: 235 QLFYQLMEDNVVPDGYILSTVLSACSILPFLEGGKQIHAHILRYGLEMDASLMNVLIDSY 294
Query: 288 ESCLCVREAQAVFDLMPHRNVITWNSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMS 347
C V A +F+ MP++N+I+W +L S Y ++ + +F M G+KPD A S
Sbjct: 295 VKCGRVIAAHKLFNGMPNKNIISWTTLLSGYKQNALHKEAMELFTSMSKFGLKPDMYACS 354
Query: 348 SILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPHR 407
SIL +C+ L L G +H + +K + D +V +L+++YA C C+ +A+ VFD+
Sbjct: 355 SILTSCASLHALGFGTQVHAYTIKANLGNDSYVTNSLIDMYAKCDCLTDARKVFDIFAAA 414
Query: 408 NVVTWNSLSSCYVNCGFP---QKGLNVFREMVLNGVKPDLVTMLSILHACSDLQDLKSGK 464
+VV +N++ Y G + LN+FR+M ++P L+T +S+L A + L L K
Sbjct: 415 DVVLFNAMIEGYSRLGTQWELHEALNIFRDMRFRLIRPSLLTFVSLLRASASLTSLGLSK 474
Query: 465 VIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREVASWNGILTAYFTNK 524
IHG ++G+ D+F +AL+ +Y+ C C++++++VFD + +++ WN + Y
Sbjct: 475 QIHGLMFKYGLNLDIFAGSALIDVYSNCYCLKDSRLVFDEMKVKDLVIWNSMFAGYVQQS 534
Query: 525 EYEKGLYMFSQMNRDEVKADEITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKPDETTIY 584
E E+ L +F + +Q +PDE T
Sbjct: 535 ENEEALNLFLE-----------------------------------LQLSRERPDEFTFA 559
Query: 585 SILRACSLSECLRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIK 644
+++ A +++G+E HC + + + + TNAL+DMYAKCG + FD +
Sbjct: 560 NMVTAAGNLASVQLGQEFHCQLLKRGLECNPYITNALLDMYAKCGSPEDAHKAFDSAASR 619
Query: 645 DVFSWNTMIFANGMHGNGKEALSLFEKMLLSMVKPDSATFTCVLSACSHSMLVEEGVQIF 704
DV WN++I + HG GK+AL + EKM+ ++P+ TF VLSACSH+ LVE+G++ F
Sbjct: 620 DVVCWNSVISSYANHGEGKKALQMLEKMMSEGIEPNYITFVGVLSACSHAGLVEDGLKQF 679
Query: 705 NSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEPTAIAWKAFLAGCRVYKN 764
M R +EPE EHY C+V + RAG L +A I++MP +P AI W++ L+GC N
Sbjct: 680 ELMLR-FGIEPETEHYVCMVSLLGRAGRLNKARELIEKMPTKPAAIVWRSLLSGCAKAGN 738
Query: 765 VELAKISAKKLFEIDPNGSANYVTLFNILVTAKLWSEASKIRKLMKERGITKTPGCSWFH 824
VELA+ +A+ DP S ++ L NI + +W+EA K+R+ MK G+ K PG SW
Sbjct: 739 VELAEHAAEMAILSDPKDSGSFTMLSNIYASKGMWTEAKKVRERMKVEGVVKEPGRSWIG 798
Query: 825 VGNRVHTFVAGDKSNMESDKIYNFLDELFAKIK 857
+ VH F++ DKS+ ++++IY LD+L +I+
Sbjct: 799 INKEVHIFLSKDKSHCKANQIYEVLDDLLVQIR 831
Score = 215 bits (548), Expect = 8e-56
Identities = 127/441 (28%), Positives = 225/441 (50%), Gaps = 5/441 (1%)
Query: 18 HGLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSI 77
+ L EA++++TS G+KPD ++ +CA+ Q H + + +D +
Sbjct: 328 NALHKEAMELFTSMSKFGLKPDMYACSSILTSCASLHALGFGTQVHAYTIKANLGNDSYV 387
Query: 78 GNAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFP---QQGLNVFRKMGL 134
N+ I Y KC C+ AR+VFD A DVV +N++ Y G + LN+FR M
Sbjct: 388 TNSLIDMYAKCDCLTDARKVFDIFAAADVVLFNAMIEGYSRLGTQWELHEALNIFRDMRF 447
Query: 135 NKVKANPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVRE 194
++ + LT S+L + L L K+IHG + ++G+ D+F SA ++ Y+ C C+++
Sbjct: 448 RLIRPSLLTFVSLLRASASLTSLGLSKQIHGLMFKYGLNLDIFAGSALIDVYSNCYCLKD 507
Query: 195 AQTVFDLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSD 254
++ VFD M +D+V WNS+ + YV ++ LN+F E+ L +PD T + +++A +
Sbjct: 508 SRLVFDEMKVKDLVIWNSMFAGYVQQSENEEALNLFLELQLSRERPDEFTFANMVTAAGN 567
Query: 255 LQDLKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSL 314
L ++ G+ H LK G+ N +++NAL+++Y C +A FD R+V+ WNS+
Sbjct: 568 LASVQLGQEFHCQLLKRGLECNPYITNALLDMYAKCGSPEDAHKAFDSAASRDVVCWNSV 627
Query: 315 ASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGM 374
S Y N G +K L + +M G++P+ + +L ACS ++ G ++ G+
Sbjct: 628 ISSYANHGEGKKALQMLEKMMSEGIEPNYITFVGVLSACSHAGLVEDGLKQFELMLRFGI 687
Query: 375 VEDVFVCTALVNLYANCLCVREAQTVFDLMPHR-NVVTWNSLSSCYVNCGFPQKGLNVFR 433
+ +V+L + +A+ + + MP + + W SL S G + +
Sbjct: 688 EPETEHYVCMVSLLGRAGRLNKARELIEKMPTKPAAIVWRSLLSGCAKAGNVELAEHAAE 747
Query: 434 EMVLNGVKPD-LVTMLSILHA 453
+L+ K TMLS ++A
Sbjct: 748 MAILSDPKDSGSFTMLSNIYA 768
Score = 61.6 bits (148), Expect = 2e-09
Identities = 49/200 (24%), Positives = 90/200 (44%), Gaps = 31/200 (15%)
Query: 586 ILRACSLSECLRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIKD 645
+L+ + + L +H + + D +N L+++Y++ GG+ +R VF+ MP ++
Sbjct: 50 LLQLRASDDLLHYQNVVHGQIIVWGLELDTYLSNILINLYSRAGGMVYARKVFEKMPERN 109
Query: 646 VFSWNTMIFANGMHGNGKEALSLF-EKMLLSMVKPDSATFTCVLSACS------------ 692
+ SW+TM+ A HG +E+L +F E P+ + + ACS
Sbjct: 110 LVSWSTMVSACNHHGIYEESLVVFLEFWRTRKDSPNEYILSSFIQACSGLDGRGRWMVFQ 169
Query: 693 -HSMLVEEGVQIFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEPTAIA 751
S LV+ G RD V T ++D Y + G ++ A +P E + +
Sbjct: 170 LQSFLVKSG------FDRDVYVG------TLLIDFYLKDGNIDYARLVFDALP-EKSTVT 216
Query: 752 WKAFLAGC----RVYKNVEL 767
W ++GC R Y +++L
Sbjct: 217 WTTMISGCVKMGRSYVSLQL 236
>At2g22070 hypothetical protein
Length = 786
Score = 473 bits (1217), Expect = e-133
Identities = 259/787 (32%), Positives = 431/787 (53%), Gaps = 78/787 (9%)
Query: 240 PDPVTVSCILSACSDLQDLKSGKA--------IHGFALKHGMVENVFVSNALVNLYESCL 291
P P+++S +L C++L K+ +H +K G++ +V++ N L+N+Y
Sbjct: 4 PVPLSLSTLLELCTNLLQKSVNKSNGRFTAQLVHCRVIKSGLMFSVYLMNNLMNVYSKTG 63
Query: 292 CVREAQAVFDLMPHRNVITWNSLASCYV-------------------------------N 320
A+ +FD MP R +WN++ S Y N
Sbjct: 64 YALHARKLFDEMPLRTAFSWNTVLSAYSKRGDMDSTCEFFDQLPQRDSVSWTTMIVGYKN 123
Query: 321 CGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDVFV 380
G K + V +M G++P ++++L + + + +++GK +H F VK G+ +V V
Sbjct: 124 IGQYHKAIRVMGDMVKEGIEPTQFTLTNVLASVAATRCMETGKKVHSFIVKLGLRGNVSV 183
Query: 381 CTALVNLYANC------------LCVRE-------------------AQTVFDLMPHRNV 409
+L+N+YA C + VR+ A F+ M R++
Sbjct: 184 SNSLLNMYAKCGDPMMAKFVFDRMVVRDISSWNAMIALHMQVGQMDLAMAQFEQMAERDI 243
Query: 410 VTWNSLSSCYVNCGFPQKGLNVFREMVLNGV-KPDLVTMLSILHACSDLQDLKSGKVIHG 468
VTWNS+ S + G+ + L++F +M+ + + PD T+ S+L AC++L+ L GK IH
Sbjct: 244 VTWNSMISGFNQRGYDLRALDIFSKMLRDSLLSPDRFTLASVLSACANLEKLCIGKQIHS 303
Query: 469 FAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHRE--VASWNGILTAYFTNKEY 526
V G V NAL+S+Y++C V A+ + + ++ + + +L Y +
Sbjct: 304 HIVTTGFDISGIVLNALISMYSRCGGVETARRLIEQRGTKDLKIEGFTALLDGYIKLGDM 363
Query: 527 EKGLYMFSQMNRDEVKADEITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKPDETTIYSI 586
+ +F + +V + W+ +I G ++ EA+ +FR M G +P+ T+ ++
Sbjct: 364 NQAKNIFVSLKDRDV----VAWTAMIVGYEQHGSYGEAINLFRSMVGGGQRPNSYTLAAM 419
Query: 587 LRACSLSECLRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPI-KD 645
L S L GK+IH + + + ++ +NAL+ MYAK G ++ + FD++ +D
Sbjct: 420 LSVASSLASLSHGKQIHGSAVKSGEIYSVSVSNALITMYAKAGNITSASRAFDLIRCERD 479
Query: 646 VFSWNTMIFANGMHGNGKEALSLFEKMLLSMVKPDSATFTCVLSACSHSMLVEEGVQIFN 705
SW +MI A HG+ +EAL LFE ML+ ++PD T+ V SAC+H+ LV +G Q F+
Sbjct: 480 TVSWTSMIIALAQHGHAEEALELFETMLMEGLRPDHITYVGVFSACTHAGLVNQGRQYFD 539
Query: 706 SMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEPTAIAWKAFLAGCRVYKNV 765
M + P HY C+VD++ RAG L+EA FI++MP+EP + W + L+ CRV+KN+
Sbjct: 540 MMKDVDKIIPTLSHYACMVDLFGRAGLLQEAQEFIEKMPIEPDVVTWGSLLSACRVHKNI 599
Query: 766 ELAKISAKKLFEIDPNGSANYVTLFNILVTAKLWSEASKIRKLMKERGITKTPGCSWFHV 825
+L K++A++L ++P S Y L N+ W EA+KIRK MK+ + K G SW V
Sbjct: 600 DLGKVAAERLLLLEPENSGAYSALANLYSACGKWEEAAKIRKSMKDGRVKKEQGFSWIEV 659
Query: 826 GNRVHTFVAGDKSNMESDKIYNFLDELFAKIKAAGYKPDTDYVLHDIDQEEKAESLCNHS 885
++VH F D ++ E ++IY + +++ +IK GY PDT VLHD+++E K + L +HS
Sbjct: 660 KHKVHVFGVEDGTHPEKNEIYMTMKKIWDEIKKMGYVPDTASVLHDLEEEVKEQILRHHS 719
Query: 886 EKLAVAFGILNLNGQSTIRVFKNLRICGDCHNAIKYMSNVVGVTIVVRDSLRFHHFKNGN 945
EKLA+AFG+++ ++T+R+ KNLR+C DCH AIK++S +VG I+VRD+ RFHHFK+G
Sbjct: 720 EKLAIAFGLISTPDKTTLRIMKNLRVCNDCHTAIKFISKLVGREIIVRDTTRFHHFKDGF 779
Query: 946 CSCKDFW 952
CSC+D+W
Sbjct: 780 CSCRDYW 786
Score = 201 bits (511), Expect = 2e-51
Identities = 137/527 (25%), Positives = 244/527 (45%), Gaps = 84/527 (15%)
Query: 79 NAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVK 138
N + AY K ++ FD L RD V+W ++ Y N G + + V M ++
Sbjct: 84 NTVLSAYSKRGDMDSTCEFFDQLPQRDSVSWTTMIVGYKNIGQYHKAIRVMGDMVKEGIE 143
Query: 139 ANPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKC--------- 189
T++++L + + +++GK++H F+V+ G+ +V VS++ +N YAKC
Sbjct: 144 PTQFTLTNVLASVAATRCMETGKKVHSFIVKLGLRGNVSVSNSLLNMYAKCGDPMMAKFV 203
Query: 190 ---LCVRE-------------------AQTVFDLMPHRDVVTWNSLSSCYVNCGFPQKGL 227
+ VR+ A F+ M RD+VTWNS+ S + G+ + L
Sbjct: 204 FDRMVVRDISSWNAMIALHMQVGQMDLAMAQFEQMAERDIVTWNSMISGFNQRGYDLRAL 263
Query: 228 NVFREMVLDG-VKPDPVTVSCILSACSDLQDLKSGKAIHGFALKHGMVENVFVSNALVNL 286
++F +M+ D + PD T++ +LSAC++L+ L GK IH + G + V NAL+++
Sbjct: 264 DIFSKMLRDSLLSPDRFTLASVLSACANLEKLCIGKQIHSHIVTTGFDISGIVLNALISM 323
Query: 287 YESCLCV---------------------------------REAQAVFDLMPHRNVITWNS 313
Y C V +A+ +F + R+V+ W +
Sbjct: 324 YSRCGGVETARRLIEQRGTKDLKIEGFTALLDGYIKLGDMNQAKNIFVSLKDRDVVAWTA 383
Query: 314 LASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHG 373
+ Y G + +N+FR M G +P+ ++++L S L L GK IHG AVK G
Sbjct: 384 MIVGYEQHGSYGEAINLFRSMVGGGQRPNSYTLAAMLSVASSLASLSHGKQIHGSAVKSG 443
Query: 374 MVEDVFVCTALVNLYANCLCVREAQTVFDLMP-HRNVVTWNSLSSCYVNCGFPQKGLNVF 432
+ V V AL+ +YA + A FDL+ R+ V+W S+ G ++ L +F
Sbjct: 444 EIYSVSVSNALITMYAKAGNITSASRAFDLIRCERDTVSWTSMIIALAQHGHAEEALELF 503
Query: 433 REMVLNGVKPDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGMVEDV---------FVCN 483
M++ G++PD +T + + AC+ + G+ M++DV + C
Sbjct: 504 ETMLMEGLRPDHITYVGVFSACTHAGLVNQGRQY------FDMMKDVDKIIPTLSHYAC- 556
Query: 484 ALLSLYAKCVCVREAQVVFDLIP-HREVASWNGILTAYFTNKEYEKG 529
++ L+ + ++EAQ + +P +V +W +L+A +K + G
Sbjct: 557 -MVDLFGRAGLLQEAQEFIEKMPIEPDVVTWGSLLSACRVHKNIDLG 602
Score = 170 bits (431), Expect = 3e-42
Identities = 128/480 (26%), Positives = 219/480 (44%), Gaps = 85/480 (17%)
Query: 19 GLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSIG 78
G ++AI++ GI+P + V + AA+R K+ H + G+ +VS+
Sbjct: 125 GQYHKAIRVMGDMVKEGIEPTQFTLTNVLASVAATRCMETGKKVHSFIVKLGLRGNVSVS 184
Query: 79 NAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVN-------------------- 118
N+ ++ Y KC A+ VFD +V RD+ +WN++ A ++
Sbjct: 185 NSLLNMYAKCGDPMMAKFVFDRMVVRDISSWNAMIALHMQVGQMDLAMAQFEQMAERDIV 244
Query: 119 ------CGFPQQG-----LNVFRKMGLNK-VKANPLTVSSILPGCSDLQDLKSGKEIHGF 166
GF Q+G L++F KM + + + T++S+L C++L+ L GK+IH
Sbjct: 245 TWNSMISGFNQRGYDLRALDIFSKMLRDSLLSPDRFTLASVLSACANLEKLCIGKQIHSH 304
Query: 167 VVRHGMVEDVFVSSAFVNFYAKCLCV---------------------------------R 193
+V G V +A ++ Y++C V
Sbjct: 305 IVTTGFDISGIVLNALISMYSRCGGVETARRLIEQRGTKDLKIEGFTALLDGYIKLGDMN 364
Query: 194 EAQTVFDLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACS 253
+A+ +F + RDVV W ++ Y G + +N+FR MV G +P+ T++ +LS S
Sbjct: 365 QAKNIFVSLKDRDVVAWTAMIVGYEQHGSYGEAINLFRSMVGGGQRPNSYTLAAMLSVAS 424
Query: 254 DLQDLKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMP-HRNVITWN 312
L L GK IHG A+K G + +V VSNAL+ +Y + A FDL+ R+ ++W
Sbjct: 425 SLASLSHGKQIHGSAVKSGEIYSVSVSNALITMYAKAGNITSASRAFDLIRCERDTVSWT 484
Query: 313 SLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKH 372
S+ G ++ L +F M + G++PD + + AC+ + G+
Sbjct: 485 SMIIALAQHGHAEEALELFETMLMEGLRPDHITYVGVFSACTHAGLVNQGRQY------F 538
Query: 373 GMVEDV---------FVCTALVNLYANCLCVREAQTVFDLMP-HRNVVTWNS-LSSCYVN 421
M++DV + C +V+L+ ++EAQ + MP +VVTW S LS+C V+
Sbjct: 539 DMMKDVDKIIPTLSHYAC--MVDLFGRAGLLQEAQEFIEKMPIEPDVVTWGSLLSACRVH 596
Score = 77.8 bits (190), Expect = 3e-14
Identities = 53/206 (25%), Positives = 93/206 (44%), Gaps = 4/206 (1%)
Query: 18 HGLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSI 77
HG EAI ++ S G +P+ A+ ++ KQ H A + G + VS+
Sbjct: 391 HGSYGEAINLFRSMVGGGQRPNSYTLAAMLSVASSLASLSHGKQIHGSAVKSGEIYSVSV 450
Query: 78 GNAFIHAYGKCKCVEGARRVFDDL-VARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNK 136
NA I Y K + A R FD + RD V+W S+ G ++ L +F M +
Sbjct: 451 SNALITMYAKAGNITSASRAFDLIRCERDTVSWTSMIIALAQHGHAEEALELFETMLMEG 510
Query: 137 VKANPLTVSSILPGCSDLQDLKSGKEIHGFVVR-HGMVEDVFVSSAFVNFYAKCLCVREA 195
++ + +T + C+ + G++ + ++ + + V+ + + ++EA
Sbjct: 511 LRPDHITYVGVFSACTHAGLVNQGRQYFDMMKDVDKIIPTLSHYACMVDLFGRAGLLQEA 570
Query: 196 QTVFDLMP-HRDVVTWNS-LSSCYVN 219
Q + MP DVVTW S LS+C V+
Sbjct: 571 QEFIEKMPIEPDVVTWGSLLSACRVH 596
>At3g02010 hypothetical protein
Length = 825
Score = 449 bits (1154), Expect = e-126
Identities = 255/779 (32%), Positives = 408/779 (51%), Gaps = 40/779 (5%)
Query: 179 SSAFVNFYAKCLCVREAQTVFDLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDG- 237
++ ++ + K V A+ +FD MP R VVTW L Y + +FR+M
Sbjct: 82 TNTMISGHVKTGDVSSARDLFDAMPDRTVVTWTILMGWYARNSHFDEAFKLFRQMCRSSS 141
Query: 238 -VKPDPVTVSCILSACSDLQDLKSGKAIHGFALKHGMVENVF--VSNALVNLYESCLCVR 294
PD VT + +L C+D + +H FA+K G N F VSN L+ Y +
Sbjct: 142 CTLPDHVTFTTLLPGCNDAVPQNAVGQVHAFAVKLGFDTNPFLTVSNVLLKSYCEVRRLD 201
Query: 295 EAQAVFDLMPHRNVITWNSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACS 354
A +F+ +P ++ +T+N+L + Y G + +++F +M +G +P S +L A
Sbjct: 202 LACVLFEEIPEKDSVTFNTLITGYEKDGLYTESIHLFLKMRQSGHQPSDFTFSGVLKAVV 261
Query: 355 QLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPHRNVVTWNS 414
L D G+ +H +V G D V +++ Y+ V E + +FD MP + V++N
Sbjct: 262 GLHDFALGQQLHALSVTTGFSRDASVGNQILDFYSKHDRVLETRMLFDEMPELDFVSYNV 321
Query: 415 LSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHACSDLQDLKSGKVIHGFAVRHG 474
+ S Y + L+ FREM G ++L ++L L+ G+ +H A+
Sbjct: 322 VISSYSQADQYEASLHFFREMQCMGFDRRNFPFATMLSIAANLSSLQMGRQLHCQALLAT 381
Query: 475 MVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMFS 534
+ V N+L+ +YAKC EA+++F +P R SW +
Sbjct: 382 ADSILHVGNSLVDMYAKCEMFEEAELIFKSLPQRTTVSWTAL------------------ 423
Query: 535 QMNRDEVKADEITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKPDETTIYSILRACSLSE 594
I G V+ +++F KM+ + D++T ++L+A +
Sbjct: 424 -----------------ISGYVQKGLHGAGLKLFTKMRGSNLRADQSTFATVLKASASFA 466
Query: 595 CLRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMIF 654
L +GK++H ++ R ++ + LVDMYAKCG + + VF+ MP ++ SWN +I
Sbjct: 467 SLLLGKQLHAFIIRSGNLENVFSGSGLVDMYAKCGSIKDAVQVFEEMPDRNAVSWNALIS 526
Query: 655 ANGMHGNGKEALSLFEKMLLSMVKPDSATFTCVLSACSHSMLVEEGVQIFNSMSRDHLVE 714
A+ +G+G+ A+ F KM+ S ++PDS + VL+ACSH VE+G + F +MS + +
Sbjct: 527 AHADNGDGEAAIGAFAKMIESGLQPDSVSILGVLTACSHCGFVEQGTEYFQAMSPIYGIT 586
Query: 715 PEAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEPTAIAWKAFLAGCRVYKNVELAKISAKK 774
P+ +HY C++D+ R G EA + MP EP I W + L CR++KN LA+ +A+K
Sbjct: 587 PKKKHYACMLDLLGRNGRFAEAEKLMDEMPFEPDEIMWSSVLNACRIHKNQSLAERAAEK 646
Query: 775 LFEIDP-NGSANYVTLFNILVTAKLWSEASKIRKLMKERGITKTPGCSWFHVGNRVHTFV 833
LF ++ +A YV++ NI A W + ++K M+ERGI K P SW V +++H F
Sbjct: 647 LFSMEKLRDAAAYVSMSNIYAAAGEWEKVRDVKKAMRERGIKKVPAYSWVEVNHKIHVFS 706
Query: 834 AGDKSNMESDKIYNFLDELFAKIKAAGYKPDTDYVLHDIDQEEKAESLCNHSEKLAVAFG 893
+ D+++ D+I ++EL A+I+ GYKPDT V+ D+D++ K ESL HSE+LAVAF
Sbjct: 707 SNDQTHPNGDEIVRKINELTAEIEREGYKPDTSSVVQDVDEQMKIESLKYHSERLAVAFA 766
Query: 894 ILNLNGQSTIRVFKNLRICGDCHNAIKYMSNVVGVTIVVRDSLRFHHFKNGNCSCKDFW 952
+++ I V KNLR C DCH AIK +S +V I VRD+ RFHHF G CSC D+W
Sbjct: 767 LISTPEGCPIVVMKNLRACRDCHAAIKLISKIVKREITVRDTSRFHHFSEGVCSCGDYW 825
Score = 237 bits (605), Expect = 2e-62
Identities = 170/658 (25%), Positives = 300/658 (44%), Gaps = 53/658 (8%)
Query: 79 NAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVK 138
N I + K V AR +FD + R VVTW L Y + +FR+M +
Sbjct: 83 NTMISGHVKTGDVSSARDLFDAMPDRTVVTWTILMGWYARNSHFDEAFKLFRQMCRSSSC 142
Query: 139 ANP--LTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVF--VSSAFVNFYAKCLCVRE 194
P +T +++LPGC+D + ++H F V+ G + F VS+ + Y + +
Sbjct: 143 TLPDHVTFTTLLPGCNDAVPQNAVGQVHAFAVKLGFDTNPFLTVSNVLLKSYCEVRRLDL 202
Query: 195 AQTVFDLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSD 254
A +F+ +P +D VT+N+L + Y G + +++F +M G +P T S +L A
Sbjct: 203 ACVLFEEIPEKDSVTFNTLITGYEKDGLYTESIHLFLKMRQSGHQPSDFTFSGVLKAVVG 262
Query: 255 LQDLKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSL 314
L D G+ +H ++ G + V N +++ Y V E + +FD MP + +++N +
