

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124970.11 + phase: 0
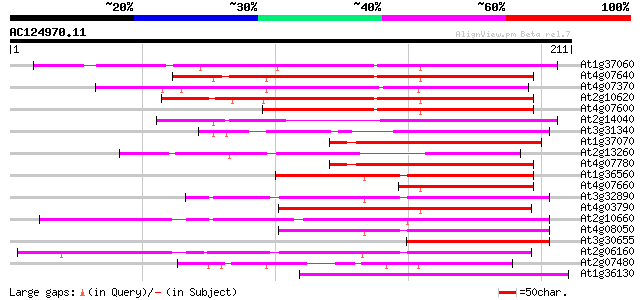
(211 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g37060 Athila retroelment ORF 1, putative 103 7e-23
At4g07640 putative athila transposon protein 99 1e-21
At4g07370 95 2e-20
At2g10620 putative Athila retroelement ORF1 protein 92 2e-19
At4g07600 83 9e-17
At2g14040 putative retroelement pol polyprotein 77 9e-15
At3g31340 Athila ORF 1, putative 71 4e-13
At1g37070 hypothetical protein 71 4e-13
At2g13260 putative Athila retroelement ORF1 protein 70 6e-13
At4g07780 putative athila transposon protein 69 1e-12
At1g36560 hypothetical protein 69 1e-12
At4g07660 putative athila transposon protein 69 2e-12
At3g32890 Athila ORF 1, putative 69 2e-12
At4g03790 putative athila-like protein 69 2e-12
At2g10660 putative Athila retroelement ORF1 protein 68 4e-12
At4g08050 67 5e-12
At3g30655 hypothetical protein, 3' partial 65 2e-11
At2g06160 putative Athila retroelement ORF1 protein 65 2e-11
At2g07480 F9A16.15 65 3e-11
At1g36130 hypothetical protein 65 3e-11
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 103 bits (257), Expect = 7e-23
Identities = 70/212 (33%), Positives = 110/212 (51%), Gaps = 22/212 (10%)
Query: 10 KEVVEKETEHGEVEKESDQGVVENERKKKKGGEKSEKLIDEDSILRKSKSQILKDGDKPQ 69
KE+ KE+ + E DQ + + G +EK I+E + + ++ L P
Sbjct: 441 KELPTKESPNQNTEDSLDQ----DGEDFCQNGNSAEKAIEEPILHQPTRP--LAPAASPL 494
Query: 70 V-------------IPSYVKLPYPHLAKNKKKEEGQFKKFMQL-FSQLQVNIPFGEALDQ 115
V IP K P P + KK ++K ++ L+V +P + L
Sbjct: 495 VEKPAAAKTKENVFIPPPYKPPLPFPGRFKKVMIQKYKALLEKQLKNLEVTMPLVDCLAL 554
Query: 116 MPVYAKFMKEMLTSRRKPKDDENIALSENCSAILQRKL-PPKLKDPGAFTIPCSIGPVDI 174
+P K++K+M+T R K + LS CSAI+Q+K+ P KL DPG+FT+PC++GP+
Sbjct: 555 IPDSNKYVKDMITERIKEVQGM-VVLSHECSAIIQQKIIPKKLGDPGSFTLPCALGPLAF 613
Query: 175 GRALCDLGASINLMPLSMMKKLGSRELSPVSV 206
+ LCDLGAS++LMPL + KKLG + P ++
Sbjct: 614 SKCLCDLGASVSLMPLPVAKKLGFNKYKPCNI 645
>At4g07640 putative athila transposon protein
Length = 866
Score = 99.4 bits (246), Expect = 1e-21
Identities = 56/139 (40%), Positives = 90/139 (64%), Gaps = 6/139 (4%)
Query: 62 LKDGDKPQVIPSYV-KLPYPHLAKNKKKEEGQFKK-FMQLFSQLQVNIPFGEALDQMPVY 119
+K+ +K V P Y +LP+P ++KK +++ F + ++++ IP +AL +P
Sbjct: 440 VKNKEKVFVPPPYKPELPFP--GRHKKALADKYRAMFAKNIKEVELRIPLVDALALIPDS 497
