

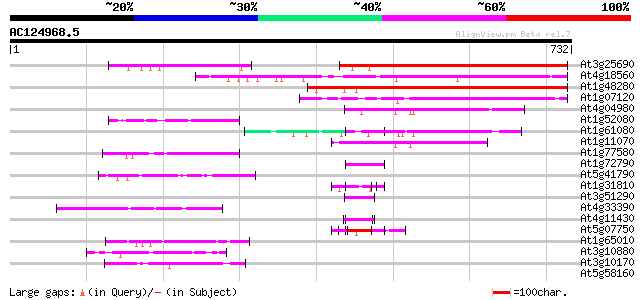
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124968.5 - phase: 0
(732 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g25690 unknown protein 329 3e-90
At4g18560 pherophorin - like protein 307 2e-83
At1g48280 unknown protein 289 4e-78
At1g07120 hypothetical protein 223 4e-58
At4g04980 81 2e-15
At1g52080 unknown protein 72 8e-13
At1g61080 hypothetical protein 70 5e-12
At1g11070 hypothetical protein 54 3e-07
At1g77580 unknown protein 50 4e-06
At1g72790 unknown protein 50 4e-06
At5g41790 myosin heavy chain-like protein 50 6e-06
At1g31810 hypothetical protein 47 4e-05
At3g51290 putative protein 45 2e-04
At4g33390 putative protein 44 2e-04
At4g11430 putative proline-rich protein 44 2e-04
At5g07750 putative protein 44 3e-04
At1g65010 hypothetical protein 44 4e-04
At3g10880 hypothetical protein 43 5e-04
At3g10170 hypothetical protein 43 5e-04
At5g58160 strong similarity to unknown protein (gb|AAD23008.1) 42 0.001
>At3g25690 unknown protein
Length = 939
Score = 329 bits (844), Expect = 3e-90
Identities = 173/319 (54%), Positives = 220/319 (68%), Gaps = 21/319 (6%)
Query: 431 LEKRALRIPNPPPRP-----SCSISSKTKQECSAQVQPPPPPP-------------PPPP 472
+EKR R+P PPPR S ++ S PPPPPP PPPP
Sbjct: 579 IEKRPPRVPRPPPRSAGGGKSTNLPSARPPLPGGGPPPPPPPPGGGPPPPPGGGPPPPPP 638
Query: 473 PMSFASRGNTA--MVKRAPQVVELYHSLMKRDSRRDSSSGGLSDAP-DVADVRSSMIGEI 529
P RG V RAP++VE Y SLMKR+S+++ + +S + + R++MIGEI
Sbjct: 639 PPGALGRGAGGGNKVHRAPELVEFYQSLMKRESKKEGAPSLISSGTGNSSAARNNMIGEI 698
Query: 530 ENRSSHLLAIKADIETQGEFVNSLIREVNDAVYENIDDVVAFVKWLDDELGFLVDERAVL 589
ENRS+ LLA+KAD+ETQG+FV SL EV + + +I+D++AFV WLD+EL FLVDERAVL
Sbjct: 699 ENRSTFLLAVKADVETQGDFVQSLATEVRASSFTDIEDLLAFVSWLDEELSFLVDERAVL 758
Query: 590 KHFDWPEKKADTLREAAFGYQDLKKLESEVSSYKDDPRLPCDIALKKMVALSEKMERTVY 649
KHFDWPE KAD LREAAF YQDL KLE +V+S+ DDP L C+ ALKKM L EK+E++VY
Sbjct: 759 KHFDWPEGKADALREAAFEYQDLMKLEKQVTSFVDDPNLSCEPALKKMYKLLEKVEQSVY 818
Query: 650 TLLRTRDSLMRNCKEFQIPVEWMLDNGIIGKIKLGSVKLAKKYMKRVAIEVQTKSAFDKD 709
LLRTRD + KEF IPV+W+ D G++GKIKL SV+LAKKYMKRVA E+ + S DKD
Sbjct: 819 ALLRTRDMAISRYKEFGIPVDWLSDTGVVGKIKLSSVQLAKKYMKRVAYELDSVSGSDKD 878
Query: 710 PAMDYMVLQGVRFAFRIHQ 728
P ++++LQGVRFAFR+HQ
Sbjct: 879 PNREFLLLQGVRFAFRVHQ 897
Score = 120 bits (300), Expect = 4e-27
Identities = 87/256 (33%), Positives = 132/256 (50%), Gaps = 69/256 (26%)
Query: 129 ELDHMRSLLQESKEREAKLNAELVE---CRKNQSEVDELVKKVAL--------------L 171
EL+ ++ L++E +ERE KL EL+E ++ +S++ EL +++ + L
Sbjct: 65 ELERLKQLVKELEEREVKLEGELLEYYGLKEQESDIVELQRQLKIKTVEIDMLNITINSL 124
Query: 172 EEEKSGLSEQL-----------VALSRSCGLERQ-------------------------- 194