Sbjct: 263 LHDFALGQQLHALSVTTGFSRDASVGNQILDFYSKHDRVLETRMLFDEMPELDFVSYNVV 322
Query: 315 ASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGM 374
S Y + L+ FREM G +++L + L L+ G+ +H A+
Sbjct: 323 ISSYSQADQYEASLHFFREMQCMGFDRRNFPFATMLSIAANLSSLQMGRQLHCQALLATA 382
Query: 375 VEDVFVCTALVNLYANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFRE 434
+ V +LV++YA C EA+ +F +P R V+W +L S YV G GL +F +
Sbjct: 383 DSILHVGNSLVDMYAKCEMFEEAELIFKSLPQRTTVSWTALISGYVQKGLHGAGLKLFTK 442
Query: 435 MVLNGVKPDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCVC 494
M + ++ D T ++L A + L GK +H F +R G +E+VF + L+ +YAKC
Sbjct: 443 MRGSNLRADQSTFATVLKASASFASLLLGKQLHAFIIRSGNLENVFSGSGLVDMYAKCGS 502
Query: 495 VREAQVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGG 554
+++A VF+ +P R SWN +++A+ N + E + F++M ++ D ++ V+
Sbjct: 503 IKDAVQVFEEMPDRNAVSWNALISAHADNGDGEAAIGAFAKMIESGLQPDSVSILGVLTA 562
Query: 555 CVKNSRIEEAMEIF---------------------------------RKMQTMGFKPDET 581
C +E+ E F + M M F+PDE
Sbjct: 563 CSHCGFVEQGTEYFQAMSPIYGITPKKKHYACMLDLLGRNGRFAEAEKLMDEMPFEPDEI 622
Query: 582 TIYSILRACSLSECLRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMM 641
S+L AC + + + + +F K D A ++ ++YA G R+V M
Sbjct: 623 MWSSVLNACRIHKNQSLAERAAEKLFSMEKLRDAAAYVSMSNIYAAAGEWEKVRDVKKAM 682
Query: 642 PIKDV-----FSW-----NTMIFANG--MHGNGKEALSLFEKMLLSM----VKPDSAT 683
+ + +SW +F++ H NG E + ++ + KPD+++
Sbjct: 683 RERGIKKVPAYSWVEVNHKIHVFSSNDQTHPNGDEIVRKINELTAEIEREGYKPDTSS 740
Score = 219 bits (558), Expect = 6e-57
Identities = 135/486 (27%), Positives = 244/486 (49%), Gaps = 4/486 (0%)
Query: 38 PDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSD--VSIGNAFIHAYGKCKCVEGAR 95
PD F + C + V Q H A + G ++ +++ N + +Y + + ++ A
Sbjct: 145 PDHVTFTTLLPGCNDAVPQNAVGQVHAFAVKLGFDTNPFLTVSNVLLKSYCEVRRLDLAC 204
Query: 96 RVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKANPLTVSSILPGCSDLQ 155
+F+++ +D VT+N+L Y G + +++F KM + + + T S +L L
Sbjct: 205 VLFEEIPEKDSVTFNTLITGYEKDGLYTESIHLFLKMRQSGHQPSDFTFSGVLKAVVGLH 264
Query: 156 DLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTWNSLSS 215
D G+++H V G D V + ++FY+K V E + +FD MP D V++N + S
Sbjct: 265 DFALGQQLHALSVTTGFSRDASVGNQILDFYSKHDRVLETRMLFDEMPELDFVSYNVVIS 324
Query: 216 CYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQDLKSGKAIHGFALKHGMVE 275
Y + L+ FREM G + +LS ++L L+ G+ +H AL
Sbjct: 325 SYSQADQYEASLHFFREMQCMGFDRRNFPFATMLSIAANLSSLQMGRQLHCQALLATADS 384
Query: 276 NVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASCYVNCGFPQKGLNVFREMG 335
+ V N+LV++Y C EA+ +F +P R ++W +L S YV G GL +F +M
Sbjct: 385 ILHVGNSLVDMYAKCEMFEEAELIFKSLPQRTTVSWTALISGYVQKGLHGAGLKLFTKMR 444
Query: 336 LNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANCLCVR 395
+ ++ D +++L A + L GK +H F ++ G +E+VF + LV++YA C ++
Sbjct: 445 GSNLRADQSTFATVLKASASFASLLLGKQLHAFIIRSGNLENVFSGSGLVDMYAKCGSIK 504
Query: 396 EAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHACS 455
+A VF+ MP RN V+WN+L S + + G + + F +M+ +G++PD V++L +L ACS
Sbjct: 505 DAVQVFEEMPDRNAVSWNALISAHADNGDGEAAIGAFAKMIESGLQPDSVSILGVLTACS 564
Query: 456 DLQDLKSG-KVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHR-EVASW 513
++ G + + +G+ +L L + EA+ + D +P + W
Sbjct: 565 HCGFVEQGTEYFQAMSPIYGITPKKKHYACMLDLLGRNGRFAEAEKLMDEMPFEPDEIMW 624
Query: 514 NGILTA 519
+ +L A
Sbjct: 625 SSVLNA 630
Score = 181 bits (458), Expect = 2e-45
Identities = 111/406 (27%), Positives = 199/406 (48%), Gaps = 3/406 (0%)
Query: 19 GLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSIG 78
GL E+I ++ R G +P F V KA D +Q H + G D S+G
Sbjct: 229 GLYTESIHLFLKMRQSGHQPSDFTFSGVLKAVVGLHDFALGQQLHALSVTTGFSRDASVG 288
Query: 79 NAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVK 138
N + Y K V R +FD++ D V++N + + Y + L+ FR+M
Sbjct: 289 NQILDFYSKHDRVLETRMLFDEMPELDFVSYNVVISSYSQADQYEASLHFFREMQCMGFD 348
Query: 139 ANPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTV 198
+++L ++L L+ G+++H + + V ++ V+ YAKC EA+ +
Sbjct: 349 RRNFPFATMLSIAANLSSLQMGRQLHCQALLATADSILHVGNSLVDMYAKCEMFEEAELI 408
Query: 199 FDLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQDL 258
F +P R V+W +L S YV G GL +F +M ++ D T + +L A + L
Sbjct: 409 FKSLPQRTTVSWTALISGYVQKGLHGAGLKLFTKMRGSNLRADQSTFATVLKASASFASL 468
Query: 259 KSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASCY 318
GK +H F ++ G +ENVF + LV++Y C +++A VF+ MP RN ++WN+L S +
Sbjct: 469 LLGKQLHAFIIRSGNLENVFSGSGLVDMYAKCGSIKDAVQVFEEMPDRNAVSWNALISAH 528
Query: 319 VNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSG-KTIHGFAVKHGMVED 377
+ G + + F +M +G++PD +++ +L ACS ++ G + + +G+
Sbjct: 529 ADNGDGEAAIGAFAKMIESGLQPDSVSILGVLTACSHCGFVEQGTEYFQAMSPIYGITPK 588
Query: 378 VFVCTALVNLYANCLCVREAQTVFDLMPHR-NVVTWNS-LSSCYVN 421
+++L EA+ + D MP + + W+S L++C ++
Sbjct: 589 KKHYACMLDLLGRNGRFAEAEKLMDEMPFEPDEIMWSSVLNACRIH 634
Score = 179 bits (454), Expect = 7e-45
Identities = 131/490 (26%), Positives = 225/490 (45%), Gaps = 47/490 (9%)
Query: 275 ENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASCYVNCGFPQKGLNVFREM 334
+N +N +++ + V A+ +FD MP R V+TW L Y + +FR+M
Sbjct: 77 KNTVSTNTMISGHVKTGDVSSARDLFDAMPDRTVVTWTILMGWYARNSHFDEAFKLFRQM 136
Query: 335 --GLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANCL 392
+ PD + +++LP C+ + +H FAVK G + F+ + V L + C
Sbjct: 137 CRSSSCTLPDHVTFTTLLPGCNDAVPQNAVGQVHAFAVKLGFDTNPFLTVSNVLLKSYCE 196
Query: 393 CVR--EAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSI 450
R A +F+ +P ++ VT+N+L + Y G + +++F +M +G +P T +
Sbjct: 197 VRRLDLACVLFEEIPEKDSVTFNTLITGYEKDGLYTESIHLFLKMRQSGHQPSDFTFSGV 256
Query: 451 LHACSDLQDLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREV 510
L A L D G+ +H +V G D V N +L Y+K V E +++FD +P +
Sbjct: 257 LKAVVGLHDFALGQQLHALSVTTGFSRDASVGNQILDFYSKHDRVLETRMLFDEMPELDF 316
Query: 511 ASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGGCVKNSRIEEAMEIFRK 570
S+N ++++Y +YE L+ FR+
Sbjct: 317 VSYNVVISSYSQADQYEASLH-----------------------------------FFRE 341
Query: 571 MQTMGFKPDETTIYSILRACSLSECLRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGG 630
MQ MGF ++L + L+MG+++HC D L N+LVDMYAKC
Sbjct: 342 MQCMGFDRRNFPFATMLSIAANLSSLQMGRQLHCQALLATADSILHVGNSLVDMYAKCEM 401
Query: 631 LSLSRNVFDMMPIKDVFSWNTMI---FANGMHGNGKEALSLFEKMLLSMVKPDSATFTCV 687
+ +F +P + SW +I G+HG G L LF KM S ++ D +TF V
Sbjct: 402 FEEAELIFKSLPQRTTVSWTALISGYVQKGLHGAG---LKLFTKMRGSNLRADQSTFATV 458
Query: 688 LSACSHSMLVEEGVQIFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEP 747
L A + + G Q+ + R +E + +VD+Y++ G +++A + MP +
Sbjct: 459 LKASASFASLLLGKQLHAFIIRSGNLE-NVFSGSGLVDMYAKCGSIKDAVQVFEEMP-DR 516
Query: 748 TAIAWKAFLA 757
A++W A ++
Sbjct: 517 NAVSWNALIS 526
Score = 80.9 bits (198), Expect = 3e-15
Identities = 50/207 (24%), Positives = 100/207 (48%), Gaps = 3/207 (1%)
Query: 16 IPHGLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDV 75
+ GL +K++T R ++ D+ F V KA A+ L KQ H R G + +V
Sbjct: 428 VQKGLHGAGLKLFTKMRGSNLRADQSTFATVLKASASFASLLLGKQLHAFIIRSGNLENV 487
Query: 76 SIGNAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLN 135
G+ + Y KC ++ A +VF+++ R+ V+WN+L + + + G + + F KM +
Sbjct: 488 FSGSGLVDMYAKCGSIKDAVQVFEEMPDRNAVSWNALISAHADNGDGEAAIGAFAKMIES 547
Query: 136 KVKANPLTVSSILPGCSDLQDLKSGKE-IHGFVVRHGMVEDVFVSSAFVNFYAKCLCVRE 194
++ + +++ +L CS ++ G E +G+ + ++ + E
Sbjct: 548 GLQPDSVSILGVLTACSHCGFVEQGTEYFQAMSPIYGITPKKKHYACMLDLLGRNGRFAE 607
Query: 195 AQTVFDLMPHR-DVVTWNS-LSSCYVN 219
A+ + D MP D + W+S L++C ++
Sbjct: 608 AEKLMDEMPFEPDEIMWSSVLNACRIH 634
Score = 79.0 bits (193), Expect = 1e-14
Identities = 70/283 (24%), Positives = 124/283 (43%), Gaps = 38/283 (13%)
Query: 495 VREAQVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGG 554
V A+ V+D +PH+ S N +++ + + +F M V +TW++++G
Sbjct: 64 VSAARKVYDEMPHKNTVSTNTMISGHVKTGDVSSARDLFDAMPDRTV----VTWTILMGW 119
Query: 555 CVKNSRIEEAMEIFRKM--QTMGFKPDETTIYSILRACSLSECLRMGKEIHCYVFRHWKD 612
+NS +EA ++FR+M + PD T ++L C+ + ++H + + D
Sbjct: 120 YARNSHFDEAFKLFRQMCRSSSCTLPDHVTFTTLLPGCNDAVPQNAVGQVHAFAVKLGFD 179
Query: 613 WD--LARTNALVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKEALSLFE 670
+ L +N L+ Y + L L+ +F+ +P KD ++NT+I G E++ LF
Sbjct: 180 TNPFLTVSNVLLKSYCEVRRLDLACVLFEEIPEKDSVTFNTLITGYEKDGLYTESIHLFL 239
Query: 671 KMLLSMVKPDSATFTCVLSAC-----------SHSMLVEEGV--------QIFNSMSR-- 709
KM S +P TF+ VL A H++ V G QI + S+
Sbjct: 240 KMRQSGHQPSDFTFSGVLKAVVGLHDFALGQQLHALSVTTGFSRDASVGNQILDFYSKHD 299
Query: 710 ---------DHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRM 743
D + E + Y V+ YS+A E + F + M
Sbjct: 300 RVLETRMLFDEMPELDFVSYNVVISSYSQADQYEASLHFFREM 342
Score = 46.2 bits (108), Expect = 9e-05
Identities = 31/100 (31%), Positives = 50/100 (50%), Gaps = 8/100 (8%)
Query: 612 DWDLARTNALVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKEALSLFEK 671
D D R+N +V+ + G +S +R V+D MP K+ S NTMI + G+ A LF+
Sbjct: 45 DTDTCRSNFIVEDLLRRGQVSAARKVYDEMPHKNTVSTNTMISGHVKTGDVSSARDLFDA 104
Query: 672 MLLSMVKPDS--ATFTCVLSACSHSMLVEEGVQIFNSMSR 709
M PD T+T ++ + + +E ++F M R
Sbjct: 105 M------PDRTVVTWTILMGWYARNSHFDEAFKLFRQMCR 138
>At1g25360
Length = 790
Score = 449 bits (1154), Expect = e-126
Identities = 258/795 (32%), Positives = 421/795 (52%), Gaps = 85/795 (10%)
Query: 238 VKPDPVTVSCI-------LSACSDLQ--DLKSGKAIHGFALKHGMVENVFVSNALVNLY- 287
++P+P V I L C L+ L+ +A+HG + G + N L+++Y
Sbjct: 1 MQPNPDLVRAIANRYAANLRLCLPLRRTSLQLARAVHGNIITFGFQPRAHILNRLIDVYC 60
Query: 288 --------------------------ESCLC----VREAQAVFDLMP--HRNVITWNSLA 315
S C + A+ VF+ P R+ + +N++
Sbjct: 61 KSSELNYARQLFDEISEPDKIARTTMVSGYCASGDITLARGVFEKAPVCMRDTVMYNAMI 120
Query: 316 SCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKD-LKSGKTIHGFAVKHGM 374
+ + + +N+F +M G KPD +S+L + + D K H A+K G
Sbjct: 121 TGFSHNNDGYSAINLFCKMKHEGFKPDNFTFASVLAGLALVADDEKQCVQFHAAALKSGA 180
Query: 375 VEDVFVCTALVNLYANCLC----VREAQTVFDLMPHRNVVTW------------------ 412
V ALV++Y+ C + A+ VFD + ++ +W
Sbjct: 181 GYITSVSNALVSVYSKCASSPSLLHSARKVFDEILEKDERSWTTMMTGYVKNGYFDLGEE 240
Query: 413 --------------NSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHACSDLQ 458
N++ S YVN GF Q+ L + R MV +G++ D T S++ AC+
Sbjct: 241 LLEGMDDNMKLVAYNAMISGYVNRGFYQEALEMVRRMVSSGIELDEFTYPSVIRACATAG 300
Query: 459 DLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREVASWNGILT 518
L+ GK +H + +R F N+L+SLY KC EA+ +F+ +P +++ SWN +L+
Sbjct: 301 LLQLGKQVHAYVLRREDFSFHFD-NSLVSLYYKCGKFDEARAIFEKMPAKDLVSWNALLS 359
Query: 519 AYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKP 578
Y ++ + +F +M + ++W ++I G +N EE +++F M+ GF+P
Sbjct: 360 GYVSSGHIGEAKLIFKEMKEKNI----LSWMIMISGLAENGFGEEGLKLFSCMKREGFEP 415
Query: 579 DETTIYSILRACSLSECLRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVF 638
+ +++C++ G++ H + + D L+ NAL+ MYAKCG + +R VF
Sbjct: 416 CDYAFSGAIKSCAVLGAYCNGQQYHAQLLKIGFDSSLSAGNALITMYAKCGVVEEARQVF 475
Query: 639 DMMPIKDVFSWNTMIFANGMHGNGKEALSLFEKMLLSMVKPDSATFTCVLSACSHSMLVE 698
MP D SWN +I A G HG+G EA+ ++E+ML ++PD T VL+ACSH+ LV+
Sbjct: 476 RTMPCLDSVSWNALIAALGQHGHGAEAVDVYEEMLKKGIRPDRITLLTVLTACSHAGLVD 535
Query: 699 EGVQIFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEPTAIAWKAFLAG 758
+G + F+SM + + P A+HY ++D+ R+G +A I+ +P +PTA W+A L+G
Sbjct: 536 QGRKYFDSMETVYRIPPGADHYARLIDLLCRSGKFSDAESVIESLPFKPTAEIWEALLSG 595
Query: 759 CRVYKNVELAKISAKKLFEIDPNGSANYVTLFNILVTAKLWSEASKIRKLMKERGITKTP 818
CRV+ N+EL I+A KLF + P Y+ L N+ W E +++RKLM++RG+ K
Sbjct: 596 CRVHGNMELGIIAADKLFGLIPEHDGTYMLLSNMHAATGQWEEVARVRKLMRDRGVKKEV 655
Query: 819 GCSWFHVGNRVHTFVAGDKSNMESDKIYNFLDELFAKIKAAGYKPDTDYVLHDIDQE-EK 877
CSW + +VHTF+ D S+ E++ +Y +L +L +++ GY PDT +VLHD++ + K
Sbjct: 656 ACSWIEMETQVHTFLVDDTSHPEAEAVYIYLQDLGKEMRRLGYVPDTSFVLHDVESDGHK 715
Query: 878 AESLCNHSEKLAVAFGILNLNGQSTIRVFKNLRICGDCHNAIKYMSNVVGVTIVVRDSLR 937
+ L HSEK+AVAFG++ L +TIR+FKNLR CGDCHN +++S VV I++RD R
Sbjct: 716 EDMLTTHSEKIAVAFGLMKLPPGTTIRIFKNLRTCGDCHNFFRFLSWVVQRDIILRDRKR 775
Query: 938 FHHFKNGNCSCKDFW 952
FHHF+NG CSC +FW
Sbjct: 776 FHHFRNGECSCGNFW 790
Score = 167 bits (423), Expect = 3e-41
Identities = 132/509 (25%), Positives = 225/509 (43%), Gaps = 73/509 (14%)
Query: 94 ARRVFDD--LVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKANPLTVSSILPGC 151
AR VF+ + RD V +N++ + + +N+F KM K + T +S+L G
Sbjct: 99 ARGVFEKAPVCMRDTVMYNAMITGFSHNNDGYSAINLFCKMKHEGFKPDNFTFASVLAGL 158
Query: 152 SDL-QDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLC----VREAQTVFDLMPHRD 206
+ + D K + H ++ G VS+A V+ Y+KC + A+ VFD + +D
Sbjct: 159 ALVADDEKQCVQFHAAALKSGAGYITSVSNALVSVYSKCASSPSLLHSARKVFDEILEKD 218
Query: 207 VVTW--------------------------------NSLSSCYVNCGFPQKGLNVFREMV 234
+W N++ S YVN GF Q+ L + R MV
Sbjct: 219 ERSWTTMMTGYVKNGYFDLGEELLEGMDDNMKLVAYNAMISGYVNRGFYQEALEMVRRMV 278
Query: 235 LDGVKPDPVTVSCILSACSDLQDLKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVR 294
G++ D T ++ AC+ L+ GK +H + L+ F N+LV+LY C
Sbjct: 279 SSGIELDEFTYPSVIRACATAGLLQLGKQVHAYVLRREDFSFHF-DNSLVSLYYKCGKFD 337
Query: 295 EAQAVFDLMPHRNVITWNSLASCYVNC-------------------------------GF 323
EA+A+F+ MP +++++WN+L S YV+ GF
Sbjct: 338 EARAIFEKMPAKDLVSWNALLSGYVSSGHIGEAKLIFKEMKEKNILSWMIMISGLAENGF 397
Query: 324 PQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTA 383
++GL +F M G +P A S + +C+ L +G+ H +K G + A
Sbjct: 398 GEEGLKLFSCMKREGFEPCDYAFSGAIKSCAVLGAYCNGQQYHAQLLKIGFDSSLSAGNA 457
Query: 384 LVNLYANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPD 443
L+ +YA C V EA+ VF MP + V+WN+L + G + ++V+ EM+ G++PD
Sbjct: 458 LITMYAKCGVVEEARQVFRTMPCLDSVSWNALIAALGQHGHGAEAVDVYEEMLKKGIRPD 517
Query: 444 LVTMLSILHACSDLQDLKSG-KVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVF 502
+T+L++L ACS + G K + + L+ L + +A+ V
Sbjct: 518 RITLLTVLTACSHAGLVDQGRKYFDSMETVYRIPPGADHYARLIDLLCRSGKFSDAESVI 577
Query: 503 DLIPHREVAS-WNGILTAYFTNKEYEKGL 530
+ +P + A W +L+ + E G+
Sbjct: 578 ESLPFKPTAEIWEALLSGCRVHGNMELGI 606
Score = 161 bits (407), Expect = 2e-39
Identities = 128/506 (25%), Positives = 221/506 (43%), Gaps = 78/506 (15%)
Query: 24 AIKIYTSSRARGIKPDKPVFMAVAKACA-ASRDALKVKQFHDDATRCGVMSDVSIGNAFI 82
AI ++ + G KPD F +V A + D + QFH A + G S+ NA +
Sbjct: 132 AINLFCKMKHEGFKPDNFTFASVLAGLALVADDEKQCVQFHAAALKSGAGYITSVSNALV 191
Query: 83 HAYGKC----KCVEGARRVFDDLVARDVVTW----------------------------- 109
Y KC + AR+VFD+++ +D +W
Sbjct: 192 SVYSKCASSPSLLHSARKVFDEILEKDERSWTTMMTGYVKNGYFDLGEELLEGMDDNMKL 251
Query: 110 ---NSLSACYVNCGFPQQGLNVFRKMGLNKVKANPLTVSSILPGCSDLQDLKSGKEIHGF 166
N++ + YVN GF Q+ L + R+M + ++ + T S++ C+ L+ GK++H +
Sbjct: 252 VAYNAMISGYVNRGFYQEALEMVRRMVSSGIELDEFTYPSVIRACATAGLLQLGKQVHAY 311
Query: 167 VVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTWNSLSSCYVNCG----- 221
V+R F +S V+ Y KC EA+ +F+ MP +D+V+WN+L S YV+ G
Sbjct: 312 VLRREDFSFHFDNS-LVSLYYKCGKFDEARAIFEKMPAKDLVSWNALLSGYVSSGHIGEA 370
Query: 222 --------------------------FPQKGLNVFREMVLDGVKPDPVTVSCILSACSDL 255
F ++GL +F M +G +P S + +C+ L
Sbjct: 371 KLIFKEMKEKNILSWMIMISGLAENGFGEEGLKLFSCMKREGFEPCDYAFSGAIKSCAVL 430
Query: 256 QDLKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLA 315
+G+ H LK G ++ NAL+ +Y C V EA+ VF MP + ++WN+L
Sbjct: 431 GAYCNGQQYHAQLLKIGFDSSLSAGNALITMYAKCGVVEEARQVFRTMPCLDSVSWNALI 490
Query: 316 SCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSG-KTIHGFAVKHGM 374
+ G + ++V+ EM G++PD + + ++L ACS + G K + +
Sbjct: 491 AALGQHGHGAEAVDVYEEMLKKGIRPDRITLLTVLTACSHAGLVDQGRKYFDSMETVYRI 550
Query: 375 VEDVFVCTALVNLYANCLCVREAQTVFDLMPHR-NVVTWNS-LSSCYVNCGFPQKGLNVF 432
L++L +A++V + +P + W + LS C V+ L +
Sbjct: 551 PPGADHYARLIDLLCRSGKFSDAESVIESLPFKPTAEIWEALLSGCRVHGNME---LGII 607
Query: 433 REMVLNGVKPD---LVTMLSILHACS 455
L G+ P+ +LS +HA +
Sbjct: 608 AADKLFGLIPEHDGTYMLLSNMHAAT 633
Score = 154 bits (388), Expect = 3e-37