Query: 120 AKFMKEMLTSRRKPKDDENIALSENCSAILQRKL-PPKLKDPGAFTIPCSIGPVDIGRAL 178
KF+K+++ R + + LS CSAI+Q+K+ P KL DPG+FT+PCS+GP+ R L
Sbjct: 498 HKFLKDLIVERIQEVQGM-VVLSHGCSAIIQKKIIPKKLSDPGSFTLPCSLGPLAFNRCL 556
Query: 179 CDLGASINLMPLSMMKKLG 197
CDLGAS++LM LS+ K+LG
Sbjct: 557 CDLGASVSLMALSVAKRLG 575
>At4g07370
Length = 531
Score = 95.1 bits (235), Expect = 2e-20
Identities = 59/171 (34%), Positives = 95/171 (55%), Gaps = 9/171 (5%)
Query: 33 NERKKKKGGEKSEKLIDEDSILRK--SKSQILK----DGDKPQVIPSYVKLPYPHLAKNK 86
NE + K E E ++D D+ + S LK ++ IP K P P ++K
Sbjct: 98 NEDQVDKPTEVLEPILDRDTRPTNPLTSSAALKLVAAKNNEIAFIPPPYKPPLPFPGRHK 157
Query: 87 KKEEGQFKK-FMQLFSQLQVNIPFGEALDQMPVYAKFMKEMLTSRRKPKDDENIALSENC 145
K+ E +++ F + ++++ IP +AL +P KF+K+++ R + + LS C
Sbjct: 158 KEVEDKYRAMFAKNIKKVELRIPLADALTHIPDSQKFLKDLIMERIQEVQKTTV-LSHEC 216
Query: 146 SAILQRK-LPPKLKDPGAFTIPCSIGPVDIGRALCDLGASINLMPLSMMKK 195
SAI+Q + KL DPG+FT+PCS+G + + LCDLG S+NLMPLS+ K+
Sbjct: 217 SAIIQENDVSEKLGDPGSFTLPCSLGSLTFNKCLCDLGPSVNLMPLSVAKR 267
>At2g10620 putative Athila retroelement ORF1 protein
Length = 451
Score = 92.4 bits (228), Expect = 2e-19
Identities = 54/143 (37%), Positives = 89/143 (61%), Gaps = 6/143 (4%)
Query: 58 KSQILKDGDKPQVIPSYVKLPYPHL-AKNKKKEEGQFK-KFMQLFSQLQVNIPFGEALDQ 115
K ++K+ +K V P Y P+P ++KK +++ F + ++++ IP +AL
Sbjct: 192 KPVVVKNKEKVFVPPPYK--PHPSFPGRHKKALAHKYRVMFAKNIKEVELQIPLVDALAL 249
Query: 116 MPVYAKFMKEMLTSRRKPKDDENIALSENCSAILQRKL-PPKLKDPGAFTIPCSIGPVDI 174
+ KF+K+++ R + + LS+ CSAI+Q+K+ KL DPG+FT+PCS+GP+
Sbjct: 250 IMDSHKFLKDLIVERIQELQGM-VVLSDECSAIIQKKIIHKKLSDPGSFTLPCSLGPLAF 308
Query: 175 GRALCDLGASINLMPLSMMKKLG 197
+ LCDLGAS +LMPLS+ K+LG
Sbjct: 309 NKCLCDLGASASLMPLSVTKRLG 331
>At4g07600
Length = 630
Score = 83.2 bits (204), Expect = 9e-17
Identities = 43/103 (41%), Positives = 66/103 (63%), Gaps = 2/103 (1%)
Query: 96 FMQLFSQLQVNIPFGEALDQMPVYAKFMKEMLTSRRKPKDDENIALSENCSAILQRKL-P 154
F + ++++ IP +AL + KF+K+++ R + + LS CSAI+Q+K+ P
Sbjct: 2 FAKNIKEVELRIPLVDALALILDTHKFLKDLIVERIQEVQGM-VVLSHECSAIIQKKIVP 60
Query: 155 PKLKDPGAFTIPCSIGPVDIGRALCDLGASINLMPLSMMKKLG 197
KL DPG+FT+PC +G V R LCDLGA ++ MPLS+ K+LG
Sbjct: 61 KKLSDPGSFTLPCFLGTVAFNRCLCDLGALVSPMPLSIAKRLG 103
>At2g14040 putative retroelement pol polyprotein
Length = 841
Score = 76.6 bits (187), Expect = 9e-15