+ E+ L E+L VA ++ L+RQ
Sbjct: 125 QAERKKLQEELSQNGIVRKELEVARNKIKELQRQIQLDANQTKGQLLLLKQHVSSLQMKE 184
Query: 195 ---------EEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSSE 245
E K + Q+LE++V+EL+R N+EL +KR L+ +L S E++++ N +E
Sbjct: 185 EEAMNKDTEVERKLKAVQDLEVQVMELKRKNRELQHEKRELSIKLDSAEARIATLSNMTE 244
Query: 246 SDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELKN----- 300
SD VAK + E + L+ NEDL KQVEGLQ +R +EVEEL YLRWVN+CLR EL+N
Sbjct: 245 SDKVAKVREEVNNLKHNNEDLLKQVEGLQMNRFSEVEELVYLRWVNACLRYELRNYQTPA 304
Query: 301 -TCSTLDSDKLSSPQS 315
S D K SP+S
Sbjct: 305 GKISARDLSKNLSPKS 320
>At4g18560 pherophorin - like protein
Length = 637
Score = 307 bits (786), Expect = 2e-83
Identities = 218/539 (40%), Positives = 293/539 (53%), Gaps = 78/539 (14%)
Query: 243 SSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLN-EVEEL----AYLRWVNSCLRTE 297
+S + +V++ + + LR + + E L+ + N E++EL A L N LR E
Sbjct: 81 ASHNGVVSELRRQVEELR--EREALLKTENLEIADKNGEIDELRKETARLAEDNERLRRE 138
Query: 298 ------LKNTCSTLDSD---KLSSPQSVVSSSGD----SISS-FSDQCGSANSFNLVKKP 343
++ C T + + ++ + +VSS D S+S F + NL++
Sbjct: 139 FDRSEEMRRECETREKEMEAEIVELRKLVSSESDDHALSVSQRFQGLMDVSAKSNLIRSL 198
Query: 344 KKW--------PITSSDQ----LSQVECTNNSIIDKNWIESISEGSNRRR----HSISGS 387
K+ PIT+ + +S + I K+ IES S SN S+S
Sbjct: 199 KRVGSLRNLPEPITNQENTNKSISSSGDADGDIYRKDEIESYSRSSNSEELTESSSLSTV 258
Query: 388 NSSEEEVSVLSKRRQSNCFDSFECLKEIEKESVPMPLFVQQCALEKRALRIPNPPPRPSC 447
S V +R + DS E + P P IP PPP P
Sbjct: 259 RSRVPRVPKPPPKRSISLGDS------TENRADPPP-----------QKSIPPPPPPPPP 301
Query: 448 SISSKTKQECSAQVQPPPPPPPPPPP-MSFASRGNTAMVKRAPQVVELYHSLMKRDS--- 503
+ + S PPPPPPPPPP +S AS A V+R P+VVE YHSLM+RDS
Sbjct: 302 PLLQQPPPPPSVSKAPPPPPPPPPPKSLSIAS----AKVRRVPEVVEFYHSLMRRDSTNS 357
Query: 504 RRDSSSGGLSDAPDVADVRSS--MIGEIENRSSHLLAIKADIETQGEFVNSLIREVNDAV 561
RRDS+ GG + A + ++ MIGEIENRS +LLAIK D+ETQG+F+ LI+EV +A
Sbjct: 358 RRDSTGGGNAAAEAILANSNARDMIGEIENRSVYLLAIKTDVETQGDFIRFLIKEVGNAA 417
Query: 562 YENIDDVVAFVKWLDDELGFL------------VDERAVLKHFDWPEKKADTLREAAFGY 609
+ +I+DVV FVKWLDDEL +L VDERAVLKHF+WPE+KAD LREAAF Y
Sbjct: 418 FSDIEDVVPFVKWLDDELSYLFKVCKFVVVSLKVDERAVLKHFEWPEQKADALREAAFCY 477
Query: 610 QDLKKLESEVSSYKDDPRLPCDIALKKMVALSEKMERTVYTLLRTRDSLMRNCKEFQIPV 669
DLKKL SE S +++DPR ALKKM AL EK+E VY+L R R+S K FQIPV
Sbjct: 478 FDLKKLISEASRFREDPRQSSSSALKKMQALFEKLEHGVYSLSRMRESAATKFKSFQIPV 537
Query: 670 EWMLDNGIIGKIKLGSVKLAKKYMKRVAIEVQTKSAFDKDPAMDYMVLQGVRFAFRIHQ 728
+WML+ GI +IKL SVKLA KYMKRV+ E++ P + +++QGVRFAFR+HQ
Sbjct: 538 DWMLETGITSQIKLASVKLAMKYMKRVSAELEAIEG--GGPEEEELIVQGVRFAFRVHQ 594
Score = 34.7 bits (78), Expect = 0.20
Identities = 25/115 (21%), Positives = 43/115 (36%)