Identities = 147/652 (22%), Positives = 268/652 (40%), Gaps = 118/652 (18%)
Query: 175 DVFVSSAFVNFYAKCLCVREAQTVFDLMP--HRDVVTWNSLSSCYVNCGFPQKGLNVFRE 232
D + V+ Y + A+ VF+ P RD V +N++ + + + +N+F +
Sbjct: 79 DKIARTTMVSGYCASGDITLARGVFEKAPVCMRDTVMYNAMITGFSHNNDGYSAINLFCK 138
Query: 233 MVLDGVKPDPVTVSCILSACSDL-QDLKSGKAIHGFALKHGMVENVFVSNALVNLYESCL 291
M +G KPD T + +L+ + + D K H ALK G VSNALV++Y C
Sbjct: 139 MKHEGFKPDNFTFASVLAGLALVADDEKQCVQFHAAALKSGAGYITSVSNALVSVYSKCA 198
Query: 292 ----CVREAQAVFDLMPHRNVITW--------------------------------NSLA 315
+ A+ VFD + ++ +W N++
Sbjct: 199 SSPSLLHSARKVFDEILEKDERSWTTMMTGYVKNGYFDLGEELLEGMDDNMKLVAYNAMI 258
Query: 316 SCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMV 375
S YVN GF Q+ L + R M +G++ D S++ AC+ L+ GK +H + ++
Sbjct: 259 SGYVNRGFYQEALEMVRRMVSSGIELDEFTYPSVIRACATAGLLQLGKQVHAYVLRREDF 318
Query: 376 EDVFVCTALVNLYANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNC------------- 422
F +LV+LY C EA+ +F+ MP +++V+WN+L S YV+
Sbjct: 319 SFHF-DNSLVSLYYKCGKFDEARAIFEKMPAKDLVSWNALLSGYVSSGHIGEAKLIFKEM 377
Query: 423 ------------------GFPQKGLNVFREMVLNGVKPDLVTMLSILHACSDLQDLKSGK 464
GF ++GL +F M G +P + +C+ L +G+
Sbjct: 378 KEKNILSWMIMISGLAENGFGEEGLKLFSCMKREGFEPCDYAFSGAIKSCAVLGAYCNGQ 437
Query: 465 VIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREVASWNGILTAYFTNK 524
H ++ G + NAL+++YAKC V EA+ VF +P + SWN ++ A +
Sbjct: 438 QYHAQLLKIGFDSSLSAGNALITMYAKCGVVEEARQVFRTMPCLDSVSWNALIAALGQHG 497
Query: 525 EYEKGLYMFSQMNRDEVKADEITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKPDETTIY 584
+ + ++ +M + ++ D IT V+ C +++ + F M+T+ P Y
Sbjct: 498 HGAEAVDVYEEMLKKGIRPDRITLLTVLTACSHAGLVDQGRKYFDSMETVYRIPPGADHY 557
Query: 585 SILRACSLSECLRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIK 644
+ L+D+ + G S + +V + +P K
Sbjct: 558 A----------------------------------RLIDLLCRSGKFSDAESVIESLPFK 583
Query: 645 DVFS-WNTMIFANGMHGNGKEALSLFEKMLLSMVKPDSATFTCVLSACSHSMLVEEGVQI 703
W ++ +HGN + + +K L ++ T+ + + + + EE ++
Sbjct: 584 PTAEIWEALLSGCRVHGNMELGIIAADK-LFGLIPEHDGTYMLLSNMHAATGQWEEVARV 642
Query: 704 FNSMSRDH---------LVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPME 746
+ RD +E E + +T +VD S E Y ++Q + E
Sbjct: 643 -RKLMRDRGVKKEVACSWIEMETQVHTFLVDDTSHPEA-EAVYIYLQDLGKE 692
Score = 107 bits (266), Expect = 4e-23
Identities = 71/269 (26%), Positives = 124/269 (45%), Gaps = 32/269 (11%)
Query: 16 IPHGLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDV 75
+ G EA+++ + GI+ D+ + +V +ACA + KQ H R S
Sbjct: 262 VNRGFYQEALEMVRRMVSSGIELDEFTYPSVIRACATAGLLQLGKQVHAYVLRREDFS-F 320
Query: 76 SIGNAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCG--------------- 120
N+ + Y KC + AR +F+ + A+D+V+WN+L + YV+ G
Sbjct: 321 HFDNSLVSLYYKCGKFDEARAIFEKMPAKDLVSWNALLSGYVSSGHIGEAKLIFKEMKEK 380
Query: 121 ----------------FPQQGLNVFRKMGLNKVKANPLTVSSILPGCSDLQDLKSGKEIH 164
F ++GL +F M + S + C+ L +G++ H
Sbjct: 381 NILSWMIMISGLAENGFGEEGLKLFSCMKREGFEPCDYAFSGAIKSCAVLGAYCNGQQYH 440
Query: 165 GFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTWNSLSSCYVNCGFPQ 224
+++ G + +A + YAKC V EA+ VF MP D V+WN+L + G
Sbjct: 441 AQLLKIGFDSSLSAGNALITMYAKCGVVEEARQVFRTMPCLDSVSWNALIAALGQHGHGA 500
Query: 225 KGLNVFREMVLDGVKPDPVTVSCILSACS 253
+ ++V+ EM+ G++PD +T+ +L+ACS
Sbjct: 501 EAVDVYEEMLKKGIRPDRITLLTVLTACS 529
Score = 78.6 bits (192), Expect = 2e-14
Identities = 55/211 (26%), Positives = 98/211 (46%), Gaps = 15/211 (7%)
Query: 18 HGLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSI 77
+G E +K+++ + G +P F K+CA +Q+H + G S +S
Sbjct: 395 NGFGEEGLKLFSCMKREGFEPCDYAFSGAIKSCAVLGAYCNGQQYHAQLLKIGFDSSLSA 454
Query: 78 GNAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKV 137
GNA I Y KC VE AR+VF + D V+WN+L A G + ++V+ +M +
Sbjct: 455 GNALITMYAKCGVVEEARQVFRTMPCLDSVSWNALIAALGQHGHGAEAVDVYEEMLKKGI 514
Query: 138 KANPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAK---CLC--- 191
+ + +T+ ++L CS + G++ +E V+ + YA+ LC
Sbjct: 515 RPDRITLLTVLTACSHAGLVDQGRKY------FDSMETVYRIPPGADHYARLIDLLCRSG 568
Query: 192 -VREAQTVFDLMPHRDVV-TWNS-LSSCYVN 219
+A++V + +P + W + LS C V+
Sbjct: 569 KFSDAESVIESLPFKPTAEIWEALLSGCRVH 599
>At5g08490 selenium-binding protein-like
Length = 849
Score = 443 bits (1139), Expect = e-124
Identities = 250/837 (29%), Positives = 454/837 (53%), Gaps = 22/837 (2%)
Query: 35 GIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSIGNAFIHAYGKCKCVEGA 94
G D VF+ V KACA+ D + H + G ++ + + ++ Y KC+ ++
Sbjct: 16 GFGTDHRVFLDVVKACASVSDLTSGRALHGCVFKLGHIACSEVSKSVLNMYAKCRRMDDC 75
Query: 95 RRVFDDLVARDVVTWNS-LSACYVNCGFPQQGLNVFRKMGL-NKVKANPLTVSSILPGCS 152
+++F + + D V WN L+ V+CG ++ + F+ M ++ K + +T + +LP C
Sbjct: 76 QKMFRQMDSLDPVVWNIVLTGLSVSCG--RETMRFFKAMHFADEPKPSSVTFAIVLPLCV 133
Query: 153 DLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKC-LCVREAQTVFDLMPHRDVVTWN 211
L D +GK +H ++++ G+ +D V +A V+ YAK +A T FD + +DVV+WN
Sbjct: 134 RLGDSYNGKSMHSYIIKAGLEKDTLVGNALVSMYAKFGFIFPDAYTAFDGIADKDVVSWN 193
Query: 212 SLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQD---LKSGKAIHGFA 268
++ + + F M+ + +P+ T++ +L C+ + +SG+ IH +
Sbjct: 194 AIIAGFSENNMMADAFRSFCLMLKEPTEPNYATIANVLPVCASMDKNIACRSGRQIHSYV 253
Query: 269 LKHGMVE-NVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASCYV-NCGFPQK 326
++ ++ +VFV N+LV+ Y + EA ++F M +++++WN + + Y NC + K
Sbjct: 254 VQRSWLQTHVFVCNSLVSFYLRVGRIEEAASLFTRMGSKDLVSWNVVIAGYASNCEW-FK 312
Query: 327 GLNVFREMGLNG-VKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHG-MVEDVFVCTAL 384
+F + G V PD + + SILP C+QL DL SGK IH + ++H ++ED V AL
Sbjct: 313 AFQLFHNLVHKGDVSPDSVTIISILPVCAQLTDLASGKEIHSYILRHSYLLEDTSVGNAL 372
Query: 385 VNLYANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDL 444
++ YA A F LM +++++WN++ + + + LN+ ++ + D
Sbjct: 373 ISFYARFGDTSAAYWAFSLMSTKDIISWNAILDAFADSPKQFQFLNLLHHLLNEAITLDS 432
Query: 445 VTMLSILHACSDLQDLKSGKVIHGFAVRHGMV---EDVFVCNALLSLYAKCVCVREAQVV 501
VT+LS+L C ++Q + K +HG++V+ G++ E+ + NALL YAKC V A +
Sbjct: 433 VTILSLLKFCINVQGIGKVKEVHGYSVKAGLLHDEEEPKLGNALLDAYAKCGNVEYAHKI 492
Query: 502 F-DLIPHREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGGCVKNSR 560
F L R + S+N +L+ Y + ++ +F++M+ D TWS+++ ++
Sbjct: 493 FLGLSERRTLVSYNSLLSGYVNSGSHDDAQMLFTEMST----TDLTTWSLMVRIYAESCC 548
Query: 561 IEEAMEIFRKMQTMGFKPDETTIYSILRACSLSECLRMGKEIHCYVFRHWKDWDLARTNA 620
EA+ +FR++Q G +P+ TI ++L C+ L + ++ H Y+ R D+
Sbjct: 549 PNEAIGVFREIQARGMRPNTVTIMNLLPVCAQLASLHLVRQCHGYIIRGGLG-DIRLKGT 607
Query: 621 LVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKEALSLFEKMLLSMVKPD 680
L+D+YAKCG L + +VF +D+ + M+ +HG GKEAL ++ M S +KPD
Sbjct: 608 LLDVYAKCGSLKHAYSVFQSDARRDLVMFTAMVAGYAVHGRGKEALMIYSHMTESNIKPD 667
Query: 681 SATFTCVLSACSHSMLVEEGVQIFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFI 740
T +L+AC H+ L+++G+QI++S+ H ++P E Y C VD+ +R G L++AY F+
Sbjct: 668 HVFITTMLTACCHAGLIQDGLQIYDSIRTVHGMKPTMEQYACAVDLIARGGRLDDAYSFV 727
Query: 741 QRMPMEPTAIAWKAFLAGCRVYKNVELAKISAKKLFEIDPNGSANYVTLFNILVTAKLWS 800
+MP+EP A W L C Y ++L A L + + + + N+V + N+ W
Sbjct: 728 TQMPVEPNANIWGTLLRACTTYNRMDLGHSVANHLLQAESDDTGNHVLISNMYAADAKWE 787
Query: 801 EASKIRKLMKERGITKTPGCSWFHVGNRVHTFVAGDKSNMESDKIYNFLDELFAKIK 857
++R LMK++ + K GCSW V + + FV+GD S+ D I++ ++ L+ ++K
Sbjct: 788 GVMELRNLMKKKEMKKPAGCSWLEVDGQRNVFVSGDCSHPRRDSIFDLVNALYLQMK 844
Score = 202 bits (513), Expect = 9e-52
Identities = 139/536 (25%), Positives = 252/536 (46%), Gaps = 48/536 (8%)
Query: 234 VLDGVKPDPVTVSCILSACSDLQDLKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCV 293
+L G D ++ AC+ + DL SG+A+HG K G + VS +++N+Y C +
Sbjct: 13 LLSGFGTDHRVFLDVVKACASVSDLTSGRALHGCVFKLGHIACSEVSKSVLNMYAKCRRM 72
Query: 294 REAQAVFDLMPHRNVITWN-SLASCYVNCGFPQKGLNVFREMGL-NGVKPDPMAMSSILP 351
+ Q +F M + + WN L V+CG ++ + F+ M + KP + + +LP
Sbjct: 73 DDCQKMFRQMDSLDPVVWNIVLTGLSVSCG--RETMRFFKAMHFADEPKPSSVTFAIVLP 130
Query: 352 ACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANC-LCVREAQTVFDLMPHRNVV 410
C +L D +GK++H + +K G+ +D V ALV++YA +A T FD + ++VV
Sbjct: 131 LCVRLGDSYNGKSMHSYIIKAGLEKDTLVGNALVSMYAKFGFIFPDAYTAFDGIADKDVV 190
Query: 411 TWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHACSDLQD---LKSGKVIH 467
+WN++ + + F M+ +P+ T+ ++L C+ + +SG+ IH
Sbjct: 191 SWNAIIAGFSENNMMADAFRSFCLMLKEPTEPNYATIANVLPVCASMDKNIACRSGRQIH 250
Query: 468 GFAVRHGMVE-DVFVCNALLSLYAKCVCVREAQVVFDLIPHREVASWNGILTAYFTNKEY 526
+ V+ ++ VFVCN+L+S Y + + EA +F + +++ SWN ++ Y +N E+
Sbjct: 251 SYVVQRSWLQTHVFVCNSLVSFYLRVGRIEEAASLFTRMGSKDLVSWNVVIAGYASNCEW 310
Query: 527 EKGLYMFSQM-NRDEVKADEITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKPDETTIYS 585
K +F + ++ +V D + TI S
Sbjct: 311 FKAFQLFHNLVHKGDVSPDSV-----------------------------------TIIS 335
Query: 586 ILRACSLSECLRMGKEIHCYVFRH-WKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIK 644
IL C+ L GKEIH Y+ RH + D + NAL+ YA+ G S + F +M K
Sbjct: 336 ILPVCAQLTDLASGKEIHSYILRHSYLLEDTSVGNALISFYARFGDTSAAYWAFSLMSTK 395
Query: 645 DVFSWNTMIFANGMHGNGKEALSLFEKMLLSMVKPDSATFTCVLSACSHSMLVEEGVQIF 704
D+ SWN ++ A + L+L +L + DS T +L C + + + ++
Sbjct: 396 DIISWNAILDAFADSPKQFQFLNLLHHLLNEAITLDSVTILSLLKFCINVQGIGKVKEVH 455
Query: 705 NSMSRDHLVEPEAEHY--TCVVDIYSRAGCLEEAYGFIQRMPMEPTAIAWKAFLAG 758
+ L+ E E ++D Y++ G +E A+ + T +++ + L+G
Sbjct: 456 GYSVKAGLLHDEEEPKLGNALLDAYAKCGNVEYAHKIFLGLSERRTLVSYNSLLSG 511
Score = 67.0 bits (162), Expect = 5e-11
Identities = 45/184 (24%), Positives = 87/184 (46%), Gaps = 2/184 (1%)
Query: 21 PNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSIGNA 80
PNEAI ++ +ARG++P+ M + CA V+Q H R G + D+ +
Sbjct: 549 PNEAIGVFREIQARGMRPNTVTIMNLLPVCAQLASLHLVRQCHGYIIR-GGLGDIRLKGT 607
Query: 81 FIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKAN 140
+ Y KC ++ A VF RD+V + ++ A Y G ++ L ++ M + +K +
Sbjct: 608 LLDVYAKCGSLKHAYSVFQSDARRDLVMFTAMVAGYAVHGRGKEALMIYSHMTESNIKPD 667
Query: 141 PLTVSSILPGCSDLQDLKSGKEIHGFV-VRHGMVEDVFVSSAFVNFYAKCLCVREAQTVF 199
+ ++++L C ++ G +I+ + HGM + + V+ A+ + +A +
Sbjct: 668 HVFITTMLTACCHAGLIQDGLQIYDSIRTVHGMKPTMEQYACAVDLIARGGRLDDAYSFV 727
Query: 200 DLMP 203
MP
Sbjct: 728 TQMP 731
>At5g13230 putative protein
Length = 822
Score = 437 bits (1123), Expect = e-122
Identities = 263/808 (32%), Positives = 417/808 (51%), Gaps = 40/808 (4%)
Query: 146 SILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHR 205
++L C D S K IH +++ G D+F ++ +N Y K ++A +FD MP R
Sbjct: 54 AMLRRCIQKNDPISAKAIHCDILKKGSCLDLFATNILLNAYVKAGFDKDALNLFDEMPER 113
Query: 206 DVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQDLKSGKAIH 265
+ V++ +L+ Y C P + ++ + +G + +P + L L + +H
Sbjct: 114 NNVSFVTLAQGYA-CQDP---IGLYSRLHREGHELNPHVFTSFLKLFVSLDKAEICPWLH 169
Query: 266 GFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASCYVNCGFPQ 325
+K G N FV AL+N Y C V A+ VF+ + ++++ W + SCYV G+ +
Sbjct: 170 SPIVKLGYDSNAFVGAALINAYSVCGSVDSARTVFEGILCKDIVVWAGIVSCYVENGYFE 229
Query: 326 KGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALV 385
L + M + G P+ + L A L K +HG +K V D V L+
Sbjct: 230 DSLKLLSCMRMAGFMPNNYTFDTALKASIGLGAFDFAKGVHGQILKTCYVLDPRVGVGLL 289
Query: 386 NLYANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLV 445
LY + +A VF+ MP +VV W+ + + + GF + +++F M V P+
Sbjct: 290 QLYTQLGDMSDAFKVFNEMPKNDVVPWSFMIARFCQNGFCNEAVDLFIRMREAFVVPNEF 349
Query: 446 TMLSILHACSDLQDLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLI 505
T+ SIL+ C+ + G+ +HG V+ G D++V NAL+ +YAKC + A +F +
Sbjct: 350 TLSSILNGCAIGKCSGLGEQLHGLVVKVGFDLDIYVSNALIDVYAKCEKMDTAVKLFAEL 409
Query: 506 PHREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGGCVKNSRIEEAM 565
+ SWN ++ Y E K MF + R++V E+T+S
Sbjct: 410 SSKNEVSWNTVIVGYENLGEGGKAFSMFREALRNQVSVTEVTFS---------------- 453
Query: 566 EIFRKMQTMGFKPDETTIYSILRACSLSECLRMGKEIHCYVFRHWKDWDLARTNALVDMY 625
S L AC+ + +G ++H + +A +N+L+DMY
Sbjct: 454 -------------------SALGACASLASMDLGVQVHGLAIKTNNAKKVAVSNSLIDMY 494
Query: 626 AKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKEALSLFEKMLLSMVKPDSATFT 685
AKCG + +++VF+ M DV SWN +I HG G++AL + + M KP+ TF
Sbjct: 495 AKCGDIKFAQSVFNEMETIDVASWNALISGYSTHGLGRQALRILDIMKDRDCKPNGLTFL 554
Query: 686 CVLSACSHSMLVEEGVQIFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPM 745
VLS CS++ L+++G + F SM RDH +EP EHYTC+V + R+G L++A I+ +P
Sbjct: 555 GVLSGCSNAGLIDQGQECFESMIRDHGIEPCLEHYTCMVRLLGRSGQLDKAMKLIEGIPY 614
Query: 746 EPTAIAWKAFLAGCRVYKNVELAKISAKKLFEIDPNGSANYVTLFNILVTAKLWSEASKI 805
EP+ + W+A L+ N E A+ SA+++ +I+P A YV + N+ AK W+ + I
Sbjct: 615 EPSVMIWRAMLSASMNQNNEEFARRSAEEILKINPKDEATYVLVSNMYAGAKQWANVASI 674
Query: 806 RKLMKERGITKTPGCSWFHVGNRVHTFVAGDKSNMESDKIYNFLDELFAKIKAAGYKPDT 865
RK MKE G+ K PG SW VH F G + + I L+ L K AGY PD
Sbjct: 675 RKSMKEMGVKKEPGLSWIEHQGDVHYFSVGLSDHPDMKLINGMLEWLNMKATRAGYVPDR 734
Query: 866 DYVLHDIDQEEKAESLCNHSEKLAVAFGILNL-NGQSTIRVFKNLRICGDCHNAIKYMSN 924
+ VL D+D EEK + L HSE+LA+A+G++ + + ++ I + KNLRIC DCH+A+K +S+
Sbjct: 735 NAVLLDMDDEEKDKRLWVHSERLALAYGLVRMPSSRNRILIMKNLRICSDCHSAMKVISS 794
Query: 925 VVGVTIVVRDSLRFHHFKNGNCSCKDFW 952
+V +V+RD RFHHF G CSC D W
Sbjct: 795 IVQRDLVIRDMNRFHHFHAGVCSCGDHW 822
Score = 242 bits (618), Expect = 6e-64
Identities = 175/650 (26%), Positives = 301/650 (45%), Gaps = 40/650 (6%)
Query: 39 DKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSIGNAFIHAYGKCKCVEGARRVF 98
D + A+ + C D + K H D + G D+ N ++AY K + A +F
Sbjct: 48 DSHAYGAMLRRCIQKNDPISAKAIHCDILKKGSCLDLFATNILLNAYVKAGFDKDALNLF 107
Query: 99 DDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKANPLTVSSILPGCSDLQDLK 158
D++ R+ V++ +L+ Y C Q + ++ ++ + NP +S L L +
Sbjct: 108 DEMPERNNVSFVTLAQGYA-C---QDPIGLYSRLHREGHELNPHVFTSFLKLFVSLDKAE 163
Query: 159 SGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTWNSLSSCYV 218
+H +V+ G + FV +A +N Y+ C V A+TVF+ + +D+V W + SCYV
Sbjct: 164 ICPWLHSPIVKLGYDSNAFVGAALINAYSVCGSVDSARTVFEGILCKDIVVWAGIVSCYV 223
Query: 219 NCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQDLKSGKAIHGFALKHGMVENVF 278
G+ + L + M + G P+ T L A L K +HG LK V +
Sbjct: 224 ENGYFEDSLKLLSCMRMAGFMPNNYTFDTALKASIGLGAFDFAKGVHGQILKTCYVLDPR 283
Query: 279 VSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASCYVNCGFPQKGLNVFREMGLNG 338
V L+ LY + +A VF+ MP +V+ W+ + + + GF + +++F M
Sbjct: 284 VGVGLLQLYTQLGDMSDAFKVFNEMPKNDVVPWSFMIARFCQNGFCNEAVDLFIRMREAF 343
Query: 339 VKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQ 398
V P+ +SSIL C+ K G+ +HG VK G D++V AL+++YA C + A
Sbjct: 344 VVPNEFTLSSILNGCAIGKCSGLGEQLHGLVVKVGFDLDIYVSNALIDVYAKCEKMDTAV 403
Query: 399 TVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHACSDLQ 458
+F + +N V+WN++ Y N G K ++FRE + N V VT S L AC+ L
Sbjct: 404 KLFAELSSKNEVSWNTVIVGYENLGEGGKAFSMFREALRNQVSVTEVTFSSALGACASLA 463
Query: 459 DLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREVASWNGILT 518
+ G +HG A++ + V V N+L+ +YAKC ++ AQ VF+ + +VASWN +++
Sbjct: 464 SMDLGVQVHGLAIKTNNAKKVAVSNSLIDMYAKCGDIKFAQSVFNEMETIDVASWNALIS 523
Query: 519 AYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKP 578
Y T+ + L + M + K + +T+ V+ GC I++ E F
Sbjct: 524 GYSTHGLGRQALRILDIMKDRDCKPNGLTFLGVLSGCSNAGLIDQGQECFE--------- 574
Query: 579 DETTIYSILRACSLSECLRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVF 638
S++R + CL H+ +V + + G L + +
Sbjct: 575 ------SMIRDHGIEPCL-----------EHY--------TCMVRLLGRSGQLDKAMKLI 609
Query: 639 DMMPIK-DVFSWNTMIFANGMHGNGKEALSLFEKMLLSMVKPDSATFTCV 687
+ +P + V W M+ A+ M+ N +E + +L + D AT+ V
Sbjct: 610 EGIPYEPSVMIWRAMLSAS-MNQNNEEFARRSAEEILKINPKDEATYVLV 658
Score = 215 bits (548), Expect = 8e-56
Identities = 149/563 (26%), Positives = 264/563 (46%), Gaps = 11/563 (1%)
Query: 23 EAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSIGNAFI 82
+ I +Y+ G + + VF + K + A H + G S+ +G A I
Sbjct: 129 DPIGLYSRLHREGHELNPHVFTSFLKLFVSLDKAEICPWLHSPIVKLGYDSNAFVGAALI 188