Identities = 49/152 (32%), Positives = 70/152 (45%), Gaps = 37/152 (24%)
Query: 56 KSKSQILKDGDKPQVIPSYV-KLPYPHLAKNKKKEEGQFKKFMQLFSQLQVNIPFGEALD 114
K + Q L++ K V+P YV KLP+P + K + E ++ F ++ QLQ
Sbjct: 2 KLRKQTLEEKAKKTVLPPYVPKLPFPG-RQRKIQREKEYSLFDEIMRQLQ---------- 50
Query: 115 QMPVYAKFMKEMLTSRRKPKDDENIALSENCSAILQRKLPPKLKDPGAFTIPCSIGPVDI 174
S CSAILQ +P K +DPG+F +P IG
Sbjct: 51 -------------------------RYSPECSAILQNVIPVKREDPGSFVLPSRIGEYTF 85
Query: 175 GRALCDLGASINLMPLSMMKKLGSRELSPVSV 206
R LCDLGA ++LMP S+ K+LG +P +
Sbjct: 86 DRCLCDLGAGVSLMPFSVAKRLGDTNFTPTKM 117
>At3g31340 Athila ORF 1, putative
Length = 781
Score = 71.2 bits (173), Expect = 4e-13
Identities = 51/136 (37%), Positives = 67/136 (48%), Gaps = 27/136 (19%)
Query: 72 PSYV-KLPYP---HLAKNKKKEEGQFKKFMQLFSQLQVNIPFGEALDQMPVYAKFMKEML 127
P YV KLP+P +++K+EE L + Q + D + V A KEM+
Sbjct: 466 PPYVPKLPFPGRERQIQSRKREE------YALLVESQRQQKEAQLTDVVEVLAG--KEMV 517
Query: 128 TSRRKPKDDENIALSENCSAILQRKLPPKLKDPGAFTIPCSIGPVDIGRALCDLGASINL 187
++ CSAI +P KL DPG+F +PC IG R LCDLGA +NL
Sbjct: 518 ST---------------CSAIPPATIPEKLGDPGSFVLPCRIGKSAFERCLCDLGAGVNL 562
Query: 188 MPLSMMKKLGSRELSP 203
MPLSM K+LG P
Sbjct: 563 MPLSMSKRLGITNFKP 578
>At1g37070 hypothetical protein
Length = 590
Score = 71.2 bits (173), Expect = 4e-13
Identities = 36/80 (45%), Positives = 51/80 (63%), Gaps = 3/80 (3%)
Query: 121 KFMKEMLTSRRKPKDDENIALSENCSAILQRKLPPKLKDPGAFTIPCSIGPVDIGRALCD 180
KF E L +K ++ E I+ + ++ + KL DPG+FT+PCS+GP+ R LCD
Sbjct: 221 KFKIEKL---KKDQNQEMISSASTQETYKKKIIQEKLDDPGSFTLPCSLGPLTFNRCLCD 277
Query: 181 LGASINLMPLSMMKKLGSRE 200
LGAS++LMPLS K+LG E
Sbjct: 278 LGASVSLMPLSTAKRLGIME 297
>At2g13260 putative Athila retroelement ORF1 protein
Length = 622
Score = 70.5 bits (171), Expect = 6e-13
Identities = 50/153 (32%), Positives = 77/153 (49%), Gaps = 31/153 (20%)
Query: 42 EKSEKLIDEDSILRKSKSQILKDGDKPQVIPSYVKLPYPH--LAKNKKKEEGQFKKFMQL 99
EK++K + + ++ KS + K + P LP+P L K KKK FK M
Sbjct: 327 EKAKKSTNGNDVVTKSVEK--NAASKVESPPYEPPLPFPGRVLTKAKKKVFSSFKSNM-- 382
Query: 100 FSQLQVNIPFGEALDQMPVYAKFMKEMLTSRRKPKDDENIALSENCSAILQRKLPPKLKD 159
S++ +P E L Q+P++ +F++ +L +R K L+D
Sbjct: 383 -SRVGAPLPCVENLSQVPLHFQFIQAILENREK------------------------LED 417
Query: 160 PGAFTIPCSIGPVDIGRALCDLGASINLMPLSM 192
PG FTIPCS+G + + ALCD GAS+N+M L+M