Query: 83 HAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKANPL 142
+AY C V+ AR VF+ ++ +D+V W + +CYV G+ + L + M + N
Sbjct: 189 NAYSVCGSVDSARTVFEGILCKDIVVWAGIVSCYVENGYFEDSLKLLSCMRMAGFMPNNY 248
Query: 143 TVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLM 202
T + L L K +HG +++ V D V + Y + + +A VF+ M
Sbjct: 249 TFDTALKASIGLGAFDFAKGVHGQILKTCYVLDPRVGVGLLQLYTQLGDMSDAFKVFNEM 308
Query: 203 PHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQDLKSGK 262
P DVV W+ + + + GF + +++F M V P+ T+S IL+ C+ + G+
Sbjct: 309 PKNDVVPWSFMIARFCQNGFCNEAVDLFIRMREAFVVPNEFTLSSILNGCAIGKCSGLGE 368
Query: 263 AIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASCYVNCG 322
+HG +K G +++VSNAL+++Y C + A +F + +N ++WN++ Y N G
Sbjct: 369 QLHGLVVKVGFDLDIYVSNALIDVYAKCEKMDTAVKLFAELSSKNEVSWNTVIVGYENLG 428
Query: 323 FPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCT 382
K ++FRE N V + SS L AC+ L + G +HG A+K + V V
Sbjct: 429 EGGKAFSMFREALRNQVSVTEVTFSSALGACASLASMDLGVQVHGLAIKTNNAKKVAVSN 488
Query: 383 ALVNLYANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKP 442
+L+++YA C ++ AQ+VF+ M +V +WN+L S Y G ++ L + M KP
Sbjct: 489 SLIDMYAKCGDIKFAQSVFNEMETIDVASWNALISGYSTHGLGRQALRILDIMKDRDCKP 548
Query: 443 DLVTMLSILHACSDLQDLKSGKVIHGFAVR-HGMVEDVFVCNALLSLYAKCVCVREAQVV 501
+ +T L +L CS+ + G+ +R HG+ + ++ L + + +A +
Sbjct: 549 NGLTFLGVLSGCSNAGLIDQGQECFESMIRDHGIEPCLEHYTCMVRLLGRSGQLDKAMKL 608
Query: 502 FDLIPHR-EVASWNGILTAYFTNKEYEKGLYMFSQMNRDEV----KADEITWSVVIGGCV 556
+ IP+ V W +L+A E F++ + +E+ DE T+ +V
Sbjct: 609 IEGIPYEPSVMIWRAMLSASMNQNNEE-----FARRSAEEILKINPKDEATYVLVSNMYA 663
Query: 557 KNSRIEEAMEIFRKMQTMGFKPD 579
+ I + M+ MG K +
Sbjct: 664 GAKQWANVASIRKSMKEMGVKKE 686
Score = 133 bits (335), Expect = 4e-31
Identities = 80/305 (26%), Positives = 153/305 (49%), Gaps = 2/305 (0%)
Query: 18 HGLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSI 77
+G NEA+ ++ R + P++ ++ CA + + +Q H + G D+ +
Sbjct: 326 NGFCNEAVDLFIRMREAFVVPNEFTLSSILNGCAIGKCSGLGEQLHGLVVKVGFDLDIYV 385
Query: 78 GNAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKV 137
NA I Y KC+ ++ A ++F +L +++ V+WN++ Y N G + ++FR+ N+V
Sbjct: 386 SNALIDVYAKCEKMDTAVKLFAELSSKNEVSWNTVIVGYENLGEGGKAFSMFREALRNQV 445
Query: 138 KANPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQT 197
+T SS L C+ L + G ++HG ++ + V VS++ ++ YAKC ++ AQ+
Sbjct: 446 SVTEVTFSSALGACASLASMDLGVQVHGLAIKTNNAKKVAVSNSLIDMYAKCGDIKFAQS 505
Query: 198 VFDLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQD 257
VF+ M DV +WN+L S Y G ++ L + M KP+ +T +LS CS+
Sbjct: 506 VFNEMETIDVASWNALISGYSTHGLGRQALRILDIMKDRDCKPNGLTFLGVLSGCSNAGL 565
Query: 258 LKSG-KAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHR-NVITWNSLA 315
+ G + HG+ + +V L + +A + + +P+ +V+ W ++
Sbjct: 566 IDQGQECFESMIRDHGIEPCLEHYTCMVRLLGRSGQLDKAMKLIEGIPYEPSVMIWRAML 625
Query: 316 SCYVN 320
S +N
Sbjct: 626 SASMN 630
Score = 110 bits (275), Expect = 4e-24
Identities = 112/487 (22%), Positives = 202/487 (40%), Gaps = 55/487 (11%)
Query: 321 CGFPQKGLNVFREMGLNGVKP--DPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDV 378
CGF K + E + + P D A ++L C Q D S K IH +K G D+
Sbjct: 26 CGFSVKTAALDLESS-DSIIPGLDSHAYGAMLRRCIQKNDPISAKAIHCDILKKGSCLDL 84
Query: 379 FVCTALVNLYANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLN 438
F L+N Y ++A +FD MP RN V++ +L+ Y C Q + ++ +
Sbjct: 85 FATNILLNAYVKAGFDKDALNLFDEMPERNNVSFVTLAQGYA-C---QDPIGLYSRLHRE 140
Query: 439 GVKPDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREA 498
G + + S L L + +H V+ G + FV AL++ Y+ C V A
Sbjct: 141 GHELNPHVFTSFLKLFVSLDKAEICPWLHSPIVKLGYDSNAFVGAALINAYSVCGSVDSA 200
Query: 499 QVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGGCVKN 558
+ VF+ I +++ W GI++ Y V+N
Sbjct: 201 RTVFEGILCKDIVVWAGIVSCY-----------------------------------VEN 225
Query: 559 SRIEEAMEIFRKMQTMGFKPDETTIYSILRACSLSECLRMGKEIHCYVFRHWKDWDLART 618
E+++++ M+ GF P+ T + L+A K +H + + D
Sbjct: 226 GYFEDSLKLLSCMRMAGFMPNNYTFDTALKASIGLGAFDFAKGVHGQILKTCYVLDPRVG 285
Query: 619 NALVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKEALSLFEKMLLSMVK 678
L+ +Y + G +S + VF+ MP DV W+ MI +G EA+ LF +M + V
Sbjct: 286 VGLLQLYTQLGDMSDAFKVFNEMPKNDVVPWSFMIARFCQNGFCNEAVDLFIRMREAFVV 345
Query: 679 PDSATFTCVLSACSHSMLVEEGVQIFNSMSRDHLVEPEAEHY--TCVVDIYSRAGCLEEA 736
P+ T + +L+ C+ G Q+ + + V + + Y ++D+Y++ ++ A
Sbjct: 346 PNEFTLSSILNGCAIGKCSGLGEQLHGLVVK---VGFDLDIYVSNALIDVYAKCEKMDTA 402
Query: 737 YGFIQRMPMEPTAIAWKAFLAGCRVYKNVELAKISAKKLFEIDPNGSANYVTLFNILVTA 796
+ + ++W + G Y+N+ K F + N V++ + ++
Sbjct: 403 VKLFAELSSK-NEVSWNTVIVG---YENLG----EGGKAFSMFREALRNQVSVTEVTFSS 454
Query: 797 KLWSEAS 803
L + AS
Sbjct: 455 ALGACAS 461
>At4g14820 hypothetical protein
Length = 722
Score = 433 bits (1113), Expect = e-121
Identities = 245/725 (33%), Positives = 396/725 (53%), Gaps = 18/725 (2%)
Query: 240 PDPVT---VSCILSACSDLQDLKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREA 296
P P+ + IL S + L K +H L+ V N +++ L NL S + +
Sbjct: 4 PPPIASTAANTILEKLSFCKSLNHIKQLHAHILR--TVINHKLNSFLFNLSVSSSSINLS 61
Query: 297 QA--VFDLMPHR-NVITWNSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPAC 353
A VF +P I +N P+ + ++ + G + D + IL A
Sbjct: 62 YALNVFSSIPSPPESIVFNPFLRDLSRSSEPRATILFYQRIRHVGGRLDQFSFLPILKAV 121
Query: 354 SQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPHRNVVTWN 413
S++ L G +HG A K + D FV T +++YA+C + A+ VFD M HR+VVTWN
Sbjct: 122 SKVSALFEGMELHGVAFKIATLCDPFVETGFMDMYASCGRINYARNVFDEMSHRDVVTWN 181
Query: 414 SLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHACSDLQDLKSGKVIHGFAVRH 473
++ Y G + +F EM + V PD + + +I+ AC +++ + I+ F + +
Sbjct: 182 TMIERYCRFGLVDEAFKLFEEMKDSNVMPDEMILCNIVSACGRTGNMRYNRAIYEFLIEN 241
Query: 474 GMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMF 533
+ D + AL+++YA C+ A+ F + R + +++ Y + +F
Sbjct: 242 DVRMDTHLLTALVTMYAGAGCMDMAREFFRKMSVRNLFVSTAMVSGYSKCGRLDDAQVIF 301
Query: 534 SQMNRDEVKADEITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKPDETTIYSILRACSLS 593
Q K D + W+ +I V++ +EA+ +F +M G KPD +++S++ AC+
Sbjct: 302 DQTE----KKDLVCWTTMISAYVESDYPQEALRVFEEMCCSGIKPDVVSMFSVISACANL 357
Query: 594 ECLRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMI 653
L K +H + + + +L+ NAL++MYAKCGGL +R+VF+ MP ++V SW++MI
Sbjct: 358 GILDKAKWVHSCIHVNGLESELSINNALINMYAKCGGLDATRDVFEKMPRRNVVSWSSMI 417
Query: 654 FANGMHGNGKEALSLFEKMLLSMVKPDSATFTCVLSACSHSMLVEEGVQIFNSMSRDHLV 713
A MHG +ALSLF +M V+P+ TF VL CSHS LVEEG +IF SM+ ++ +
Sbjct: 418 NALSMHGEASDALSLFARMKQENVEPNEVTFVGVLYGCSHSGLVEEGKKIFASMTDEYNI 477
Query: 714 EPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEPTAIAWKAFLAGCRVYKNVELAKISAK 773
P+ EHY C+VD++ RA L EA I+ MP+ + W + ++ CR++ +EL K +AK
Sbjct: 478 TPKLEHYGCMVDLFGRANLLREALEVIESMPVASNVVIWGSLMSACRIHGELELGKFAAK 537
Query: 774 KLFEIDPNGSANYVTLFNILVTAKLWSEASKIRKLMKERGITKTPGCSWFHVGNRVHTFV 833
++ E++P+ V + NI + W + IR++M+E+ + K G S + H F+
Sbjct: 538 RILELEPDHDGALVLMSNIYAREQRWEDVRNIRRVMEEKNVFKEKGLSRIDQNGKSHEFL 597
Query: 834 AGDKSNMESDKIYNFLDELFAKIKAAGYKPDTDYVLHDIDQEEKAESLCNHSEKLAVAFG 893
GDK + +S++IY LDE+ +K+K AGY PD VL D+++EEK + + HSEKLA+ FG
Sbjct: 598 IGDKRHKQSNEIYAKLDEVVSKLKLAGYVPDCGSVLVDVEEEEKKDLVLWHSEKLALCFG 657
Query: 894 ILNLNGQ------STIRVFKNLRICGDCHNAIKYMSNVVGVTIVVRDSLRFHHFKNGNCS 947
++N + IR+ KNLR+C DCH K +S V I+VRD RFH +KNG CS
Sbjct: 658 LMNEEKEEEKDSCGVIRIVKNLRVCEDCHLFFKLVSKVYEREIIVRDRTRFHCYKNGLCS 717
Query: 948 CKDFW 952
C+D+W
Sbjct: 718 CRDYW 722
Score = 209 bits (532), Expect = 6e-54
Identities = 142/556 (25%), Positives = 262/556 (46%), Gaps = 81/556 (14%)
Query: 145 SSILPGCSDLQDLKSGKEIHGFVVR-------HGMVEDVFVSSAFVNFYAKCLCVREAQT 197
++IL S + L K++H ++R + + ++ VSS+ +N + A
Sbjct: 13 NTILEKLSFCKSLNHIKQLHAHILRTVINHKLNSFLFNLSVSSSSIN-------LSYALN 65
Query: 198 VFDLMPHR-DVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQ 256
VF +P + + +N P+ + ++ + G + D + IL A S +
Sbjct: 66 VFSSIPSPPESIVFNPFLRDLSRSSEPRATILFYQRIRHVGGRLDQFSFLPILKAVSKVS 125
Query: 257 DLKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLAS 316
L G +HG A K + + FV +++Y SC + A+ VFD M HR+V+TWN++
Sbjct: 126 ALFEGMELHGVAFKIATLCDPFVETGFMDMYASCGRINYARNVFDEMSHRDVVTWNTMIE 185
Query: 317 CYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGM-- 374
Y G + +F EM + V PD M + +I+ AC + +++ + I+ F +++ +
Sbjct: 186 RYCRFGLVDEAFKLFEEMKDSNVMPDEMILCNIVSACGRTGNMRYNRAIYEFLIENDVRM 245
Query: 375 -----------------------------VEDVFVCTALVNLYANCLCVREAQTVFDLMP 405
V ++FV TA+V+ Y+ C + +AQ +FD
Sbjct: 246 DTHLLTALVTMYAGAGCMDMAREFFRKMSVRNLFVSTAMVSGYSKCGRLDDAQVIFDQTE 305
Query: 406 HRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHACSDLQDLKSGKV 465
+++V W ++ S YV +PQ+ L VF EM +G+KPD+V+M S++ AC++L L K
Sbjct: 306 KKDLVCWTTMISAYVESDYPQEALRVFEEMCCSGIKPDVVSMFSVISACANLGILDKAKW 365
Query: 466 IHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREVASWNGILTAYFTNKE 525
+H +G+ ++ + NAL+++YAKC + + VF+ +P R V SW+ ++ A + E
Sbjct: 366 VHSCIHVNGLESELSINNALINMYAKCGGLDATRDVFEKMPRRNVVSWSSMINALSMHGE 425
Query: 526 YEKGLYMFSQMNRDEVKADEITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKPDETTIYS 585
L +F++M ++ V+ +E+T+ V+ GC + +EE +IF M T Y+
Sbjct: 426 ASDALSLFARMKQENVEPNEVTFVGVLYGCSHSGLVEEGKKIFASM---------TDEYN 476
Query: 586 ILRACSLSECLRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPI-K 644
I L +VD++ + L + V + MP+
Sbjct: 477 ITP-------------------------KLEHYGCMVDLFGRANLLREALEVIESMPVAS 511
Query: 645 DVFSWNTMIFANGMHG 660
+V W +++ A +HG
Sbjct: 512 NVVIWGSLMSACRIHG 527
Score = 194 bits (492), Expect = 3e-49
Identities = 118/435 (27%), Positives = 209/435 (47%), Gaps = 34/435 (7%)
Query: 21 PNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSIGNA 80
P I Y R G + D+ F+ + KA + + + H A + + D +
Sbjct: 92 PRATILFYQRIRHVGGRLDQFSFLPILKAVSKVSALFEGMELHGVAFKIATLCDPFVETG 151
Query: 81 FIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKAN 140
F+ Y C + AR VFD++ RDVVTWN++ Y G + +F +M + V +
Sbjct: 152 FMDMYASCGRINYARNVFDEMSHRDVVTWNTMIERYCRFGLVDEAFKLFEEMKDSNVMPD 211
Query: 141 PLTVSSILPGCSDLQDLKSGKEIHGFVVRHGM---------------------------- 172
+ + +I+ C +++ + I+ F++ + +
Sbjct: 212 EMILCNIVSACGRTGNMRYNRAIYEFLIENDVRMDTHLLTALVTMYAGAGCMDMAREFFR 271
Query: 173 ---VEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTWNSLSSCYVNCGFPQKGLNV 229
V ++FVS+A V+ Y+KC + +AQ +FD +D+V W ++ S YV +PQ+ L V
Sbjct: 272 KMSVRNLFVSTAMVSGYSKCGRLDDAQVIFDQTEKKDLVCWTTMISAYVESDYPQEALRV 331
Query: 230 FREMVLDGVKPDPVTVSCILSACSDLQDLKSGKAIHGFALKHGMVENVFVSNALVNLYES 289
F EM G+KPD V++ ++SAC++L L K +H +G+ + ++NAL+N+Y
Sbjct: 332 FEEMCCSGIKPDVVSMFSVISACANLGILDKAKWVHSCIHVNGLESELSINNALINMYAK 391
Query: 290 CLCVREAQAVFDLMPHRNVITWNSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSI 349
C + + VF+ MP RNV++W+S+ + G L++F M V+P+ + +
Sbjct: 392 CGGLDATRDVFEKMPRRNVVSWSSMINALSMHGEASDALSLFARMKQENVEPNEVTFVGV 451
Query: 350 LPACSQLKDLKSGKTIH-GFAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMP-HR 407
L CS ++ GK I ++ + + +V+L+ +REA V + MP
Sbjct: 452 LYGCSHSGLVEEGKKIFASMTDEYNITPKLEHYGCMVDLFGRANLLREALEVIESMPVAS 511
Query: 408 NVVTWNSL-SSCYVN 421
NVV W SL S+C ++
Sbjct: 512 NVVIWGSLMSACRIH 526
Score = 129 bits (325), Expect = 6e-30
Identities = 80/331 (24%), Positives = 154/331 (46%), Gaps = 33/331 (9%)
Query: 19 GLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSD---- 74
GL +EA K++ + + PD+ + + AC + + + ++ V D
Sbjct: 191 GLVDEAFKLFEEMKDSNVMPDEMILCNIVSACGRTGNMRYNRAIYEFLIENDVRMDTHLL 250
Query: 75 ---------------------------VSIGNAFIHAYGKCKCVEGARRVFDDLVARDVV 107
+ + A + Y KC ++ A+ +FD +D+V
Sbjct: 251 TALVTMYAGAGCMDMAREFFRKMSVRNLFVSTAMVSGYSKCGRLDDAQVIFDQTEKKDLV 310
Query: 108 TWNSLSACYVNCGFPQQGLNVFRKMGLNKVKANPLTVSSILPGCSDLQDLKSGKEIHGFV 167
W ++ + YV +PQ+ L VF +M + +K + +++ S++ C++L L K +H +
Sbjct: 311 CWTTMISAYVESDYPQEALRVFEEMCCSGIKPDVVSMFSVISACANLGILDKAKWVHSCI 370
Query: 168 VRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTWNSLSSCYVNCGFPQKGL 227
+G+ ++ +++A +N YAKC + + VF+ MP R+VV+W+S+ + G L
Sbjct: 371 HVNGLESELSINNALINMYAKCGGLDATRDVFEKMPRRNVVSWSSMINALSMHGEASDAL 430
Query: 228 NVFREMVLDGVKPDPVTVSCILSACSDLQDLKSGKAIH-GFALKHGMVENVFVSNALVNL 286
++F M + V+P+ VT +L CS ++ GK I ++ + + +V+L
Sbjct: 431 SLFARMKQENVEPNEVTFVGVLYGCSHSGLVEEGKKIFASMTDEYNITPKLEHYGCMVDL 490
Query: 287 YESCLCVREAQAVFDLMP-HRNVITWNSLAS 316
+ +REA V + MP NV+ W SL S
Sbjct: 491 FGRANLLREALEVIESMPVASNVVIWGSLMS 521
Score = 35.4 bits (80), Expect = 0.15
Identities = 23/104 (22%), Positives = 48/104 (46%), Gaps = 3/104 (2%)
Query: 18 HGLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDAT-RCGVMSDVS 76
HG ++A+ ++ + ++P++ F+ V C+ S + K+ T + +
Sbjct: 423 HGEASDALSLFARMKQENVEPNEVTFVGVLYGCSHSGLVEEGKKIFASMTDEYNITPKLE 482
Query: 77 IGNAFIHAYGKCKCVEGARRVFDDL-VARDVVTWNSL-SACYVN 118
+ +G+ + A V + + VA +VV W SL SAC ++
Sbjct: 483 HYGCMVDLFGRANLLREALEVIESMPVASNVVIWGSLMSACRIH 526
>At3g15130 hypothetical protein
Length = 689
Score = 432 bits (1112), Expect = e-121
Identities = 250/714 (35%), Positives = 379/714 (53%), Gaps = 45/714 (6%)
Query: 248 ILSACSDLQDLKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRN 307
IL C+ G +H + LK G N+ SN L+++Y C A VFD MP RN
Sbjct: 12 ILRVCTRKGLSDQGGQVHCYLLKSGSGLNLITSNYLIDMYCKCREPLMAYKVFDSMPERN 71
Query: 308 VITWNSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHG 367
V++W++L S +V G + L++F EMG G+ P+ S+ L AC L L+ G IHG
Sbjct: 72 VVSWSALMSGHVLNGDLKGSLSLFSEMGRQGIYPNEFTFSTNLKACGLLNALEKGLQIHG 131
Query: 368 FAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQK 427
F +K G V V +LV++Y+ C + EA+ VF + R++++WN++ + +V+ G+ K
Sbjct: 132 FCLKIGFEMMVEVGNSLVDMYSKCGRINEAEKVFRRIVDRSLISWNAMIAGFVHAGYGSK 191
Query: 428 GLNVFREMVLNGVK--PDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGM--VEDVFVCN 483
L+ F M +K PD T+ S+L ACS + +GK IHGF VR G +
Sbjct: 192 ALDTFGMMQEANIKERPDEFTLTSLLKACSSTGMIYAGKQIHGFLVRSGFHCPSSATITG 251
Query: 484 ALLSLYAKCVCVREAQVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKA 543
+L+ LY KC + A+ FD I + + SW+ ++ Y E+ + + +F
Sbjct: 252 SLVDLYVKCGYLFSARKAFDQIKEKTMISWSSLILGYAQEGEFVEAMGLF---------- 301
Query: 544 DEITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKPDETTIYSILRACSLSECLRMGKEIH 603
+++Q + + D + SI+ + LR GK++
Sbjct: 302 -------------------------KRLQELNSQIDSFALSSIIGVFADFALLRQGKQMQ 336
Query: 604 CYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGK 663
+ + + N++VDMY KCG + + F M +KDV SW +I G HG GK
Sbjct: 337 ALAVKLPSGLETSVLNSVVDMYLKCGLVDEAEKCFAEMQLKDVISWTVVITGYGKHGLGK 396
Query: 664 EALSLFEKMLLSMVKPDSATFTCVLSACSHSMLVEEGVQIFNSMSRDHLVEPEAEHYTCV 723
+++ +F +ML ++PD + VLSACSHS +++EG ++F+ + H ++P EHY CV
Sbjct: 397 KSVRIFYEMLRHNIEPDEVCYLAVLSACSHSGMIKEGEELFSKLLETHGIKPRVEHYACV 456
Query: 724 VDIYSRAGCLEEAYGFIQRMPMEPTAIAWKAFLAGCRVYKNVELAKISAKKLFEIDPNGS 783
VD+ RAG L+EA I MP++P W+ L+ CRV+ ++EL K K L ID
Sbjct: 457 VDLLGRAGRLKEAKHLIDTMPIKPNVGIWQTLLSLCRVHGDIELGKEVGKILLRIDAKNP 516
Query: 784 ANYVTLFNILVTAKLWSEASKIRKLMKERGITKTPGCSWFHVGNRVHTFVAGDKSNMESD 843
ANYV + N+ A W+E R+L +G+ K G SW + VH F +G+ S+ +
Sbjct: 517 ANYVMMSNLYGQAGYWNEQGNARELGNIKGLKKEAGMSWVEIEREVHFFRSGEDSHPLTP 576
Query: 844 KIYNFLDELFAKIK-AAGYKPDTDYVLHDIDQEEKAESLCNHSEKLAV----AFGILNLN 898
I L E +++ GY + LHDID E K E+L HSEKLA+ A G LN
Sbjct: 577 VIQETLKEAERRLREELGYVYGLKHELHDIDDESKEENLRAHSEKLAIGLALATGGLNQK 636
Query: 899 GQSTIRVFKNLRICGDCHNAIKYMSNVVGVTIVVRDSLRFHHFKNGNCSCKDFW 952
G+ TIRVFKNLR+C DCH IK +S + + VVRD++RFH F++G CSC D+W
Sbjct: 637 GK-TIRVFKNLRVCVDCHEFIKGLSKITKIAYVVRDAVRFHSFEDGCCSCGDYW 689
Score = 233 bits (595), Expect = 3e-61
Identities = 155/528 (29%), Positives = 263/528 (49%), Gaps = 44/528 (8%)
Query: 146 SILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHR 205
SIL C+ G ++H ++++ G ++ S+ ++ Y KC A VFD MP R
Sbjct: 11 SILRVCTRKGLSDQGGQVHCYLLKSGSGLNLITSNYLIDMYCKCREPLMAYKVFDSMPER 70
Query: 206 DVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQDLKSGKAIH 265
+VV+W++L S +V G + L++F EM G+ P+ T S L AC L L+ G IH
Sbjct: 71 NVVSWSALMSGHVLNGDLKGSLSLFSEMGRQGIYPNEFTFSTNLKACGLLNALEKGLQIH 130
Query: 266 GFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASCYVNCGFPQ 325
GF LK G V V N+LV++Y C + EA+ VF + R++I+WN++ + +V+ G+
Sbjct: 131 GFCLKIGFEMMVEVGNSLVDMYSKCGRINEAEKVFRRIVDRSLISWNAMIAGFVHAGYGS 190
Query: 326 KGLNVFREMGLNGVK--PDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGM--VEDVFVC 381
K L+ F M +K PD ++S+L ACS + +GK IHGF V+ G +
Sbjct: 191 KALDTFGMMQEANIKERPDEFTLTSLLKACSSTGMIYAGKQIHGFLVRSGFHCPSSATIT 250
Query: 382 TALVNLYANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVK 441
+LV+LY C + A+ FD + + +++W+SL Y G + + +F+ + +
Sbjct: 251 GSLVDLYVKCGYLFSARKAFDQIKEKTMISWSSLILGYAQEGEFVEAMGLFKRLQELNSQ 310
Query: 442 PDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVV 501
D + SI+ +D L+ GK + AV+ + V N+++ +Y KC V EA+
Sbjct: 311 IDSFALSSIIGVFADFALLRQGKQMQALAVKLPSGLETSVLNSVVDMYLKCGLVDEAEKC 370
Query: 502 FDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGGCVKNSRI 561
F + ++V SW ++T Y + +K + +F +M R ++ DE+ + V+ C + I
Sbjct: 371 FAEMQLKDVISWTVVITGYGKHGLGKKSVRIFYEMLRHNIEPDEVCYLAVLSACSHSGMI 430
Query: 562 EEAMEIFRK-MQTMGFKPDETTIYSILRACSLSECLRMGKEIHCYVFRHWKDWDLARTNA 620
+E E+F K ++T G KP + Y
Sbjct: 431 KEGEELFSKLLETHGIKP----------------------RVEHYA-------------C 455
Query: 621 LVDMYAKCGGLSLSRNVFDMMPIK-DVFSWNTMIFANGMHGN---GKE 664
+VD+ + G L ++++ D MPIK +V W T++ +HG+ GKE
Sbjct: 456 VVDLLGRAGRLKEAKHLIDTMPIKPNVGIWQTLLSLCRVHGDIELGKE 503
Score = 211 bits (537), Expect = 2e-54
Identities = 137/492 (27%), Positives = 252/492 (50%), Gaps = 6/492 (1%)
Query: 44 MAVAKACAASRDALKVKQFHDDATRCGVMSDVSIGNAFIHAYGKCKCVEGARRVFDDLVA 103
+++ + C + + Q H + G ++ N I Y KC+ A +VFD +
Sbjct: 10 VSILRVCTRKGLSDQGGQVHCYLLKSGSGLNLITSNYLIDMYCKCREPLMAYKVFDSMPE 69
Query: 104 RDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKANPLTVSSILPGCSDLQDLKSGKEI 163
R+VV+W++L + +V G + L++F +MG + N T S+ L C L L+ G +I
Sbjct: 70 RNVVSWSALMSGHVLNGDLKGSLSLFSEMGRQGIYPNEFTFSTNLKACGLLNALEKGLQI 129
Query: 164 HGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTWNSLSSCYVNCGFP 223
HGF ++ G V V ++ V+ Y+KC + EA+ VF + R +++WN++ + +V+ G+
Sbjct: 130 HGFCLKIGFEMMVEVGNSLVDMYSKCGRINEAEKVFRRIVDRSLISWNAMIAGFVHAGYG 189
Query: 224 QKGLNVFREMVLDGVK--PDPVTVSCILSACSDLQDLKSGKAIHGFALKHGM--VENVFV 279
K L+ F M +K PD T++ +L ACS + +GK IHGF ++ G + +
Sbjct: 190 SKALDTFGMMQEANIKERPDEFTLTSLLKACSSTGMIYAGKQIHGFLVRSGFHCPSSATI 249
Query: 280 SNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASCYVNCGFPQKGLNVFREMGLNGV 339
+ +LV+LY C + A+ FD + + +I+W+SL Y G + + +F+ +
Sbjct: 250 TGSLVDLYVKCGYLFSARKAFDQIKEKTMISWSSLILGYAQEGEFVEAMGLFKRLQELNS 309
Query: 340 KPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQT 399
+ D A+SSI+ + L+ GK + AVK + V ++V++Y C V EA+
Sbjct: 310 QIDSFALSSIIGVFADFALLRQGKQMQALAVKLPSGLETSVLNSVVDMYLKCGLVDEAEK 369
Query: 400 VFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHACSDLQD 459
F M ++V++W + + Y G +K + +F EM+ + ++PD V L++L ACS
Sbjct: 370 CFAEMQLKDVISWTVVITGYGKHGLGKKSVRIFYEMLRHNIEPDEVCYLAVLSACSHSGM 429
Query: 460 LKSGKVIHGFAVR-HGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHR-EVASWNGIL 517
+K G+ + + HG+ V ++ L + ++EA+ + D +P + V W +L
Sbjct: 430 IKEGEELFSKLLETHGIKPRVEHYACVVDLLGRAGRLKEAKHLIDTMPIKPNVGIWQTLL 489
Query: 518 TAYFTNKEYEKG 529
+ + + E G
Sbjct: 490 SLCRVHGDIELG 501
Score = 176 bits (446), Expect = 6e-44
Identities = 110/400 (27%), Positives = 200/400 (49%), Gaps = 6/400 (1%)
Query: 24 AIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSIGNAFIH 83
++ +++ +GI P++ F KAC K Q H + G V +GN+ +
Sbjct: 91 SLSLFSEMGRQGIYPNEFTFSTNLKACGLLNALEKGLQIHGFCLKIGFEMMVEVGNSLVD 150
Query: 84 AYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKANP-- 141
Y KC + A +VF +V R +++WN++ A +V+ G+ + L+ F M +K P
Sbjct: 151 MYSKCGRINEAEKVFRRIVDRSLISWNAMIAGFVHAGYGSKALDTFGMMQEANIKERPDE 210
Query: 142 LTVSSILPGCSDLQDLKSGKEIHGFVVRHGM--VEDVFVSSAFVNFYAKCLCVREAQTVF 199
T++S+L CS + +GK+IHGF+VR G ++ + V+ Y KC + A+ F
Sbjct: 211 FTLTSLLKACSSTGMIYAGKQIHGFLVRSGFHCPSSATITGSLVDLYVKCGYLFSARKAF 270
Query: 200 DLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQDLK 259
D + + +++W+SL Y G + + +F+ + + D +S I+ +D L+
Sbjct: 271 DQIKEKTMISWSSLILGYAQEGEFVEAMGLFKRLQELNSQIDSFALSSIIGVFADFALLR 330
Query: 260 SGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASCYV 319
GK + A+K V N++V++Y C V EA+ F M ++VI+W + + Y
Sbjct: 331 QGKQMQALAVKLPSGLETSVLNSVVDMYLKCGLVDEAEKCFAEMQLKDVISWTVVITGYG 390
Query: 320 NCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVK-HGMVEDV 378
G +K + +F EM + ++PD + ++L ACS +K G+ + ++ HG+ V
Sbjct: 391 KHGLGKKSVRIFYEMLRHNIEPDEVCYLAVLSACSHSGMIKEGEELFSKLLETHGIKPRV 450
Query: 379 FVCTALVNLYANCLCVREAQTVFDLMPHR-NVVTWNSLSS 417
+V+L ++EA+ + D MP + NV W +L S
Sbjct: 451 EHYACVVDLLGRAGRLKEAKHLIDTMPIKPNVGIWQTLLS 490
Score = 120 bits (301), Expect = 4e-27
Identities = 88/315 (27%), Positives = 146/315 (45%), Gaps = 39/315 (12%)
Query: 447 MLSILHACSDLQDLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIP 506
++SIL C+ G +H + ++ G ++ N L+ +Y KC A VFD +P
Sbjct: 9 LVSILRVCTRKGLSDQGGQVHCYLLKSGSGLNLITSNYLIDMYCKCREPLMAYKVFDSMP 68
Query: 507 HREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGGCVKNSRIEEAME 566
R V SW+ +++ + N + + L +FS+M R + +E T+S + C + +E+ ++
Sbjct: 69 ERNVVSWSALMSGHVLNGDLKGSLSLFSEMGRQGIYPNEFTFSTNLKACGLLNALEKGLQ 128
Query: 567 IFRKMQTMGFKPDETTIYSILRACSLSECLRMGKEIHCYVFRHWKDWDLARTNALVDMYA 626
I GF CL++G E+ V N+LVDMY+
Sbjct: 129 IH------GF------------------CLKIGFEMMVEV-----------GNSLVDMYS 153
Query: 627 KCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKEALSLFEKMLLSMVK--PDSATF 684
KCG ++ + VF + + + SWN MI G G +AL F M + +K PD T
Sbjct: 154 KCGRINEAEKVFRRIVDRSLISWNAMIAGFVHAGYGSKALDTFGMMQEANIKERPDEFTL 213
Query: 685 TCVLSACSHSMLVEEGVQIFNSMSRDHLVEPEAEHYT-CVVDIYSRAGCLEEAYGFIQRM 743
T +L ACS + ++ G QI + R P + T +VD+Y + G L A ++
Sbjct: 214 TSLLKACSSTGMIYAGKQIHGFLVRSGFHCPSSATITGSLVDLYVKCGYLFSARKAFDQI 273
Query: 744 PMEPTAIAWKAFLAG 758
E T I+W + + G
Sbjct: 274 -KEKTMISWSSLILG 287
Score = 99.0 bits (245), Expect = 1e-20
Identities = 57/176 (32%), Positives = 97/176 (54%), Gaps = 2/176 (1%)
Query: 583 IYSILRACSLSECLRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMP 642
+ SILR C+ G ++HCY+ + +L +N L+DMY KC ++ VFD MP
Sbjct: 9 LVSILRVCTRKGLSDQGGQVHCYLLKSGSGLNLITSNYLIDMYCKCREPLMAYKVFDSMP 68
Query: 643 IKDVFSWNTMIFANGMHGNGKEALSLFEKMLLSMVKPDSATFTCVLSACSHSMLVEEGVQ 702
++V SW+ ++ + ++G+ K +LSLF +M + P+ TF+ L AC +E+G+Q
Sbjct: 69 ERNVVSWSALMSGHVLNGDLKGSLSLFSEMGRQGIYPNEFTFSTNLKACGLLNALEKGLQ 128
Query: 703 IFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEPTAIAWKAFLAG 758
I + E E +VD+YS+ G + EA +R+ ++ + I+W A +AG
Sbjct: 129 IHGFCLKIGF-EMMVEVGNSLVDMYSKCGRINEAEKVFRRI-VDRSLISWNAMIAG 182
Score = 31.2 bits (69), Expect = 2.9
Identities = 23/104 (22%), Positives = 48/104 (46%), Gaps = 3/104 (2%)
Query: 18 HGLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRC-GVMSDVS 76
HGL ++++I+ I+PD+ ++AV AC+ S + ++ G+ V
Sbjct: 392 HGLGKKSVRIFYEMLRHNIEPDEVCYLAVLSACSHSGMIKEGEELFSKLLETHGIKPRVE 451
Query: 77 IGNAFIHAYGKCKCVEGARRVFDDL-VARDVVTWNS-LSACYVN 118
+ G+ ++ A+ + D + + +V W + LS C V+
Sbjct: 452 HYACVVDLLGRAGRLKEAKHLIDTMPIKPNVGIWQTLLSLCRVH 495
>At5g13270 putative protein
Length = 752
Score = 431 bits (1109), Expect = e-121
Identities = 238/727 (32%), Positives = 384/727 (52%), Gaps = 51/727 (7%)
Query: 231 REMVLDGVKPDPVTVSCILSACSDLQDLKSGKAIHGFALKHGMVEN--VFVSNALVNLYE 288
+EM GV + C+ AC +L+ L G+ +H ++ G +EN V + N ++ +Y
Sbjct: 72 QEMDKAGVSVSSYSYQCLFEACRELRSLSHGRLLHD-RMRMG-IENPSVLLQNCVLQMYC 129
Query: 289 SCLCVREAQAVFDLMPHRNVITWNSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSS 348
C + +A +FD M N ++ ++ S Y G K + +F M +G KP ++
Sbjct: 130 ECRSLEDADKLFDEMSELNAVSRTTMISAYAEQGILDKAVGLFSGMLASGDKPPSSMYTT 189
Query: 349 ILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPHRN 408
+L + + L G+ IH ++ G+ + + T +VN+Y C + A+ VFD M +
Sbjct: 190 LLKSLVNPRALDFGRQIHAHVIRAGLCSNTSIETGIVNMYVKCGWLVGAKRVFDQMAVKK 249
Query: 409 VVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHACSDLQDLKSGKVIHG 468
V L Y G + L +F ++V GV+ D +L AC+ L++L GK IH
Sbjct: 250 PVACTGLMVGYTQAGRARDALKLFVDLVTEGVEWDSFVFSVVLKACASLEELNLGKQIHA 309
Query: 469 FAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREVASWNGILTAYFTNKEYEK 528
+ G+ +V V L+ Y KC +E
Sbjct: 310 CVAKLGLESEVSVGTPLVDFYIKC-------------------------------SSFES 338
Query: 529 GLYMFSQMNRDEVKADEITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKPDETTIY-SIL 587
F ++ + ++++WS +I G + S+ EEA++ F+ +++ + Y SI
Sbjct: 339 ACRAFQEIR----EPNDVSWSAIISGYCQMSQFEEAVKTFKSLRSKNASILNSFTYTSIF 394
Query: 588 RACSLSECLRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIKDVF 647
+ACS+ +G ++H + +AL+ MY+KCG L + VF+ M D+
Sbjct: 395 QACSVLADCNIGGQVHADAIKRSLIGSQYGESALITMYSKCGCLDDANEVFESMDNPDIV 454
Query: 648 SWNTMIFANGMHGNGKEALSLFEKMLLSMVKPDSATFTCVLSACSHSMLVEEGVQIFNSM 707
+W I + +GN EAL LFEKM+ +KP+S TF VL+ACSH+ LVE+G ++M
Sbjct: 455 AWTAFISGHAYYGNASEALRLFEKMVSCGMKPNSVTFIAVLTACSHAGLVEQGKHCLDTM 514
Query: 708 SRDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEPTAIAWKAFLAGCRVYKNVEL 767
R + V P +HY C++DIY+R+G L+EA F++ MP EP A++WK FL+GC +KN+EL
Sbjct: 515 LRKYNVAPTIDHYDCMIDIYARSGLLDEALKFMKNMPFEPDAMSWKCFLSGCWTHKNLEL 574
Query: 768 AKISAKKLFEIDPNGSANYVTLFNILVTAKLWSEASKIRKLMKERGITKTPGCSWFHVGN 827
+I+ ++L ++DP +A YV FN+ A W EA+++ KLM ER + K CSW
Sbjct: 575 GEIAGEELRQLDPEDTAGYVLPFNLYTWAGKWEEAAEMMKLMNERMLKKELSCSWIQEKG 634
Query: 828 RVHTFVAGDKSNMESDKIYNFLDELFAKIKAAGYKPDTDYVLHDIDQEEKAESLCNHSEK 887
++H F+ GDK + ++ +IY L E ++ + + E+ E L +HSE+
Sbjct: 635 KIHRFIVGDKHHPQTQEIYEKLKEFDGFMEGD---------MFQCNMTERREQLLDHSER 685
Query: 888 LAVAFGILNLNGQ--STIRVFKNLRICGDCHNAIKYMSNVVGVTIVVRDSLRFHHFKNGN 945
LA+AFG+++++G + I+VFKNLR C DCH K++S V G IV+RDS RFHHFK G
Sbjct: 686 LAIAFGLISVHGNAPAPIKVFKNLRACPDCHEFAKHVSLVTGHEIVIRDSRRFHHFKEGK 745
Query: 946 CSCKDFW 952
CSC D+W
Sbjct: 746 CSCNDYW 752
Score = 203 bits (516), Expect = 4e-52
Identities = 131/516 (25%), Positives = 240/516 (46%), Gaps = 5/516 (0%)
Query: 18 HGLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMS-DVS 76
H NEA + G+ + + +AC R + HD R G+ + V
Sbjct: 61 HRKLNEAFEFLQEMDKAGVSVSSYSYQCLFEACRELRSLSHGRLLHD-RMRMGIENPSVL 119
Query: 77 IGNAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNK 136
+ N + Y +C+ +E A ++FD++ + V+ ++ + Y G + + +F M +
Sbjct: 120 LQNCVLQMYCECRSLEDADKLFDEMSELNAVSRTTMISAYAEQGILDKAVGLFSGMLASG 179
Query: 137 VKANPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQ 196
K +++L + + L G++IH V+R G+ + + + VN Y KC + A+
Sbjct: 180 DKPPSSMYTTLLKSLVNPRALDFGRQIHAHVIRAGLCSNTSIETGIVNMYVKCGWLVGAK 239
Query: 197 TVFDLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQ 256
VFD M + V L Y G + L +F ++V +GV+ D S +L AC+ L+
Sbjct: 240 RVFDQMAVKKPVACTGLMVGYTQAGRARDALKLFVDLVTEGVEWDSFVFSVVLKACASLE 299
Query: 257 DLKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLAS 316
+L GK IH K G+ V V LV+ Y C A F + N ++W+++ S
Sbjct: 300 ELNLGKQIHACVAKLGLESEVSVGTPLVDFYIKCSSFESACRAFQEIREPNDVSWSAIIS 359
Query: 317 CYVNCGFPQKGLNVFREM-GLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMV 375
Y ++ + F+ + N + +SI ACS L D G +H A+K ++
Sbjct: 360 GYCQMSQFEEAVKTFKSLRSKNASILNSFTYTSIFQACSVLADCNIGGQVHADAIKRSLI 419
Query: 376 EDVFVCTALVNLYANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREM 435
+ +AL+ +Y+ C C+ +A VF+ M + ++V W + S + G + L +F +M
Sbjct: 420 GSQYGESALITMYSKCGCLDDANEVFESMDNPDIVAWTAFISGHAYYGNASEALRLFEKM 479
Query: 436 VLNGVKPDLVTMLSILHACSDLQDLKSGK-VIHGFAVRHGMVEDVFVCNALLSLYAKCVC 494
V G+KP+ VT +++L ACS ++ GK + ++ + + + ++ +YA+
Sbjct: 480 VSCGMKPNSVTFIAVLTACSHAGLVEQGKHCLDTMLRKYNVAPTIDHYDCMIDIYARSGL 539
Query: 495 VREAQVVFDLIPHR-EVASWNGILTAYFTNKEYEKG 529
+ EA +P + SW L+ +T+K E G
Sbjct: 540 LDEALKFMKNMPFEPDAMSWKCFLSGCWTHKNLELG 575
Score = 178 bits (451), Expect = 1e-44
Identities = 122/540 (22%), Positives = 229/540 (41%), Gaps = 36/540 (6%)
Query: 124 QGLNVFRKMGLNKVKANPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFV 183
+ ++M V + + + C +L+ L G+ +H + V + + +
Sbjct: 66 EAFEFLQEMDKAGVSVSSYSYQCLFEACRELRSLSHGRLLHDRMRMGIENPSVLLQNCVL 125
Query: 184 NFYAKCLCVREAQTVFDLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPV 243
Y +C + +A +FD M + V+ ++ S Y G K + +F M+ G KP
Sbjct: 126 QMYCECRSLEDADKLFDEMSELNAVSRTTMISAYAEQGILDKAVGLFSGMLASGDKPPSS 185
Query: 244 TVSCILSACSDLQDLKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLM 303
+ +L + + + L G+ IH ++ G+ N + +VN+Y C + A+ VFD M
Sbjct: 186 MYTTLLKSLVNPRALDFGRQIHAHVIRAGLCSNTSIETGIVNMYVKCGWLVGAKRVFDQM 245
Query: 304 PHRNVITWNSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGK 363
+ + L Y G + L +F ++ GV+ D S +L AC+ L++L GK
Sbjct: 246 AVKKPVACTGLMVGYTQAGRARDALKLFVDLVTEGVEWDSFVFSVVLKACASLEELNLGK 305
Query: 364 TIHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCG 423
IH K G+ +V V T LV+ Y C A F + N V+W+++ S Y
Sbjct: 306 QIHACVAKLGLESEVSVGTPLVDFYIKCSSFESACRAFQEIREPNDVSWSAIISGYCQMS 365
Query: 424 FPQKGLNVFREM-VLNGVKPDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGMVEDVFVC 482
++ + F+ + N + T SI ACS L D G +H A++ ++ +
Sbjct: 366 QFEEAVKTFKSLRSKNASILNSFTYTSIFQACSVLADCNIGGQVHADAIKRSLIGSQYGE 425
Query: 483 NALLSLYAKCVCVREAQVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVK 542
+AL+++Y+KC C+ +A VF+ + + ++ +W ++ + + L +F +M +K
Sbjct: 426 SALITMYSKCGCLDDANEVFESMDNPDIVAWTAFISGHAYYGNASEALRLFEKMVSCGMK 485
Query: 543 ADEITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKPDETTIYSILRACSLSECLRMGKEI 602
+ +T+ V+ C +E+ + TM K Y++ +C
Sbjct: 486 PNSVTFIAVLTACSHAGLVEQGKHC---LDTMLRK------YNVAPTIDHYDC------- 529
Query: 603 HCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIK-DVFSWNTMIFANGMHGN 661
++D+YA+ G L + MP + D SW + H N
Sbjct: 530 ------------------MIDIYARSGLLDEALKFMKNMPFEPDAMSWKCFLSGCWTHKN 571
Score = 30.0 bits (66), Expect = 6.5
Identities = 19/87 (21%), Positives = 35/87 (39%), Gaps = 1/87 (1%)
Query: 650 NTMIFANGMHGNGKEALSLFEKMLLSMVKPDSATFTCVLSACSHSMLVEEGVQIFNSMSR 709
N + + H EA ++M + V S ++ C+ AC + G ++ + R
Sbjct: 52 NLHLVSLSKHRKLNEAFEFLQEMDKAGVSVSSYSYQCLFEACRELRSLSHG-RLLHDRMR 110
Query: 710 DHLVEPEAEHYTCVVDIYSRAGCLEEA 736
+ P CV+ +Y LE+A
Sbjct: 111 MGIENPSVLLQNCVLQMYCECRSLEDA 137
>At1g71420 hypothetical protein
Length = 745
Score = 429 bits (1103), Expect = e-120
Identities = 250/727 (34%), Positives = 391/727 (53%), Gaps = 64/727 (8%)
Query: 246 SCILSACSDLQDLKSGKAIHGFALKHGMV--ENVFVSNALVNLYESCLCVREAQAVFDLM 303
+ + AC++ ++L G +H L H +NV ++N L+N+Y C + A+ VFD M
Sbjct: 63 AALFQACAEQRNLLDGINLHHHMLSHPYCYSQNVILANFLINMYAKCGNILYARQVFDTM 122
Query: 304 PHRNVITWNSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGK 363
P RNV++W +L + YV G Q+G +F M L+ P+ +SS+L +C + GK
Sbjct: 123 PERNVVSWTALITGYVQAGNEQEGFCLFSSM-LSHCFPNEFTLSSVLTSCRY----EPGK 177
Query: 364 TIHGFAVKHGMVEDVFVCTALVNLYANC---LCVREAQTVFDLMPHRNVVTWNSLSSCYV 420
+HG A+K G+ ++V A++++Y C EA TVF+ + +N+VTWNS+ + +
Sbjct: 178 QVHGLALKLGLHCSIYVANAVISMYGRCHDGAAAYEAWTVFEAIKFKNLVTWNSMIAAFQ 237
Query: 421 NCGFPQKGLNVFREMVLNGVKPDLVTMLSILHACSDLQDLKSGKV------IHGFAVRHG 474
C +K + VF M +GV D T+L+I + DL +V +H V+ G
Sbjct: 238 CCNLGKKAIGVFMRMHSDGVGFDRATLLNICSSLYKSSDLVPNEVSKCCLQLHSLTVKSG 297
Query: 475 MVEDVFVCNALLSLYAKCV-CVREAQVVFDLIPH-REVASWNGILTAYFTNKEYEKGLYM 532
+V V AL+ +Y++ + + +F + H R++ +WNGI+TA F + E+ +++