Sbjct: 418 PGKFTIPCSLGDLQLDDALCDSGASVNVMSLAM 450
>At4g07780 putative athila transposon protein
Length = 446
Score = 69.3 bits (168), Expect = 1e-12
Identities = 35/77 (45%), Positives = 49/77 (63%), Gaps = 3/77 (3%)
Query: 121 KFMKEMLTSRRKPKDDENIALSENCSAILQRKLPPKLKDPGAFTIPCSIGPVDIGRALCD 180
KF E L +K ++ E I+ ++ + KL DPG+FT+PCS+GP+ R LCD
Sbjct: 248 KFKIEKL---KKDQNQEMISRVSTQETYKKKFIQEKLDDPGSFTLPCSLGPLTFNRCLCD 304
Query: 181 LGASINLMPLSMMKKLG 197
LGAS++LMPLS K+LG
Sbjct: 305 LGASVSLMPLSTAKRLG 321
>At1g36560 hypothetical protein
Length = 524
Score = 69.3 bits (168), Expect = 1e-12
Identities = 37/100 (37%), Positives = 62/100 (62%), Gaps = 5/100 (5%)
Query: 101 SQLQVNIPFGEALDQMPVYAKFMKEMLTSRRK---PKDDENIALSENCSAILQRKLPPKL 157
++++ +P E L+ +P K ++ ++ K DDE+ A + +R + KL
Sbjct: 62 NEIEAVMPLMEVLNLIPNPHKDVRNLILEWIKMYHDSDDESDATPSRAAD--KRIVQEKL 119
Query: 158 KDPGAFTIPCSIGPVDIGRALCDLGASINLMPLSMMKKLG 197
+DPG+FT+PCS+G + + LCDLGAS++LMPLS+ K+LG
Sbjct: 120 EDPGSFTLPCSLGSLTFNKCLCDLGASVSLMPLSVAKRLG 159
>At4g07660 putative athila transposon protein
Length = 724
Score = 68.9 bits (167), Expect = 2e-12
Identities = 32/52 (61%), Positives = 42/52 (80%), Gaps = 1/52 (1%)
Query: 147 AILQRKL-PPKLKDPGAFTIPCSIGPVDIGRALCDLGASINLMPLSMMKKLG 197
AI Q+K+ P KL DPG+FT+PCS+GP+ R LCDLGA ++LMPLS+ K+LG
Sbjct: 3 AITQKKIVPKKLIDPGSFTLPCSLGPLAFKRCLCDLGALVSLMPLSVAKRLG 54
>At3g32890 Athila ORF 1, putative
Length = 755
Score = 68.9 bits (167), Expect = 2e-12
Identities = 47/140 (33%), Positives = 75/140 (53%), Gaps = 9/140 (6%)
Query: 67 KPQVIPSYVKLPYPHLAKNKKKEEGQFKKFMQLFSQLQVNIPFGEALDQMPVYAKFMKEM 126
KP +PS LP+ +K + K + +++ +P E L+ +P K ++ +
Sbjct: 436 KPTPLPSSA-LPWTFRKAWMEKYKSVASKQLD---EIEAVMPLIEVLNLIPDPHKDVRNL 491
Query: 127 LTSRRK---PKDDENIALSENCSAILQRKLPPKLKDPGAFTIPCSIGPVDIGRALCDLGA 183
+ R K DDE+ A S +R + KL+DPG+FT+PCSIG + LCDLGA
Sbjct: 492 ILERIKMYHDSDDESDATPSRASD--KRIVQEKLEDPGSFTLPCSIGELAFSDCLCDLGA 549
Query: 184 SINLMPLSMMKKLGSRELSP 203
S++LMPLS+ ++L + P
Sbjct: 550 SVSLMPLSVARRLEFIQYKP 569
>At4g03790 putative athila-like protein
Length = 1064
Score = 68.6 bits (166), Expect = 2e-12
Identities = 34/96 (35%), Positives = 60/96 (62%), Gaps = 1/96 (1%)
Query: 102 QLQVNIPFGEALDQMPVYAKFMKEMLTSRRKPKDDENIALSENCSAILQRKL-PPKLKDP 160
+++ +P E L+ +P + K ++ ++ + K D + S ++ +++ KL+DP