Sbjct: 298 LVTQTEVATALIKVYSEMLEDYTDCYKLFMEMSHCRDIVAWNGIITA-FAVYDPERAIHL 356
Query: 533 FSQMNRDEVKADEITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKPDETTIYSILRACSL 592
F Q+ ++++ PD T S+L+AC+
Sbjct: 357 FGQLRQEKLS-----------------------------------PDWYTFSSVLKACAG 381
Query: 593 SECLRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTM 652
R IH V + D N+L+ YAKCG L L VFD M +DV SWN+M
Sbjct: 382 LVTARHALSIHAQVIKGGFLADTVLNNSLIHAYAKCGSLDLCMRVFDDMDSRDVVSWNSM 441
Query: 653 IFANGMHGNGKEALSLFEKMLLSMVKPDSATFTCVLSACSHSMLVEEGVQIFNSMSRDHL 712
+ A +HG L +F+KM + PDSATF +LSACSH+ VEEG++IF SM
Sbjct: 442 LKAYSLHGQVDSILPVFQKM---DINPDSATFIALLSACSHAGRVEEGLRIFRSMFEKPE 498
Query: 713 VEPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEPTAIAWKAFLAGCRVYKNVELAKISA 772
P+ HY CV+D+ SRA EA I++MPM+P A+ W A L CR + N L K++A
Sbjct: 499 TLPQLNHYACVIDMLSRAERFAEAEEVIKQMPMDPDAVVWIALLGSCRKHGNTRLGKLAA 558
Query: 773 KKLFE-IDPNGSANYVTLFNILVTAKLWSEASKIRKLMKERGITKTPGCSWFHVGNRVHT 831
KL E ++P S +Y+ + NI ++EA+ K M+ + K P SW +GN+VH
Sbjct: 559 DKLKELVEPTNSMSYIQMSNIYNAEGSFNEANLSIKEMETWRVRKEPDLSWTEIGNKVHE 618
Query: 832 FVAGDKSNMESDKIYNFLDELFAKIKAAGYKPDTDYVLHDI-DQEEKAESLCNHSEKLAV 890
F +G + + + +Y L L + +K GY P+ DI D+E++ ++L +HSEKLA+
Sbjct: 619 FASGGRHRPDKEAVYRELKRLISWLKEMGYVPEMRSASQDIEDEEQEEDNLLHHSEKLAL 678
Query: 891 AFGILNLN-----GQSTIRVFKNLRICGDCHNAIKYMSNVVGVTIVVRDSLRFHHFKNGN 945
AF ++ G + I++ KN RIC DCHN +K S ++G I++RDS RFHHFK+ +
Sbjct: 679 AFAVMEGRKSSDCGVNLIQIMKNTRICIDCHNFMKLASKLLGKEILMRDSNRFHHFKDSS 738
Query: 946 CSCKDFW 952
CSC D+W
Sbjct: 739 CSCNDYW 745
Score = 218 bits (556), Expect = 1e-56
Identities = 150/552 (27%), Positives = 267/552 (48%), Gaps = 58/552 (10%)
Query: 145 SSILPGCSDLQDLKSGKEIHGFVVRHGMV--EDVFVSSAFVNFYAKCLCVREAQTVFDLM 202
+++ C++ ++L G +H ++ H ++V +++ +N YAKC + A+ VFD M
Sbjct: 63 AALFQACAEQRNLLDGINLHHHMLSHPYCYSQNVILANFLINMYAKCGNILYARQVFDTM 122
Query: 203 PHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQDLKSGK 262
P R+VV+W +L + YV G Q+G +F M L P+ T+S +L++C + GK
Sbjct: 123 PERNVVSWTALITGYVQAGNEQEGFCLFSSM-LSHCFPNEFTLSSVLTSCR----YEPGK 177
Query: 263 AIHGFALKHGMVENVFVSNALVNLYESC---LCVREAQAVFDLMPHRNVITWNSLASCYV 319
+HG ALK G+ +++V+NA++++Y C EA VF+ + +N++TWNS+ + +
Sbjct: 178 QVHGLALKLGLHCSIYVANAVISMYGRCHDGAAAYEAWTVFEAIKFKNLVTWNSMIAAFQ 237
Query: 320 NCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDL------KSGKTIHGFAVKHG 373
C +K + VF M +GV D + +I + + DL K +H VK G
Sbjct: 238 CCNLGKKAIGVFMRMHSDGVGFDRATLLNICSSLYKSSDLVPNEVSKCCLQLHSLTVKSG 297
Query: 374 MVEDVFVCTALVNLYANCL-CVREAQTVFDLMPH-RNVVTWNSLSSCYVNCGFPQKGLNV 431
+V V TAL+ +Y+ L + +F M H R++V WN + + + P++ +++
Sbjct: 298 LVTQTEVATALIKVYSEMLEDYTDCYKLFMEMSHCRDIVAWNGIITAFAVYD-PERAIHL 356
Query: 432 FREMVLNGVKPDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAK 491
F ++ + PD T S+L AC+ L + IH ++ G + D + N+L+ YAK
Sbjct: 357 FGQLRQEKLSPDWYTFSSVLKACAGLVTARHALSIHAQVIKGGFLADTVLNNSLIHAYAK 416
Query: 492 CVCVREAQVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVV 551
C + VFD + R+V SWN +L AY + + + L +F +M ++ D T+ +
Sbjct: 417 CGSLDLCMRVFDDMDSRDVVSWNSMLKAYSLHGQVDSILPVFQKM---DINPDSATFIAL 473
Query: 552 IGGCVKNSRIEEAMEIFRKMQTMGFKPDETTIYSILRACSLSECLRMGKEIHCYVFRHWK 611
+ C R+EE + IFR M F+ ET AC
Sbjct: 474 LSACSHAGRVEEGLRIFRSM----FEKPETLPQLNHYAC--------------------- 508
Query: 612 DWDLARTNALVDMYAKCGGLSLSRNVFDMMPI-KDVFSWNTMIFANGMHGNGKEALSLFE 670
++DM ++ + + V MP+ D W ++ + HGN + +
Sbjct: 509 ---------VIDMLSRAERFAEAEEVIKQMPMDPDAVVWIALLGSCRKHGNTRLGKLAAD 559
Query: 671 KMLLSMVKPDSA 682
K L +V+P ++
Sbjct: 560 K-LKELVEPTNS 570
Score = 200 bits (509), Expect = 3e-51
Identities = 135/449 (30%), Positives = 223/449 (49%), Gaps = 29/449 (6%)
Query: 43 FMAVAKACAASRDALKVKQFHDDATR---CGVMSDVSIGNAFIHAYGKCKCVEGARRVFD 99
+ A+ +ACA R+ L H C +V + N I+ Y KC + AR+VFD
Sbjct: 62 YAALFQACAEQRNLLDGINLHHHMLSHPYC-YSQNVILANFLINMYAKCGNILYARQVFD 120
Query: 100 DLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKANPLTVSSILPGCSDLQDLKS 159
+ R+VV+W +L YV G Q+G +F M L+ N T+SS+L C +
Sbjct: 121 TMPERNVVSWTALITGYVQAGNEQEGFCLFSSM-LSHCFPNEFTLSSVLTSCR----YEP 175
Query: 160 GKEIHGFVVRHGMVEDVFVSSAFVNFYAKC---LCVREAQTVFDLMPHRDVVTWNSLSSC 216
GK++HG ++ G+ ++V++A ++ Y +C EA TVF+ + +++VTWNS+ +
Sbjct: 176 GKQVHGLALKLGLHCSIYVANAVISMYGRCHDGAAAYEAWTVFEAIKFKNLVTWNSMIAA 235
Query: 217 YVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQDL------KSGKAIHGFALK 270
+ C +K + VF M DGV D T+ I S+ DL K +H +K
Sbjct: 236 FQCCNLGKKAIGVFMRMHSDGVGFDRATLLNICSSLYKSSDLVPNEVSKCCLQLHSLTVK 295
Query: 271 HGMVENVFVSNALVNLYESCL-CVREAQAVFDLMPH-RNVITWNSLASCYVNCGFPQKGL 328
G+V V+ AL+ +Y L + +F M H R+++ WN + + + P++ +
Sbjct: 296 SGLVTQTEVATALIKVYSEMLEDYTDCYKLFMEMSHCRDIVAWNGIITAFAVYD-PERAI 354
Query: 329 NVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLY 388
++F ++ + PD SS+L AC+ L + +IH +K G + D + +L++ Y
Sbjct: 355 HLFGQLRQEKLSPDWYTFSSVLKACAGLVTARHALSIHAQVIKGGFLADTVLNNSLIHAY 414
Query: 389 ANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTML 448
A C + VFD M R+VV+WNS+ Y G L VF++M +N PD T +
Sbjct: 415 AKCGSLDLCMRVFDDMDSRDVVSWNSMLKAYSLHGQVDSILPVFQKMDIN---PDSATFI 471
Query: 449 SILHACSDLQDLKSGKVIHGFAVRHGMVE 477
++L ACS +G+V G + M E
Sbjct: 472 ALLSACS-----HAGRVEEGLRIFRSMFE 495
Score = 140 bits (353), Expect = 3e-33
Identities = 104/380 (27%), Positives = 183/380 (47%), Gaps = 32/380 (8%)
Query: 60 KQFHDDATRCGVMSDVSIGNAFIHAYGKCK---CVEGARRVFDDLVARDVVTWNSLSACY 116
KQ H A + G+ + + NA I YG+C A VF+ + +++VTWNS+ A +
Sbjct: 177 KQVHGLALKLGLHCSIYVANAVISMYGRCHDGAAAYEAWTVFEAIKFKNLVTWNSMIAAF 236
Query: 117 VNCGFPQQGLNVFRKM---GLNKVKANPLTVSSILPGCSDLQDLKSGK---EIHGFVVRH 170
C ++ + VF +M G+ +A L + S L SDL + K ++H V+
Sbjct: 237 QCCNLGKKAIGVFMRMHSDGVGFDRATLLNICSSLYKSSDLVPNEVSKCCLQLHSLTVKS 296
Query: 171 GMVEDVFVSSAFVNFYAKCL-CVREAQTVFDLMPH-RDVVTWNSLSSCYVNCGFPQKGLN 228
G+V V++A + Y++ L + +F M H RD+V WN + + + P++ ++
Sbjct: 297 GLVTQTEVATALIKVYSEMLEDYTDCYKLFMEMSHCRDIVAWNGIITAFAVYD-PERAIH 355
Query: 229 VFREMVLDGVKPDPVTVSCILSACSDLQDLKSGKAIHGFALKHGMVENVFVSNALVNLYE 288
+F ++ + + PD T S +L AC+ L + +IH +K G + + ++N+L++ Y
Sbjct: 356 LFGQLRQEKLSPDWYTFSSVLKACAGLVTARHALSIHAQVIKGGFLADTVLNNSLIHAYA 415
Query: 289 SCLCVREAQAVFDLMPHRNVITWNSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSS 348
C + VFD M R+V++WNS+ Y G L VF++M +N PD +
Sbjct: 416 KCGSLDLCMRVFDDMDSRDVVSWNSMLKAYSLHGQVDSILPVFQKMDIN---PDSATFIA 472
Query: 349 ILPACSQLKDLKSGKTIHGFAVKHGMVE--------DVFVCTALVNLYANCLCVREAQTV 400
+L ACS +G+ G + M E + + C ++++ + EA+ V
Sbjct: 473 LLSACSH-----AGRVEEGLRIFRSMFEKPETLPQLNHYAC--VIDMLSRAERFAEAEEV 525
Query: 401 FDLMP-HRNVVTWNS-LSSC 418
MP + V W + L SC
Sbjct: 526 IKQMPMDPDAVVWIALLGSC 545
Score = 87.8 bits (216), Expect = 3e-17
Identities = 61/199 (30%), Positives = 92/199 (45%), Gaps = 6/199 (3%)
Query: 21 PNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSIGNA 80
P AI ++ R + PD F +V KACA A H + G ++D + N+
Sbjct: 350 PERAIHLFGQLRQEKLSPDWYTFSSVLKACAGLVTARHALSIHAQVIKGGFLADTVLNNS 409
Query: 81 FIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKAN 140
IHAY KC ++ RVFDD+ +RDVV+WNS+ Y G L VF+KM +N A
Sbjct: 410 LIHAYAKCGSLDLCMRVFDDMDSRDVVSWNSMLKAYSLHGQVDSILPVFQKMDINPDSA- 468
Query: 141 PLTVSSILPGCSDLQDLKSGKEI-HGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVF 199
T ++L CS ++ G I + + + + ++ ++ EA+ V
Sbjct: 469 --TFIALLSACSHAGRVEEGLRIFRSMFEKPETLPQLNHYACVIDMLSRAERFAEAEEVI 526
Query: 200 DLMP-HRDVVTWNS-LSSC 216
MP D V W + L SC
Sbjct: 527 KQMPMDPDAVVWIALLGSC 545
>At3g02330 hypothetical protein
Length = 861
Score = 428 bits (1100), Expect = e-120
Identities = 253/824 (30%), Positives = 427/824 (51%), Gaps = 21/824 (2%)
Query: 35 GIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSIGNAFIHAYGKCKCVEGA 94
G +P V + + SRD + D + DV N I+ Y K + A
Sbjct: 36 GFRPTTFVLNCLLQVYTNSRDFVSASMVFDKMP----LRDVVSWNKMINGYSKSNDMFKA 91
Query: 95 RRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKANPLTVSSILPGCSDL 154
F+ + RDVV+WNS+ + Y+ G + + VF MG ++ + T + IL CS L
Sbjct: 92 NSFFNMMPVRDVVSWNSMLSGYLQNGESLKSIEVFVDMGREGIEFDGRTFAIILKVCSFL 151
Query: 155 QDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTWNSLS 214
+D G +IHG VVR G DV +SA ++ YAK E+ VF +P ++ V+W+++
Sbjct: 152 EDTSLGMQIHGIVVRVGCDTDVVAASALLDMYAKGKRFVESLRVFQGIPEKNSVSWSAII 211
Query: 215 SCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQDLKSGKAIHGFALKHGMV 274
+ V L F+EM + +L +C+ L +L+ G +H ALK
Sbjct: 212 AGCVQNNLLSLALKFFKEMQKVNAGVSQSIYASVLRSCAALSELRLGGQLHAHALKSDFA 271
Query: 275 ENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASCYVNCGFPQKGLNVFREM 334
+ V A +++Y C +++AQ +FD + N ++N++ + Y K L +F +
Sbjct: 272 ADGIVRTATLDMYAKCDNMQDAQILFDNSENLNRQSYNAMITGYSQEEHGFKALLLFHRL 331
Query: 335 GLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANCLCV 394
+G+ D +++S + AC+ +K L G I+G A+K + DV V A +++Y C +
Sbjct: 332 MSSGLGFDEISLSGVFRACALVKGLSEGLQIYGLAIKSSLSLDVCVANAAIDMYGKCQAL 391
Query: 395 REAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHAC 454
EA VFD M R+ V+WN++ + + G + L +F M+ + ++PD T SIL AC
Sbjct: 392 AEAFRVFDEMRRRDAVSWNAIIAAHEQNGKGYETLFLFVSMLRSRIEPDEFTFGSILKAC 451
Query: 455 SDLQDLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREVASWN 514
+ L G IH V+ GM + V +L+ +Y+KC + EA+ + R S
Sbjct: 452 TG-GSLGYGMEIHSSIVKSGMASNSSVGCSLIDMYSKCGMIEEAEKIHSRFFQRANVSG- 509
Query: 515 GILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGGCVKNSRIEEAMEIFRKMQTM 574
T +E EK M+ ++ ++W+ +I G V + E+A +F +M M
Sbjct: 510 -------TMEELEK-------MHNKRLQEMCVSWNSIISGYVMKEQSEDAQMLFTRMMEM 555
Query: 575 GFKPDETTIYSILRACSLSECLRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGGLSLS 634
G PD+ T ++L C+ +GK+IH V + D+ + LVDMY+KCG L S
Sbjct: 556 GITPDKFTYATVLDTCANLASAGLGKQIHAQVIKKELQSDVYICSTLVDMYSKCGDLHDS 615
Query: 635 RNVFDMMPIKDVFSWNTMIFANGMHGNGKEALSLFEKMLLSMVKPDSATFTCVLSACSHS 694
R +F+ +D +WN MI HG G+EA+ LFE+M+L +KP+ TF +L AC+H
Sbjct: 616 RLMFEKSLRRDFVTWNAMICGYAHHGKGEEAIQLFERMILENIKPNHVTFISILRACAHM 675
Query: 695 MLVEEGVQIFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEPTAIAWKA 754
L+++G++ F M RD+ ++P+ HY+ +VDI ++G ++ A I+ MP E + W+
Sbjct: 676 GLIDKGLEYFYMMKRDYGLDPQLPHYSNMVDILGKSGKVKRALELIREMPFEADDVIWRT 735
Query: 755 FLAGCRVYK-NVELAKISAKKLFEIDPNGSANYVTLFNILVTAKLWSEASKIRKLMKERG 813
L C +++ NVE+A+ + L +DP S+ Y L N+ A +W + S +R+ M+
Sbjct: 736 LLGVCTIHRNNVEVAEEATAALLRLDPQDSSAYTLLSNVYADAGMWEKVSDLRRNMRGFK 795
Query: 814 ITKTPGCSWFHVGNRVHTFVAGDKSNMESDKIYNFLDELFAKIK 857
+ K PGCSW + + +H F+ GDK++ ++IY L +++++K
Sbjct: 796 LKKEPGCSWVELKDELHVFLVGDKAHPRWEEIYEELGLIYSEMK 839
Score = 249 bits (637), Expect = 4e-66
Identities = 169/691 (24%), Positives = 318/691 (45%), Gaps = 56/691 (8%)
Query: 16 IPHGLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDV 75
+ +G ++I+++ GI+ D F + K C+ D Q H R G +DV
Sbjct: 114 LQNGESLKSIEVFVDMGREGIEFDGRTFAIILKVCSFLEDTSLGMQIHGIVVRVGCDTDV 173
Query: 76 SIGNAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLN 135
+A + Y K K + RVF + ++ V+W+++ A V L F++M
Sbjct: 174 VAASALLDMYAKGKRFVESLRVFQGIPEKNSVSWSAIIAGCVQNNLLSLALKFFKEMQKV 233
Query: 136 KVKANPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREA 195
+ +S+L C+ L +L+ G ++H ++ D V +A ++ YAKC +++A
Sbjct: 234 NAGVSQSIYASVLRSCAALSELRLGGQLHAHALKSDFAADGIVRTATLDMYAKCDNMQDA 293
Query: 196 QTVFDLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDL 255
Q +FD + + ++N++ + Y K L +F ++ G+ D +++S + AC+ +
Sbjct: 294 QILFDNSENLNRQSYNAMITGYSQEEHGFKALLLFHRLMSSGLGFDEISLSGVFRACALV 353
Query: 256 QDLKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLA 315
+ L G I+G A+K + +V V+NA +++Y C + EA VFD M R+ ++WN++
Sbjct: 354 KGLSEGLQIYGLAIKSSLSLDVCVANAAIDMYGKCQALAEAFRVFDEMRRRDAVSWNAII 413
Query: 316 SCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMV 375
+ + G + L +F M + ++PD SIL AC+ L G IH VK GM
Sbjct: 414 AAHEQNGKGYETLFLFVSMLRSRIEPDEFTFGSILKACTG-GSLGYGMEIHSSIVKSGMA 472
Query: 376 EDVFVCTALVNLYANCLCVREAQTVFDLMPHR-NV-------------------VTWNSL 415
+ V +L+++Y+ C + EA+ + R NV V+WNS+
Sbjct: 473 SNSSVGCSLIDMYSKCGMIEEAEKIHSRFFQRANVSGTMEELEKMHNKRLQEMCVSWNSI 532
Query: 416 SSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGM 475
S YV + +F M+ G+ PD T ++L C++L GK IH ++ +
Sbjct: 533 ISGYVMKEQSEDAQMLFTRMMEMGITPDKFTYATVLDTCANLASAGLGKQIHAQVIKKEL 592
Query: 476 VEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQ 535
DV++C+ L+ +Y+KC + +++++F+ R+ +WN ++ Y + + E+ + +F +
Sbjct: 593 QSDVYICSTLVDMYSKCGDLHDSRLMFEKSLRRDFVTWNAMICGYAHHGKGEEAIQLFER 652
Query: 536 MNRDEVKADEITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKPDETTIYSILRACSLSEC 595
M + +K + +T+ ++ C I++ +E F Y + R L
Sbjct: 653 MILENIKPNHVTFISILRACAHMGLIDKGLEYF---------------YMMKRDYGL--- 694
Query: 596 LRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIK-DVFSWNTMIF 654
D L + +VD+ K G + + + MP + D W T++
Sbjct: 695 ----------------DPQLPHYSNMVDILGKSGKVKRALELIREMPFEADDVIWRTLLG 738
Query: 655 ANGMHGNGKEALSLFEKMLLSMVKPDSATFT 685
+H N E LL + DS+ +T
Sbjct: 739 VCTIHRNNVEVAEEATAALLRLDPQDSSAYT 769
Score = 235 bits (600), Expect = 8e-62
Identities = 176/678 (25%), Positives = 307/678 (44%), Gaps = 59/678 (8%)
Query: 151 CSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDV--- 207
C+ L+ GK+ H ++ G FV + + Y A VFD MP RDV
Sbjct: 16 CAKQGALELGKQAHAHMIISGFRPTTFVLNCLLQVYTNSRDFVSASMVFDKMPLRDVVSW 75
Query: 208 ----------------------------VTWNSLSSCYVNCGFPQKGLNVFREMVLDGVK 239
V+WNS+ S Y+ G K + VF +M +G++
Sbjct: 76 NKMINGYSKSNDMFKANSFFNMMPVRDVVSWNSMLSGYLQNGESLKSIEVFVDMGREGIE 135
Query: 240 PDPVTVSCILSACSDLQDLKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAV 299
D T + IL CS L+D G IHG ++ G +V ++AL+++Y E+ V
Sbjct: 136 FDGRTFAIILKVCSFLEDTSLGMQIHGIVVRVGCDTDVVAASALLDMYAKGKRFVESLRV 195
Query: 300 FDLMPHRNVITWNSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDL 359
F +P +N ++W+++ + V L F+EM +S+L +C+ L +L
Sbjct: 196 FQGIPEKNSVSWSAIIAGCVQNNLLSLALKFFKEMQKVNAGVSQSIYASVLRSCAALSEL 255
Query: 360 KSGKTIHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPHRNVVTWNSLSSCY 419
+ G +H A+K D V TA +++YA C +++AQ +FD + N ++N++ + Y
Sbjct: 256 RLGGQLHAHALKSDFAADGIVRTATLDMYAKCDNMQDAQILFDNSENLNRQSYNAMITGY 315
Query: 420 VNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGMVEDV 479
K L +F ++ +G+ D +++ + AC+ ++ L G I+G A++ + DV
Sbjct: 316 SQEEHGFKALLLFHRLMSSGLGFDEISLSGVFRACALVKGLSEGLQIYGLAIKSSLSLDV 375
Query: 480 FVCNALLSLYAKCVCVREAQVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQMNRD 539
V NA + +Y KC + EA VFD + R+ SWN I+ A+ N + + L++F M R
Sbjct: 376 CVANAAIDMYGKCQALAEAFRVFDEMRRRDAVSWNAIIAAHEQNGKGYETLFLFVSMLRS 435
Query: 540 EVKADEITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKPDETTIYSILRACSLSECLRMG 599
++ DE T+ ++ C S + MEI + G + + S++ S +
Sbjct: 436 RIEPDEFTFGSILKACTGGS-LGYGMEIHSSIVKSGMASNSSVGCSLIDMYSKCGMIEEA 494
Query: 600 KEIHCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMH 659
++IH F+ A + ++ K L + SWN++I M
Sbjct: 495 EKIHSRFFQR------ANVSGTMEELEKMHNKRLQ---------EMCVSWNSIISGYVMK 539
Query: 660 GNGKEALSLFEKMLLSMVKPDSATFTCVLSACSHSMLVEEGVQIFNSMSRDHLVEPEAEH 719
++A LF +M+ + PD T+ VL C++ G QI + + L +++
Sbjct: 540 EQSEDAQMLFTRMMEMGITPDKFTYATVLDTCANLASAGLGKQIHAQVIKKEL---QSDV 596
Query: 720 YTC--VVDIYSRAGCLEEAYGFIQRMPMEPTAIAWKAFLAGCRVYKNVELAKISAKKLFE 777
Y C +VD+YS+ G L ++ ++ + + W A + G + E A +LFE
Sbjct: 597 YICSTLVDMYSKCGDLHDSRLMFEK-SLRRDFVTWNAMICGYAHHGKGE----EAIQLFE 651
Query: 778 --IDPNGSANYVTLFNIL 793
I N N+VT +IL
Sbjct: 652 RMILENIKPNHVTFISIL 669
Score = 209 bits (531), Expect = 8e-54
Identities = 159/579 (27%), Positives = 258/579 (44%), Gaps = 91/579 (15%)
Query: 252 CSDLQDLKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNV--- 308