Sbjct: 352 EIEAVMPLMEVLNLIPDHHKDVRNLILEKIKMYHDSDDESDATPSRVVDKRIVQEKLEDP 411
Query: 161 GAFTIPCSIGPVDIGRALCDLGASINLMPLSMMKKL 196
G+FT+PCSIG + LCDLGAS++LMPLSM ++L
Sbjct: 412 GSFTLPCSIGELAFSDFLCDLGASVSLMPLSMGRRL 447
>At2g10660 putative Athila retroelement ORF1 protein
Length = 1303
Score = 67.8 bits (164), Expect = 4e-12
Identities = 51/193 (26%), Positives = 93/193 (47%), Gaps = 10/193 (5%)
Query: 12 VVEKETEHGEVEKESDQGVVENERKKKKGGEKSEKLIDEDSILRKSKSQILKDGDKPQVI 71
+V+ + + GE + + VV + + + L ++ +I+ + + KP +
Sbjct: 291 IVDSDVQEGEACTQIEISVVGLDHSAESCFQIHSNLDEKAAIIDRMVKRF-----KPAPL 345
Query: 72 PSYVKLPYPHLAKNKKKEEGQFKKFMQLFSQLQVNIPFGEALDQMPVYAKFMKEMLTSRR 131
PS LP+ ++ +K + + + I + L+ +P K ++ + R
Sbjct: 346 PSRA-LPWKFRKAWIERYNSLAEKQLDEMEAVMLLI---KVLNLIPDPHKDVRNSILERI 401
Query: 132 KPKDDENIALSENCSAI-LQRKLPPKLKDPGAFTIPCSIGPVDIGRALCDLGASINLMPL 190
K D N S + ++R + KL+DPG+FT+PCSIG + LCDLGAS++LMPL
Sbjct: 402 KMYQDSEDECDANPSRVTVKRSVQEKLEDPGSFTLPCSIGQLVFSNCLCDLGASVSLMPL 461
Query: 191 SMMKKLGSRELSP 203
S+ +KL + P
Sbjct: 462 SVARKLEFTQYRP 474
>At4g08050
Length = 1428
Score = 67.4 bits (163), Expect = 5e-12
Identities = 39/105 (37%), Positives = 60/105 (57%), Gaps = 5/105 (4%)
Query: 102 QLQVNIPFGEALDQMPVYAKFMKEMLTSRRK---PKDDENIALSENCSAILQRKLPPKLK 158
+++ +P E L+ + K ++ ++ R K DDE+ A + +R + KL+
Sbjct: 515 EIEAVMPLMEVLNLILDPHKVVRNLILERIKMYHDSDDESDATPSRAAD--KRIVQEKLE 572
Query: 159 DPGAFTIPCSIGPVDIGRALCDLGASINLMPLSMMKKLGSRELSP 203
DPG+FT+PCSIG LCDLGAS+NLMPLSM ++L + P
Sbjct: 573 DPGSFTLPCSIGEFAFSDCLCDLGASVNLMPLSMARRLEFIQYKP 617
>At3g30655 hypothetical protein, 3' partial
Length = 660
Score = 65.5 bits (158), Expect = 2e-11
Identities = 28/54 (51%), Positives = 40/54 (73%)
Query: 150 QRKLPPKLKDPGAFTIPCSIGPVDIGRALCDLGASINLMPLSMMKKLGSRELSP 203
Q+ +P KL+DPG+FT+PCSI + LCDLGAS+++MPLS+ +KLG + P
Sbjct: 584 QKIVPEKLEDPGSFTLPCSIRQLTFSNCLCDLGASVSIMPLSVARKLGFVQYKP 637
>At2g06160 putative Athila retroelement ORF1 protein
Length = 750
Score = 65.5 bits (158), Expect = 2e-11
Identities = 53/201 (26%), Positives = 94/201 (46%), Gaps = 19/201 (9%)
Query: 4 DKDVEVKEVVEKETE-----HGEVEKESDQGVVENERKKKKGGEKSEKLIDEDSILRKSK 58
D+++ + V TE GE + + V E + + + L ++ +I+ +
Sbjct: 455 DRELSTQHVSTSNTEDSVIQEGEASVQIEVSVAEIDYAAQTFYQAQSDLEEKATIIERMV 514
Query: 59 SQILKDGDKPQVIPSYVKLPYPHLAKNKKKEEGQFKKFMQLFSQLQVNIPFGEALDQMPV 118