C+ L+ GK H + G FV N L+ +Y + A VFD MP R+V
Sbjct: 16 CAKQGALELGKQAHAHMIISGFRPTTFVLNCLLQVYTNSRDFVSASMVFDKMPLRDVVSW 75
Query: 309 ----------------------------ITWNSLASCYVNCGFPQKGLNVFREMGLNGVK 340
++WNS+ S Y+ G K + VF +MG G++
Sbjct: 76 NKMINGYSKSNDMFKANSFFNMMPVRDVVSWNSMLSGYLQNGESLKSIEVFVDMGREGIE 135
Query: 341 PDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQTV 400
D + IL CS L+D G IHG V+ G DV +AL+++YA E+ V
Sbjct: 136 FDGRTFAIILKVCSFLEDTSLGMQIHGIVVRVGCDTDVVAASALLDMYAKGKRFVESLRV 195
Query: 401 FDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREM--VLNGVKPDLVTMLSILHACSDLQ 458
F +P +N V+W+++ + V L F+EM V GV + S+L +C+ L
Sbjct: 196 FQGIPEKNSVSWSAIIAGCVQNNLLSLALKFFKEMQKVNAGVSQSIYA--SVLRSCAALS 253
Query: 459 DLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREVASWNGILT 518
+L+ G +H A++ D V A L +YAKC +++AQ++FD + S+N ++T
Sbjct: 254 ELRLGGQLHAHALKSDFAADGIVRTATLDMYAKCDNMQDAQILFDNSENLNRQSYNAMIT 313
Query: 519 AYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKP 578
Y + K L +F ++ + DEI+ S V
Sbjct: 314 GYSQEEHGFKALLLFHRLMSSGLGFDEISLSGV--------------------------- 346
Query: 579 DETTIYSILRACSLSECLRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVF 638
RAC+L + L G +I+ + D+ NA +DMY KC L+ + VF
Sbjct: 347 --------FRACALVKGLSEGLQIYGLAIKSSLSLDVCVANAAIDMYGKCQALAEAFRVF 398
Query: 639 DMMPIKDVFSWNTMIFANGMHGNGKEALSLFEKMLLSMVKPDSATFTCVLSACSHSMLVE 698
D M +D SWN +I A+ +G G E L LF ML S ++PD TF +L AC+ L
Sbjct: 399 DEMRRRDAVSWNAIIAAHEQNGKGYETLFLFVSMLRSRIEPDEFTFGSILKACTGGSL-G 457
Query: 699 EGVQIFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEA----YGFIQRMPMEPT------ 748
G++I +S+ + + + + ++D+YS+ G +EEA F QR + T
Sbjct: 458 YGMEIHSSIVKSGMASNSSVGCS-LIDMYSKCGMIEEAEKIHSRFFQRANVSGTMEELEK 516
Query: 749 ---------AIAWKAFLAGCRVYKNVELAKISAKKLFEI 778
++W + ++G + + E A++ ++ E+
Sbjct: 517 MHNKRLQEMCVSWNSIISGYVMKEQSEDAQMLFTRMMEM 555
Score = 151 bits (382), Expect = 1e-36
Identities = 122/441 (27%), Positives = 188/441 (41%), Gaps = 78/441 (17%)
Query: 353 CSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPHRNV--- 409
C++ L+ GK H + G FV L+ +Y N A VFD MP R+V
Sbjct: 16 CAKQGALELGKQAHAHMIISGFRPTTFVLNCLLQVYTNSRDFVSASMVFDKMPLRDVVSW 75
Query: 410 ----------------------------VTWNSLSSCYVNCGFPQKGLNVFREMVLNGVK 441
V+WNS+ S Y+ G K + VF +M G++
Sbjct: 76 NKMINGYSKSNDMFKANSFFNMMPVRDVVSWNSMLSGYLQNGESLKSIEVFVDMGREGIE 135
Query: 442 PDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVV 501
D T IL CS L+D G IHG VR G DV +ALL +YAK E+ V
Sbjct: 136 FDGRTFAIILKVCSFLEDTSLGMQIHGIVVRVGCDTDVVAASALLDMYAKGKRFVESLRV 195
Query: 502 FDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGGCVKNSRI 561
F IP + SW+ I I GCV+N+ +
Sbjct: 196 FQGIPEKNSVSWSAI-----------------------------------IAGCVQNNLL 220
Query: 562 EEAMEIFRKMQTMGFKPDETTIYSILRACSLSECLRMGKEIHCYVFRHWKDWDLARTNAL 621
A++ F++MQ + ++ S+LR+C+ LR+G ++H + + D A
Sbjct: 221 SLALKFFKEMQKVNAGVSQSIYASVLRSCAALSELRLGGQLHAHALKSDFAADGIVRTAT 280
Query: 622 VDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKEALSLFEKMLLSMVKPDS 681
+DMYAKC + ++ +FD + S+N MI +G +AL LF +++ S + D
Sbjct: 281 LDMYAKCDNMQDAQILFDNSENLNRQSYNAMITGYSQEEHGFKALLLFHRLMSSGLGFDE 340
Query: 682 ATFTCVLSACSHSMLVEEGVQIF-----NSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEA 736
+ + V AC+ + EG+QI+ +S+S D V A +D+Y + L EA
Sbjct: 341 ISLSGVFRACALVKGLSEGLQIYGLAIKSSLSLDVCVANAA------IDMYGKCQALAEA 394
Query: 737 YGFIQRMPMEPTAIAWKAFLA 757
+ M A++W A +A
Sbjct: 395 FRVFDEM-RRRDAVSWNAIIA 414
>At4g02750 hypothetical protein
Length = 781
Score = 426 bits (1096), Expect = e-119
Identities = 248/754 (32%), Positives = 404/754 (52%), Gaps = 42/754 (5%)
Query: 206 DVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQDLKSGKAIH 265
D+ WN S Y+ G + L VF+ M + V+ + ++S L++G+
Sbjct: 63 DIKEWNVAISSYMRTGRCNEALRVFKRMP----RWSSVSYNGMISGY-----LRNGEFEL 113
Query: 266 GFALKHGMVENVFVS-NALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASCYVNCGFP 324
L M E VS N ++ Y + +A+ +F++MP R+V +WN++ S Y G
Sbjct: 114 ARKLFDEMPERDLVSWNVMIKGYVRNRNLGKARELFEIMPERDVCSWNTMLSGYAQNGCV 173
Query: 325 QKGLNVF------REMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDV 378
+VF ++ N + + S + AC K ++ + + G V+
Sbjct: 174 DDARSVFDRMPEKNDVSWNALLSAYVQNSKMEEACMLFKSRENWALVSWNCLLGGFVKKK 233
Query: 379 FVCTALVNLYANCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLN 438
+ EA+ FD M R+VV+WN++ + Y G + +F E +
Sbjct: 234 KIV--------------EARQFFDSMNVRDVVSWNTIITGYAQSGKIDEARQLFDESPVQ 279
Query: 439 GVKPDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREA 498
D+ T +++ + ++ + + V NA+L+ Y + + A
Sbjct: 280 ----DVFTWTAMVSGYIQNRMVEEARELFDKMPERNEVS----WNAMLAGYVQGERMEMA 331
Query: 499 QVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGGCVKN 558
+ +FD++P R V++WN ++T Y + + +F +M K D ++W+ +I G ++
Sbjct: 332 KELFDVMPCRNVSTWNTMITGYAQCGKISEAKNLFDKMP----KRDPVSWAAMIAGYSQS 387
Query: 559 SRIEEAMEIFRKMQTMGFKPDETTIYSILRACSLSECLRMGKEIHCYVFRHWKDWDLART 618
EA+ +F +M+ G + + ++ S L C+ L +GK++H + + +
Sbjct: 388 GHSFEALRLFVQMEREGGRLNRSSFSSALSTCADVVALELGKQLHGRLVKGGYETGCFVG 447
Query: 619 NALVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKEALSLFEKMLLSMVK 678
NAL+ MY KCG + + ++F M KD+ SWNTMI HG G+ AL FE M +K
Sbjct: 448 NALLLMYCKCGSIEEANDLFKEMAGKDIVSWNTMIAGYSRHGFGEVALRFFESMKREGLK 507
Query: 679 PDSATFTCVLSACSHSMLVEEGVQIFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYG 738
PD AT VLSACSH+ LV++G Q F +M++D+ V P ++HY C+VD+ RAG LE+A+
Sbjct: 508 PDDATMVAVLSACSHTGLVDKGRQYFYTMTQDYGVMPNSQHYACMVDLLGRAGLLEDAHN 567
Query: 739 FIQRMPMEPTAIAWKAFLAGCRVYKNVELAKISAKKLFEIDPNGSANYVTLFNILVTAKL 798
++ MP EP A W L RV+ N ELA+ +A K+F ++P S YV L N+ ++
Sbjct: 568 LMKNMPFEPDAAIWGTLLGASRVHGNTELAETAADKIFAMEPENSGMYVLLSNLYASSGR 627
Query: 799 WSEASKIRKLMKERGITKTPGCSWFHVGNRVHTFVAGDKSNMESDKIYNFLDELFAKIKA 858
W + K+R M+++G+ K PG SW + N+ HTF GD+ + E D+I+ FL+EL ++K
Sbjct: 628 WGDVGKLRVRMRDKGVKKVPGYSWIEIQNKTHTFSVGDEFHPEKDEIFAFLEELDLRMKK 687
Query: 859 AGYKPDTDYVLHDIDQEEKAESLCNHSEKLAVAFGILNLNGQSTIRVFKNLRICGDCHNA 918
AGY T VLHD+++EEK + HSE+LAVA+GI+ ++ IRV KNLR+C DCHNA
Sbjct: 688 AGYVSKTSVVLHDVEEEEKERMVRYHSERLAVAYGIMRVSSGRPIRVIKNLRVCEDCHNA 747
Query: 919 IKYMSNVVGVTIVVRDSLRFHHFKNGNCSCKDFW 952
IKYM+ + G I++RD+ RFHHFK+G+CSC D+W
Sbjct: 748 IKYMARITGRLIILRDNNRFHHFKDGSCSCGDYW 781
Score = 146 bits (368), Expect = 6e-35
Identities = 94/361 (26%), Positives = 176/361 (48%), Gaps = 14/361 (3%)
Query: 57 LKVKQFHDDATRCGVMSDVSIGNAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACY 116
++ +QF D + DV N I Y + ++ AR++FD+ +DV TW ++ + Y
Sbjct: 236 VEARQFFDSMN----VRDVVSWNTIITGYAQSGKIDEARQLFDESPVQDVFTWTAMVSGY 291
Query: 117 VNCGFPQQGLNVFRKMGLNKVKANPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDV 176
+ ++ +F KM + N ++ +++L G + ++ KE+ + +V
Sbjct: 292 IQNRMVEEARELFDKMP----ERNEVSWNAMLAGYVQGERMEMAKELFDVMP----CRNV 343
Query: 177 FVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLD 236
+ + YA+C + EA+ +FD MP RD V+W ++ + Y G + L +F +M +
Sbjct: 344 STWNTMITGYAQCGKISEAKNLFDKMPKRDPVSWAAMIAGYSQSGHSFEALRLFVQMERE 403
Query: 237 GVKPDPVTVSCILSACSDLQDLKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREA 296
G + + + S LS C+D+ L+ GK +HG +K G FV NAL+ +Y C + EA
Sbjct: 404 GGRLNRSSFSSALSTCADVVALELGKQLHGRLVKGGYETGCFVGNALLLMYCKCGSIEEA 463
Query: 297 QAVFDLMPHRNVITWNSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQL 356
+F M +++++WN++ + Y GF + L F M G+KPD M ++L ACS
Sbjct: 464 NDLFKEMAGKDIVSWNTMIAGYSRHGFGEVALRFFESMKREGLKPDDATMVAVLSACSHT 523
Query: 357 KDLKSGKT-IHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPHR-NVVTWNS 414
+ G+ + +G++ + +V+L + +A + MP + W +
Sbjct: 524 GLVDKGRQYFYTMTQDYGVMPNSQHYACMVDLLGRAGLLEDAHNLMKNMPFEPDAAIWGT 583
Query: 415 L 415
L
Sbjct: 584 L 584
Score = 128 bits (322), Expect = 1e-29
Identities = 130/528 (24%), Positives = 216/528 (40%), Gaps = 126/528 (23%)
Query: 79 NAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVK 138
N I Y + E AR++FD++ RD+V+WN + YV R L K +
Sbjct: 99 NGMISGYLRNGEFELARKLFDEMPERDLVSWNVMIKGYV------------RNRNLGKAR 146
Query: 139 ANPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTV 198
+ I+P DV + ++ YA+ CV +A++V
Sbjct: 147 E----LFEIMPE-----------------------RDVCSWNTMLSGYAQNGCVDDARSV 179
Query: 199 FDLMPHRDVVTWNSLSSCYV-----------------------NC---GFPQKGLNVFRE 232
FD MP ++ V+WN+L S YV NC GF +K V
Sbjct: 180 FDRMPEKNDVSWNALLSAYVQNSKMEEACMLFKSRENWALVSWNCLLGGFVKKKKIVEAR 239
Query: 233 MVLDGVK-PDPVTVSCILSAC---------------SDLQDLKSGKAIHGFALKHGMVE- 275
D + D V+ + I++ S +QD+ + A+ +++ MVE
Sbjct: 240 QFFDSMNVRDVVSWNTIITGYAQSGKIDEARQLFDESPVQDVFTWTAMVSGYIQNRMVEE 299
Query: 276 -----------NVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASCYVNC--- 321
N NA++ Y + A+ +FD+MP RNV TWN++ + Y C
Sbjct: 300 ARELFDKMPERNEVSWNAMLAGYVQGERMEMAKELFDVMPCRNVSTWNTMITGYAQCGKI 359
Query: 322 -----------------------GFPQKG-----LNVFREMGLNGVKPDPMAMSSILPAC 353
G+ Q G L +F +M G + + + SS L C
Sbjct: 360 SEAKNLFDKMPKRDPVSWAAMIAGYSQSGHSFEALRLFVQMEREGGRLNRSSFSSALSTC 419
Query: 354 SQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPHRNVVTWN 413
+ + L+ GK +HG VK G FV AL+ +Y C + EA +F M +++V+WN
Sbjct: 420 ADVVALELGKQLHGRLVKGGYETGCFVGNALLLMYCKCGSIEEANDLFKEMAGKDIVSWN 479
Query: 414 SLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHACSDLQDLKSGK-VIHGFAVR 472
++ + Y GF + L F M G+KPD TM+++L ACS + G+ +
Sbjct: 480 TMIAGYSRHGFGEVALRFFESMKREGLKPDDATMVAVLSACSHTGLVDKGRQYFYTMTQD 539
Query: 473 HGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHR-EVASWNGILTA 519
+G++ + ++ L + + +A + +P + A W +L A
Sbjct: 540 YGVMPNSQHYACMVDLLGRAGLLEDAHNLMKNMPFEPDAAIWGTLLGA 587
Score = 42.7 bits (99), Expect = 0.001
Identities = 34/146 (23%), Positives = 65/146 (44%), Gaps = 10/146 (6%)
Query: 612 DWDLARTNALVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKEALSLFEK 671
D D+ N + Y + G + + VF MP S+N MI +G NG+ L K
Sbjct: 61 DSDIKEWNVAISSYMRTGRCNEALRVFKRMPRWSSVSYNGMI--SGYLRNGE--FELARK 116
Query: 672 MLLSMVKPDSATFTCVLSACSHSMLVEEGVQIFNSMSRDHLVEPEAEHYTCVVDIYSRAG 731
+ M + D ++ ++ + + + ++F M E + + ++ Y++ G
Sbjct: 117 LFDEMPERDLVSWNVMIKGYVRNRNLGKARELFEIMP-----ERDVCSWNTMLSGYAQNG 171
Query: 732 CLEEAYGFIQRMPMEPTAIAWKAFLA 757
C+++A RMP E ++W A L+
Sbjct: 172 CVDDARSVFDRMP-EKNDVSWNALLS 196
Score = 37.0 bits (84), Expect = 0.053
Identities = 24/97 (24%), Positives = 43/97 (43%), Gaps = 2/97 (2%)
Query: 18 HGLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATR-CGVMSDVS 76
HG A++ + S + G+KPD +AV AC+ + K +Q+ T+ GVM +
Sbjct: 488 HGFGEVALRFFESMKREGLKPDDATMVAVLSACSHTGLVDKGRQYFYTMTQDYGVMPNSQ 547
Query: 77 IGNAFIHAYGKCKCVEGARRVFDDL-VARDVVTWNSL 112
+ G+ +E A + ++ D W +L
Sbjct: 548 HYACMVDLLGRAGLLEDAHNLMKNMPFEPDAAIWGTL 584
>At4g16840 hypothetical protein
Length = 661
Score = 424 bits (1089), Expect = e-118
Identities = 250/724 (34%), Positives = 371/724 (50%), Gaps = 99/724 (13%)
Query: 240 PDPVTV-SCILSACSDLQDLKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVRE--- 295
PD + V SC S+CS + + K + +F N ++ CVR
Sbjct: 26 PDTILVESCSSSSCSSPEP---SLVRSDYLTKPSDQDQIFPLNKII-----ARCVRSGDI 77
Query: 296 --AQAVFDLMPHRNVITWNSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPAC 353
A VF M +N ITWNSL L G+ DP M
Sbjct: 78 DGALRVFHGMRAKNTITWNSL---------------------LIGISKDPSRMMEAHQLF 116
Query: 354 SQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFDLMPHRNVVTWN 413
++ + D F +++ Y + +AQ+ FD MP ++ +WN
Sbjct: 117 DEIPE-----------------PDTFSYNIMLSCYVRNVNFEKAQSFFDRMPFKDAASWN 159
Query: 414 SLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHACSDLQDLKSGKVIHGFAVRH 473
++ + Y G +K +F M
Sbjct: 160 TMITGYARRGEMEKARELFYSM-------------------------------------- 181
Query: 474 GMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMF 533
M ++ NA++S Y +C + +A F + P R V +W ++T Y K+ E MF
Sbjct: 182 -MEKNEVSWNAMISGYIECGDLEKASHFFKVAPVRGVVAWTAMITGYMKAKKVELAEAMF 240
Query: 534 SQMNRDEVKADEITWSVVIGGCVKNSRIEEAMEIFRKMQTMGFKPDETTIYSILRACSLS 593
M V + +TW+ +I G V+NSR E+ +++FR M G +P+ + + S L CS
Sbjct: 241 KDMT---VNKNLVTWNAMISGYVENSRPEDGLKLFRAMLEEGIRPNSSGLSSALLGCSEL 297
Query: 594 ECLRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMI 653
L++G++IH V + D+ +L+ MY KCG L + +F++M KDV +WN MI
Sbjct: 298 SALQLGRQIHQIVSKSTLCNDVTALTSLISMYCKCGELGDAWKLFEVMKKKDVVAWNAMI 357
Query: 654 FANGMHGNGKEALSLFEKMLLSMVKPDSATFTCVLSACSHSMLVEEGVQIFNSMSRDHLV 713
HGN +AL LF +M+ + ++PD TF VL AC+H+ LV G+ F SM RD+ V
Sbjct: 358 SGYAQHGNADKALCLFREMIDNKIRPDWITFVAVLLACNHAGLVNIGMAYFESMVRDYKV 417
Query: 714 EPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEPTAIAWKAFLAGCRVYKNVELAKISAK 773
EP+ +HYTC+VD+ RAG LEEA I+ MP P A + L CRV+KNVELA+ +A+
Sbjct: 418 EPQPDHYTCMVDLLGRAGKLEEALKLIRSMPFRPHAAVFGTLLGACRVHKNVELAEFAAE 477
Query: 774 KLFEIDPNGSANYVTLFNILVTAKLWSEASKIRKLMKERGI-----TKTPGCSWFHVGNR 828
KL +++ +A YV L NI + W + +++RK MKE + K PG SW + N+
Sbjct: 478 KLLQLNSQNAAGYVQLANIYASKNRWEDVARVRKRMKESNVVKVERVKVPGYSWIEIRNK 537
Query: 829 VHTFVAGDKSNMESDKIYNFLDELFAKIKAAGYKPDTDYVLHDIDQEEKAESLCNHSEKL 888
VH F + D+ + E D I+ L EL K+K AGYKP+ ++ LH++++E+K + L HSEKL
Sbjct: 538 VHHFRSSDRIHPELDSIHKKLKELEKKMKLAGYKPELEFALHNVEEEQKEKLLLWHSEKL 597
Query: 889 AVAFGILNLNGQSTIRVFKNLRICGDCHNAIKYMSNVVGVTIVVRDSLRFHHFKNGNCSC 948
AVAFG + L S I+VFKNLRICGDCH AIK++S + I+VRD+ RFHHFK+G+CSC
Sbjct: 598 AVAFGCIKLPQGSQIQVFKNLRICGDCHKAIKFISEIEKREIIVRDTTRFHHFKDGSCSC 657
Query: 949 KDFW 952
D+W
Sbjct: 658 GDYW 661
Score = 164 bits (415), Expect = 2e-40
Identities = 118/431 (27%), Positives = 206/431 (47%), Gaps = 52/431 (12%)
Query: 91 VEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKANPLTVSSILPG 150
++GA RVF + A++ +TWNSL +G++K + + +
Sbjct: 77 IDGALRVFHGMRAKNTITWNSLL------------------IGISKDPSRMMEAHQLFDE 118
Query: 151 CSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTW 210
+ D F + ++ Y + + +AQ+ FD MP +D +W
Sbjct: 119 IPE--------------------PDTFSYNIMLSCYVRNVNFEKAQSFFDRMPFKDAASW 158
Query: 211 NSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQDLKSGKAIHGFALK 270
N++ + Y G +K +F M+ + + V+ + ++S + DL+ KA H F K
Sbjct: 159 NTMITGYARRGEMEKARELFYSMM----EKNEVSWNAMISGYIECGDLE--KASHFF--K 210
Query: 271 HGMVENVFVSNALVNLYESCLCVREAQAVF-DLMPHRNVITWNSLASCYVNCGFPQKGLN 329
V V A++ Y V A+A+F D+ ++N++TWN++ S YV P+ GL
Sbjct: 211 VAPVRGVVAWTAMITGYMKAKKVELAEAMFKDMTVNKNLVTWNAMISGYVENSRPEDGLK 270
Query: 330 VFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYA 389
+FR M G++P+ +SS L CS+L L+ G+ IH K + DV T+L+++Y
Sbjct: 271 LFRAMLEEGIRPNSSGLSSALLGCSELSALQLGRQIHQIVSKSTLCNDVTALTSLISMYC 330
Query: 390 NCLCVREAQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLS 449
C + +A +F++M ++VV WN++ S Y G K L +FREM+ N ++PD +T ++
Sbjct: 331 KCGELGDAWKLFEVMKKKDVVAWNAMISGYAQHGNADKALCLFREMIDNKIRPDWITFVA 390
Query: 450 ILHACSDLQDLKSGKVIHGFAVRHGMVE---DVFVCNALLSLYAKCVCVREAQVVFDLIP 506
+L AC+ + G VR VE D + C ++ L + + EA + +P
Sbjct: 391 VLLACNHAGLVNIGMAYFESMVRDYKVEPQPDHYTC--MVDLLGRAGKLEEALKLIRSMP 448
Query: 507 HREVASWNGIL 517
R A+ G L
Sbjct: 449 FRPHAAVFGTL 459
Score = 134 bits (336), Expect = 3e-31
Identities = 95/336 (28%), Positives = 166/336 (49%), Gaps = 20/336 (5%)
Query: 79 NAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVK 138
N + Y + E A+ FD + +D +WN++ Y G ++ +F M ++
Sbjct: 128 NIMLSCYVRNVNFEKAQSFFDRMPFKDAASWNTMITGYARRGEMEKARELFYSM----ME 183
Query: 139 ANPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTV 198
N ++ ++++ G + DL+ K H F V V V +A + Y K V A+ +
Sbjct: 184 KNEVSWNAMISGYIECGDLE--KASHFFKV--APVRGVVAWTAMITGYMKAKKVELAEAM 239
Query: 199 F-DLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQD 257
F D+ ++++VTWN++ S YV P+ GL +FR M+ +G++P+ +S L CS+L