+ KP PS + LP+ K E +K + +++ +P E L +
Sbjct: 515 KRF-----KPAPSPS-LALPWTFRKAWKSIYESLAEKQLD---EMESVMPLVEVLKLISD 565
Query: 119 YAKFMKEMLTSRR---KPKDDENIALSENCSAILQRKLPPKLKDPGAFTIPCSIGPVDIG 175
K ++ ++ R + DDE + + +R + KL+DPG+FT+PCSI +
Sbjct: 566 PHKDVRNLILERLNIYQDSDDEEDVIMHQAAG--ERIIQEKLEDPGSFTLPCSISQLTFS 623
Query: 176 RALCDLGASINLMPLSMMKKL 196
LCDLGAS++LMPLS+ +KL
Sbjct: 624 NCLCDLGASVSLMPLSVARKL 644
>At2g07480 F9A16.15
Length = 1012
Score = 64.7 bits (156), Expect = 3e-11
Identities = 49/134 (36%), Positives = 74/134 (54%), Gaps = 22/134 (16%)
Query: 64 DGDKPQVIPS-YVKLP-YPHLAKNKKKEEGQFKK----FMQLFSQLQVNIPFGEALDQMP 117
D P IPS YV P +P + + +E F+K F++ +L+ +P +
Sbjct: 477 DPSNPIHIPSPYVPKPAFPE--RIAQIDEMIFQKHKMMFIKCIKELEEKVPLVDT----- 529
Query: 118 VYAKFMKEMLTSRRKPKDDENIA-LSENCSAILQRK-LPPKLKDPGAFTIPCSIGPVDIG 175
KEM+ R P++ + I LS CSAI+Q+K +P KL DPG+FT+PCS+ P+
Sbjct: 530 -----PKEMIMER--PQEAQQIVELSFECSAIIQKKVIPKKLGDPGSFTLPCSLAPLVFN 582
Query: 176 RALCDLGASINLMP 189
+LCDLGAS++ P
Sbjct: 583 NSLCDLGASMDEEP 596
>At1g36130 hypothetical protein
Length = 914
Score = 64.7 bits (156), Expect = 3e-11
Identities = 32/101 (31%), Positives = 54/101 (52%)
Query: 110 GEALDQMPVYAKFMKEMLTSRRKPKDDENIALSENCSAILQRKLPPKLKDPGAFTIPCSI 169
G +LD+ + + + ++ + K + + ++E I + KL DPG+F + CSI
Sbjct: 546 GISLDEAKLLTRDISAIMLPKAKKEKAHRVDIAEYIRTITPGQTTEKLPDPGSFVLDCSI 605
Query: 170 GPVDIGRALCDLGASINLMPLSMMKKLGSRELSPVSVFIEF 210
R+LCDLG+SINLMP S+ ++LG P + + F
Sbjct: 606 STSRFSRSLCDLGSSINLMPKSVAERLGMTHYRPTRITLLF 646
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.134 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,018,562
Number of Sequences: 26719
Number of extensions: 233193
Number of successful extensions: 1902
Number of sequences better than 10.0: 207
Number of HSP's better than 10.0 without gapping: 104
Number of HSP's successfully gapped in prelim test: 105
Number of HSP's that attempted gapping in prelim test: 1284
Number of HSP's gapped (non-prelim): 520
length of query: 211
length of database: 11,318,596
effective HSP length: 95
effective length of query: 116
effective length of database: 8,780,291
effective search space: 1018513756
effective search space used: 1018513756
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC124970.11