Sbjct: 240 FKDMTVNKNLVTWNAMISGYVENSRPEDGLKLFRAMLEEGIRPNSSGLSSALLGCSELSA 299
Query: 258 LKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASC 317
L+ G+ IH K + +V +L+++Y C + +A +F++M ++V+ WN++ S
Sbjct: 300 LQLGRQIHQIVSKSTLCNDVTALTSLISMYCKCGELGDAWKLFEVMKKKDVVAWNAMISG 359
Query: 318 YVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVED 377
Y G K L +FREM N ++PD + ++L AC+ +G G A MV D
Sbjct: 360 YAQHGNADKALCLFREMIDNKIRPDWITFVAVLLACNH-----AGLVNIGMAYFESMVRD 414
Query: 378 VFV------CTALVNLYANCLCVREAQTVFDLMPHR 407
V T +V+L + EA + MP R
Sbjct: 415 YKVEPQPDHYTCMVDLLGRAGKLEEALKLIRSMPFR 450
Score = 62.8 bits (151), Expect = 9e-10
Identities = 37/159 (23%), Positives = 76/159 (47%)
Query: 16 IPHGLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDV 75
+ + P + +K++ + GI+P+ + C+ +Q H ++ + +DV
Sbjct: 260 VENSRPEDGLKLFRAMLEEGIRPNSSGLSSALLGCSELSALQLGRQIHQIVSKSTLCNDV 319
Query: 76 SIGNAFIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLN 135
+ + I Y KC + A ++F+ + +DVV WN++ + Y G + L +FR+M N
Sbjct: 320 TALTSLISMYCKCGELGDAWKLFEVMKKKDVVAWNAMISGYAQHGNADKALCLFREMIDN 379
Query: 136 KVKANPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVE 174
K++ + +T ++L C+ + G +VR VE
Sbjct: 380 KIRPDWITFVAVLLACNHAGLVNIGMAYFESMVRDYKVE 418
>At1g74630 hypothetical protein
Length = 643
Score = 406 bits (1043), Expect = e-113
Identities = 228/637 (35%), Positives = 348/637 (53%), Gaps = 35/637 (5%)
Query: 348 SILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANCL--CVREAQTVFDLMP 405
S+L +C L+ L IHG +K+G+ D + L+ A + + A+ + P
Sbjct: 10 SLLNSCKNLRALTQ---IHGLFIKYGVDTDSYFTGKLILHCAISISDALPYARRLLLCFP 66
Query: 406 HRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNG-VKPDLVTMLSILHACSDLQDLKSGK 464
+ +N+L Y P + VF EM+ G V PD + ++ A + + L++G
Sbjct: 67 EPDAFMFNTLVRGYSESDEPHNSVAVFVEMMRKGFVFPDSFSFAFVIKAVENFRSLRTGF 126
Query: 465 VIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREVASWNGILTAYFTNK 524
+H A++HG+ +FV L+ +Y C CV A+ VFD + + +WN ++TA F
Sbjct: 127 QMHCQALKHGLESHLFVGTTLIGMYGGCGCVEFARKVFDEMHQPNLVAWNAVITACFRGN 186
Query: 525 EYEKGLYMFSQM---NRDE--------VKA----------------DEITWSVVIGGCVK 557
+ +F +M N +KA D+++WS +I G
Sbjct: 187 DVAGAREIFDKMLVRNHTSWNVMLAGYIKAGELESAKRIFSEMPHRDDVSWSTMIVGIAH 246
Query: 558 NSRIEEAMEIFRKMQTMGFKPDETTIYSILRACSLSECLRMGKEIHCYVFRHWKDWDLAR 617
N E+ FR++Q G P+E ++ +L ACS S GK +H +V + W ++
Sbjct: 247 NGSFNESFLYFRELQRAGMSPNEVSLTGVLSACSQSGSFEFGKILHGFVEKAGYSWIVSV 306
Query: 618 TNALVDMYAKCGGLSLSRNVFDMMPIKD-VFSWNTMIFANGMHGNGKEALSLFEKMLLSM 676
NAL+DMY++CG + ++R VF+ M K + SW +MI MHG G+EA+ LF +M
Sbjct: 307 NNALIDMYSRCGNVPMARLVFEGMQEKRCIVSWTSMIAGLAMHGQGEEAVRLFNEMTAYG 366
Query: 677 VKPDSATFTCVLSACSHSMLVEEGVQIFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEA 736
V PD +F +L ACSH+ L+EEG F+ M R + +EPE EHY C+VD+Y R+G L++A
Sbjct: 367 VTPDGISFISLLHACSHAGLIEEGEDYFSEMKRVYHIEPEIEHYGCMVDLYGRSGKLQKA 426
Query: 737 YGFIQRMPMEPTAIAWKAFLAGCRVYKNVELAKISAKKLFEIDPNGSANYVTLFNILVTA 796
Y FI +MP+ PTAI W+ L C + N+ELA+ ++L E+DPN S + V L N TA
Sbjct: 427 YDFICQMPIPPTAIVWRTLLGACSSHGNIELAEQVKQRLNELDPNNSGDLVLLSNAYATA 486
Query: 797 KLWSEASKIRKLMKERGITKTPGCSWFHVGNRVHTFVAGDKSNMESDKIYNFLDELFAKI 856
W + + IRK M + I KT S VG ++ F AG+K + + L E+ ++
Sbjct: 487 GKWKDVASIRKSMIVQRIKKTTAWSLVEVGKTMYKFTAGEKKKGIDIEAHEKLKEIILRL 546
Query: 857 K-AAGYKPDTDYVLHDIDQEEKAESLCNHSEKLAVAFGILNLNGQSTIRVFKNLRICGDC 915
K AGY P+ L+D+++EEK + + HSEKLA+AF + L+ + IR+ KNLRIC DC
Sbjct: 547 KDEAGYTPEVASALYDVEEEEKEDQVSKHSEKLALAFALARLSKGANIRIVKNLRICRDC 606
Query: 916 HNAIKYMSNVVGVTIVVRDSLRFHHFKNGNCSCKDFW 952
H +K S V GV I+VRD RFH FK+G+CSC+D+W
Sbjct: 607 HAVMKLTSKVYGVEILVRDRNRFHSFKDGSCSCRDYW 643
Score = 143 bits (361), Expect = 4e-34
Identities = 118/495 (23%), Positives = 210/495 (41%), Gaps = 102/495 (20%)
Query: 146 SILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCL--CVREAQTVFDLMP 203
S+L C +L+ L +IHG +++G+ D + + + A + + A+ + P
Sbjct: 10 SLLNSCKNLRALT---QIHGLFIKYGVDTDSYFTGKLILHCAISISDALPYARRLLLCFP 66
Query: 204 HRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDG-VKPDPVTVSCILSACSDLQDLKSGK 262
D +N+L Y P + VF EM+ G V PD + + ++ A + + L++G
Sbjct: 67 EPDAFMFNTLVRGYSESDEPHNSVAVFVEMMRKGFVFPDSFSFAFVIKAVENFRSLRTGF 126
Query: 263 AIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPHRNVITWNSLASC----- 317
+H ALKHG+ ++FV L+ +Y C CV A+ VFD M N++ WN++ +
Sbjct: 127 QMHCQALKHGLESHLFVGTTLIGMYGGCGCVEFARKVFDEMHQPNLVAWNAVITACFRGN 186
Query: 318 --------------------------YVNCGFPQKGLNVFREM-------------GL-- 336
Y+ G + +F EM G+
Sbjct: 187 DVAGAREIFDKMLVRNHTSWNVMLAGYIKAGELESAKRIFSEMPHRDDVSWSTMIVGIAH 246
Query: 337 ----------------NGVKPDPMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDVFV 380
G+ P+ ++++ +L ACSQ + GK +HGF K G V V
Sbjct: 247 NGSFNESFLYFRELQRAGMSPNEVSLTGVLSACSQSGSFEFGKILHGFVEKAGYSWIVSV 306
Query: 381 CTALVNLYANCLCVREAQTVFD-LMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNG 439
AL+++Y+ C V A+ VF+ + R +V+W S+ + G ++ + +F EM G
Sbjct: 307 NNALIDMYSRCGNVPMARLVFEGMQEKRCIVSWTSMIAGLAMHGQGEEAVRLFNEMTAYG 366
Query: 440 VKPDLVTMLSILHACSDLQDLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQ 499
V PD ++ +S+LHACS H + G ED F E +
Sbjct: 367 VTPDGISFISLLHACS-----------HAGLIEEG--EDYF---------------SEMK 398
Query: 500 VVFDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGGCVKNS 559
V+ + P E+ + ++ Y + + +K QM + I W ++G C +
Sbjct: 399 RVYHIEP--EIEHYGCMVDLYGRSGKLQKAYDFICQM---PIPPTAIVWRTLLGACSSHG 453
Query: 560 RIEEAMEIFRKMQTM 574
IE A ++ +++ +
Sbjct: 454 NIELAEQVKQRLNEL 468
Score = 95.9 bits (237), Expect = 1e-19
Identities = 73/306 (23%), Positives = 127/306 (40%), Gaps = 64/306 (20%)
Query: 21 PNEAIKIYTSSRARG-IKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSIGN 79
P+ ++ ++ +G + PD F V KA R Q H A + G+ S + +G
Sbjct: 86 PHNSVAVFVEMMRKGFVFPDSFSFAFVIKAVENFRSLRTGFQMHCQALKHGLESHLFVGT 145
Query: 80 AFIHAYGKCKCVE-------------------------------GARRVFDDLVARDVVT 108
I YG C CVE GAR +FD ++ R+ +
Sbjct: 146 TLIGMYGGCGCVEFARKVFDEMHQPNLVAWNAVITACFRGNDVAGAREIFDKMLVRNHTS 205
Query: 109 WNSLSACYVNCGFPQQGLNVFRKM-------------------GLNK------------V 137
WN + A Y+ G + +F +M N+ +
Sbjct: 206 WNVMLAGYIKAGELESAKRIFSEMPHRDDVSWSTMIVGIAHNGSFNESFLYFRELQRAGM 265
Query: 138 KANPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQT 197
N ++++ +L CS + GK +HGFV + G V V++A ++ Y++C V A+
Sbjct: 266 SPNEVSLTGVLSACSQSGSFEFGKILHGFVEKAGYSWIVSVNNALIDMYSRCGNVPMARL 325
Query: 198 VFD-LMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACSDLQ 256
VF+ + R +V+W S+ + G ++ + +F EM GV PD ++ +L ACS
Sbjct: 326 VFEGMQEKRCIVSWTSMIAGLAMHGQGEEAVRLFNEMTAYGVTPDGISFISLLHACSHAG 385
Query: 257 DLKSGK 262
++ G+
Sbjct: 386 LIEEGE 391
Score = 57.8 bits (138), Expect = 3e-08
Identities = 50/203 (24%), Positives = 89/203 (43%), Gaps = 4/203 (1%)
Query: 15 GIPH-GLPNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMS 73
GI H G NE+ + + G+ P++ V AC+ S K H + G
Sbjct: 243 GIAHNGSFNESFLYFRELQRAGMSPNEVSLTGVLSACSQSGSFEFGKILHGFVEKAGYSW 302
Query: 74 DVSIGNAFIHAYGKCKCVEGARRVFDDL-VARDVVTWNSLSACYVNCGFPQQGLNVFRKM 132
VS+ NA I Y +C V AR VF+ + R +V+W S+ A G ++ + +F +M
Sbjct: 303 IVSVNNALIDMYSRCGNVPMARLVFEGMQEKRCIVSWTSMIAGLAMHGQGEEAVRLFNEM 362
Query: 133 GLNKVKANPLTVSSILPGCSDLQDLKSGKEIHGFVVRHGMVE-DVFVSSAFVNFYAKCLC 191
V + ++ S+L CS ++ G++ + R +E ++ V+ Y +
Sbjct: 363 TAYGVTPDGISFISLLHACSHAGLIEEGEDYFSEMKRVYHIEPEIEHYGCMVDLYGRSGK 422
Query: 192 VREAQTVFDLMP-HRDVVTWNSL 213
+++A MP + W +L
Sbjct: 423 LQKAYDFICQMPIPPTAIVWRTL 445
>At5g06540 selenium-binding protein-like
Length = 622
Score = 405 bits (1040), Expect = e-113
Identities = 217/618 (35%), Positives = 341/618 (55%), Gaps = 25/618 (4%)
Query: 348 SILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANCLCVRE----------- 396
++L +CS DLK IHGF ++ ++ DVFV + L+ LCV +
Sbjct: 17 ALLQSCSSFSDLK---IIHGFLLRTHLISDVFVASRLL-----ALCVDDSTFNKPTNLLG 68
Query: 397 -AQTVFDLMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHACS 455
A +F + + N+ +N L C+ P K + +M+ + + PD +T ++ A S
Sbjct: 69 YAYGIFSQIQNPNLFVFNLLIRCFSTGAEPSKAFGFYTQMLKSRIWPDNITFPFLIKASS 128
Query: 456 DLQDLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCVCVREAQVVFDLIPHREVASWNG 515
+++ + G+ H VR G DV+V N+L+ +YA C + A +F + R+V SW
Sbjct: 129 EMECVLVGEQTHSQIVRFGFQNDVYVENSLVHMYANCGFIAAAGRIFGQMGFRDVVSWTS 188
Query: 516 ILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGGCVKNSRIEEAMEIFRKMQTMG 575
++ Y E MF +M + TWS++I G KN+ E+A+++F M+ G
Sbjct: 189 MVAGYCKCGMVENAREMFDEMPHRNL----FTWSIMINGYAKNNCFEKAIDLFEFMKREG 244
Query: 576 FKPDETTIYSILRACSLSECLRMGKEIHCYVFRHWKDWDLARTNALVDMYAKCGGLSLSR 635
+ET + S++ +C+ L G+ + YV + +L ALVDM+ +CG + +
Sbjct: 245 VVANETVMVSVISSCAHLGALEFGERAYEYVVKSHMTVNLILGTALVDMFWRCGDIEKAI 304
Query: 636 NVFDMMPIKDVFSWNTMIFANGMHGNGKEALSLFEKMLLSMVKPDSATFTCVLSACSHSM 695
+VF+ +P D SW+++I +HG+ +A+ F +M+ P TFT VLSACSH
Sbjct: 305 HVFEGLPETDSLSWSSIIKGLAVHGHAHKAMHYFSQMISLGFIPRDVTFTAVLSACSHGG 364
Query: 696 LVEEGVQIFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLEEAYGFIQRMPMEPTAIAWKAF 755
LVE+G++I+ +M +DH +EP EHY C+VD+ RAG L EA FI +M ++P A A
Sbjct: 365 LVEKGLEIYENMKKDHGIEPRLEHYGCIVDMLGRAGKLAEAENFILKMHVKPNAPILGAL 424
Query: 756 LAGCRVYKNVELAKISAKKLFEIDPNGSANYVTLFNILVTAKLWSEASKIRKLMKERGIT 815
L C++YKN E+A+ L ++ P S YV L NI A W + +R +MKE+ +
Sbjct: 425 LGACKIYKNTEVAERVGNMLIKVKPEHSGYYVLLSNIYACAGQWDKIESLRDMMKEKLVK 484
Query: 816 KTPGCSWFHVGNRVHTFVAG-DKSNMESDKIYNFLDELFAKIKAAGYKPDTDYVLHDIDQ 874
K PG S + +++ F G D+ + E KI +E+ KI+ GYK +T D+D+
Sbjct: 485 KPPGWSLIEIDGKINKFTMGDDQKHPEMGKIRRKWEEILGKIRLIGYKGNTGDAFFDVDE 544
Query: 875 EEKAESLCNHSEKLAVAFGILNLNGQSTIRVFKNLRICGDCHNAIKYMSNVVGVTIVVRD 934
EEK S+ HSEKLA+A+G++ +TIR+ KNLR+C DCH K +S V G ++VRD
Sbjct: 545 EEKESSIHMHSEKLAIAYGMMKTKPGTTIRIVKNLRVCEDCHTVTKLISEVYGRELIVRD 604
Query: 935 SLRFHHFKNGNCSCKDFW 952
RFHHF+NG CSC+D+W
Sbjct: 605 RNRFHHFRNGVCSCRDYW 622
Score = 144 bits (362), Expect = 3e-34
Identities = 97/376 (25%), Positives = 175/376 (45%), Gaps = 56/376 (14%)
Query: 146 SILPGCSDLQDLKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVRE----------- 194
++L CS DLK IHGF++R ++ DVFV+S + LCV +
Sbjct: 17 ALLQSCSSFSDLKI---IHGFLLRTHLISDVFVASRLL-----ALCVDDSTFNKPTNLLG 68
Query: 195 -AQTVFDLMPHRDVVTWNSLSSCYVNCGFPQKGLNVFREMVLDGVKPDPVTVSCILSACS 253
A +F + + ++ +N L C+ P K + +M+ + PD +T ++ A S
Sbjct: 69 YAYGIFSQIQNPNLFVFNLLIRCFSTGAEPSKAFGFYTQMLKSRIWPDNITFPFLIKASS 128
Query: 254 DLQDLKSGKAIHGFALKHGMVENVFVSNALVNLYESC----------------------- 290
+++ + G+ H ++ G +V+V N+LV++Y +C
Sbjct: 129 EMECVLVGEQTHSQIVRFGFQNDVYVENSLVHMYANCGFIAAAGRIFGQMGFRDVVSWTS 188
Query: 291 ----LC----VREAQAVFDLMPHRNVITWNSLASCYVNCGFPQKGLNVFREMGLNGVKPD 342
C V A+ +FD MPHRN+ TW+ + + Y +K +++F M GV +
Sbjct: 189 MVAGYCKCGMVENAREMFDEMPHRNLFTWSIMINGYAKNNCFEKAIDLFEFMKREGVVAN 248
Query: 343 PMAMSSILPACSQLKDLKSGKTIHGFAVKHGMVEDVFVCTALVNLYANCLCVREAQTVFD 402
M S++ +C+ L L+ G+ + + VK M ++ + TALV+++ C + +A VF+
Sbjct: 249 ETVMVSVISSCAHLGALEFGERAYEYVVKSHMTVNLILGTALVDMFWRCGDIEKAIHVFE 308
Query: 403 LMPHRNVVTWNSLSSCYVNCGFPQKGLNVFREMVLNGVKPDLVTMLSILHACSDLQDLKS 462
+P + ++W+S+ G K ++ F +M+ G P VT ++L ACS
Sbjct: 309 GLPETDSLSWSSIIKGLAVHGHAHKAMHYFSQMISLGFIPRDVTFTAVLSACS-----HG 363
Query: 463 GKVIHGFAVRHGMVED 478
G V G + M +D
Sbjct: 364 GLVEKGLEIYENMKKD 379
Score = 115 bits (287), Expect = 2e-25
Identities = 79/361 (21%), Positives = 165/361 (44%), Gaps = 41/361 (11%)
Query: 44 MAVAKACAASRDALKVKQFHDDATRCGVMSDVSIGNAFI------HAYGKCKCVEG-ARR 96
+A+ ++C++ D +K H R ++SDV + + + + K + G A
Sbjct: 16 LALLQSCSSFSD---LKIIHGFLLRTHLISDVFVASRLLALCVDDSTFNKPTNLLGYAYG 72
Query: 97 VFDDLVARDVVTWNSLSACYVNCGFPQQGLNVFRKMGLNKVKANPLTVSSILPGCSDLQD 156
+F + ++ +N L C+ P + + +M +++ + +T ++ S+++
Sbjct: 73 IFSQIQNPNLFVFNLLIRCFSTGAEPSKAFGFYTQMLKSRIWPDNITFPFLIKASSEMEC 132
Query: 157 LKSGKEIHGFVVRHGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTWNSLSSC 216
+ G++ H +VR G DV+V ++ V+ YA C + A +F M RDVV+W S+ +
Sbjct: 133 VLVGEQTHSQIVRFGFQNDVYVENSLVHMYANCGFIAAAGRIFGQMGFRDVVSWTSMVAG 192
Query: 217 YVNCGFP-------------------------------QKGLNVFREMVLDGVKPDPVTV 245
Y CG +K +++F M +GV + +
Sbjct: 193 YCKCGMVENAREMFDEMPHRNLFTWSIMINGYAKNNCFEKAIDLFEFMKREGVVANETVM 252
Query: 246 SCILSACSDLQDLKSGKAIHGFALKHGMVENVFVSNALVNLYESCLCVREAQAVFDLMPH 305
++S+C+ L L+ G+ + + +K M N+ + ALV+++ C + +A VF+ +P
Sbjct: 253 VSVISSCAHLGALEFGERAYEYVVKSHMTVNLILGTALVDMFWRCGDIEKAIHVFEGLPE 312
Query: 306 RNVITWNSLASCYVNCGFPQKGLNVFREMGLNGVKPDPMAMSSILPACSQLKDLKSGKTI 365
+ ++W+S+ G K ++ F +M G P + +++L ACS ++ G I
Sbjct: 313 TDSLSWSSIIKGLAVHGHAHKAMHYFSQMISLGFIPRDVTFTAVLSACSHGGLVEKGLEI 372
Query: 366 H 366
+
Sbjct: 373 Y 373
Score = 114 bits (285), Expect = 3e-25
Identities = 70/264 (26%), Positives = 119/264 (44%), Gaps = 31/264 (11%)
Query: 21 PNEAIKIYTSSRARGIKPDKPVFMAVAKACAASRDALKVKQFHDDATRCGVMSDVSIGNA 80
P++A YT I PD F + KA + L +Q H R G +DV + N+
Sbjct: 98 PSKAFGFYTQMLKSRIWPDNITFPFLIKASSEMECVLVGEQTHSQIVRFGFQNDVYVENS 157
Query: 81 FIHAYGKCKCVEGARRVFDDLVARDVVTWNSLSACYVNCGFPQ----------------- 123
+H Y C + A R+F + RDVV+W S+ A Y CG +
Sbjct: 158 LVHMYANCGFIAAAGRIFGQMGFRDVVSWTSMVAGYCKCGMVENAREMFDEMPHRNLFTW 217
Query: 124 --------------QGLNVFRKMGLNKVKANPLTVSSILPGCSDLQDLKSGKEIHGFVVR 169
+ +++F M V AN + S++ C+ L L+ G+ + +VV+
Sbjct: 218 SIMINGYAKNNCFEKAIDLFEFMKREGVVANETVMVSVISSCAHLGALEFGERAYEYVVK 277
Query: 170 HGMVEDVFVSSAFVNFYAKCLCVREAQTVFDLMPHRDVVTWNSLSSCYVNCGFPQKGLNV 229
M ++ + +A V+ + +C + +A VF+ +P D ++W+S+ G K ++
Sbjct: 278 SHMTVNLILGTALVDMFWRCGDIEKAIHVFEGLPETDSLSWSSIIKGLAVHGHAHKAMHY 337
Query: 230 FREMVLDGVKPDPVTVSCILSACS 253
F +M+ G P VT + +LSACS
Sbjct: 338 FSQMISLGFIPRDVTFTAVLSACS 361
Score = 106 bits (265), Expect = 5e-23
Identities = 83/334 (24%), Positives = 154/334 (45%), Gaps = 28/334 (8%)
Query: 448 LSILHACSDLQDLKSGKVIHGFAVRHGMVEDVFVCNALLSLYAKCV----------CVRE 497
L++L +CS DL K+IHGF +R ++ DVFV + LL+L CV +
Sbjct: 16 LALLQSCSSFSDL---KIIHGFLLRTHLISDVFVASRLLAL---CVDDSTFNKPTNLLGY 69
Query: 498 AQVVFDLIPHREVASWNGILTAYFTNKEYEKGLYMFSQMNRDEVKADEITWSVVIGGCVK 557
A +F I + + +N ++ + T E K ++QM + + D IT+ +I +
Sbjct: 70 AYGIFSQIQNPNLFVFNLLIRCFSTGAEPSKAFGFYTQMLKSRIWPDNITFPFLIKASSE 129
Query: 558 NSRIEEAMEIFRKMQTMGFKPD---ETTIYSILRACSLSECLRMGKEIHCYVFRHWKDWD 614
+ + ++ GF+ D E ++ + C G+ FR W
Sbjct: 130 MECVLVGEQTHSQIVRFGFQNDVYVENSLVHMYANCGF--IAAAGRIFGQMGFRDVVSW- 186
Query: 615 LARTNALVDMYAKCGGLSLSRNVFDMMPIKDVFSWNTMIFANGMHGNGKEALSLFEKMLL 674
++V Y KCG + +R +FD MP +++F+W+ MI + ++A+ LFE M
Sbjct: 187 ----TSMVAGYCKCGMVENAREMFDEMPHRNLFTWSIMINGYAKNNCFEKAIDLFEFMKR 242
Query: 675 SMVKPDSATFTCVLSACSHSMLVEEGVQIFNSMSRDHLVEPEAEHYTCVVDIYSRAGCLE 734
V + V+S+C+H +E G + + + + H+ T +VD++ R G +E
Sbjct: 243 EGVVANETVMVSVISSCAHLGALEFGERAYEYVVKSHMT-VNLILGTALVDMFWRCGDIE 301
Query: 735 EAYGFIQRMPMEPTAIAWKAFLAGCRVYKNVELA 768
+A + +P E +++W + + G V+ + A
Sbjct: 302 KAIHVFEGLP-ETDSLSWSSIIKGLAVHGHAHKA 334
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.137 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,937,877
Number of Sequences: 26719
Number of extensions: 946777
Number of successful extensions: 15994
Number of sequences better than 10.0: 445
Number of HSP's better than 10.0 without gapping: 418
Number of HSP's successfully gapped in prelim test: 27
Number of HSP's that attempted gapping in prelim test: 2707
Number of HSP's gapped (non-prelim): 3420
length of query: 952
length of database: 11,318,596
effective HSP length: 109
effective length of query: 843
effective length of database: 8,406,225
effective search space: 7086447675
effective search space used: 7086447675
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 65 (29.6 bits)
Medicago: description of AC124970.5