Query: 450 SSKTKQECSAQVQPPPPPPPPPPPMSFASRGNTAMVKRAPQVVELYHSLMKRDSRRDSSS 509
+ KTK S PPPPPPP PP S ++ + P + R S + ++
Sbjct: 18 TKKTKDMPSPLPLPPPPPPPLKPPSSGSATTKPPINPSKPGFTRSFGVYFPRASAQVHAT 77
Query: 510 GGLSDAPDVADVRSSMIGEIENRSSHLLAIKADIETQGEFVNSLIREVNDAVYEN 564
+ V + E+ R + L +I + ++ L +E +N
Sbjct: 78 AAAASHNGVVSELRRQVEELREREALLKTENLEIADKNGEIDELRKETARLAEDN 132
>At1g48280 unknown protein
Length = 558
Score = 289 bits (739), Expect = 4e-78
Identities = 159/354 (44%), Positives = 226/354 (62%), Gaps = 14/354 (3%)
Query: 389 SSEEEVSVLS------KRRQSNCFDSFECL--KEIEKESVPMPLFVQQCALEKRA---LR 437
S+E ++S LS K Q++ F + L ++E+ V + V+ L + R
Sbjct: 169 SAEAKISSLSSNDKPAKEHQNSRFKDIQRLIASKLEQPKVKKEVAVESSRLSPPSPSPSR 228
Query: 438 IPNPPPRPSCSIS---SKTKQECSAQVQPPPPPPPPPPPMSFASRGNTAMVKRAPQVVEL 494
+P PP P +S S K++ ++ PP PPPPPPP A +++P V +L
Sbjct: 229 LPPTPPLPKFLVSPASSLGKRDENSSPFAPPTPPPPPPPPPPRPLAKAARAQKSPPVSQL 288
Query: 495 YHSLMKRDSRRDSSSGGLSDAPDVADVRSSMIGEIENRSSHLLAIKADIETQGEFVNSLI 554
+ L K+D+ R+ S + V +S++GEI+NRS+HL+AIKADIET+GEF+N LI
Sbjct: 289 FQLLNKQDNSRNLSQSVNGNKSQVNSAHNSIVGEIQNRSAHLIAIKADIETKGEFINDLI 348
Query: 555 REVNDAVYENIDDVVAFVKWLDDELGFLVDERAVLKHFDWPEKKADTLREAAFGYQDLKK 614
++V + +++DV+ FV WLD EL L DERAVLKHF WPEKKADTL+EAA Y++LKK
Sbjct: 349 QKVLTTCFSDMEDVMKFVDWLDKELATLADERAVLKHFKWPEKKADTLQEAAVEYRELKK 408
Query: 615 LESEVSSYKDDPRLPCDIALKKMVALSEKMERTVYTLLRTRDSLMRNCKEFQIPVEWMLD 674
LE E+SSY DDP + +ALKKM L +K E+ + L+R R S MR+ ++F+IPVEWMLD
Sbjct: 409 LEKELSSYSDDPNIHYGVALKKMANLLDKSEQRIRRLVRLRGSSMRSYQDFKIPVEWMLD 468
Query: 675 NGIIGKIKLGSVKLAKKYMKRVAIEVQTKSAFDKDPAMDYMVLQGVRFAFRIHQ 728
+G+I KIK S+KLAK YM RVA E+Q+ D++ + ++LQGVRFA+R HQ
Sbjct: 469 SGMICKIKRASIKLAKTYMNRVANELQSARNLDRESTKEALLLQGVRFAYRTHQ 522
>At1g07120 hypothetical protein
Length = 392
Score = 223 bits (567), Expect = 4e-58
Identities = 140/355 (39%), Positives = 207/355 (57%), Gaps = 34/355 (9%)
Query: 379 RRRHSISGSNSSEEE-VSVLSKRRQSNCFDSFECLKEIEKESVPMPLFVQQCALEKRALR 437
R R +S S E E S+L K+ QS+ S + ++ P V+ +
Sbjct: 41 RLRAQVSNLKSHENERKSMLWKKLQSSYDGS-----NTDGSNLKAPESVKS---NTKGQE 92
Query: 438 IPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFASRGNTAMVKRAPQVVELYHS 497
+ NP P+P+ S + PPPPPP P + R V+RAP+VVE Y +
Sbjct: 93 VRNPNPKPTIQGQSTATK--------PPPPPPLPSKRTLGKRS----VRRAPEVVEFYRA 140
Query: 498 LMKRDSR---RDSSSGGLSDAPDVADVRSSMIGEIENRSSHLLAIKADIETQGEFVNSLI 554
L KR+S + + +G LS A +MIGEIENRS +L IK+D + + ++ LI
Sbjct: 141 LTKRESHMGNKINQNGVLSPA-----FNRNMIGEIENRSKYLSDIKSDTDRHRDHIHILI 195
Query: 555 REVNDAVYENIDDVVAFVKWLDDELGFLVDERAVLKHFD-WPEKKADTLREAAFGYQDLK 613
+V A + +I +V FVKW+D+EL LVDERAVLKHF WPE+K D+LREAA Y+ K
Sbjct: 196 SKVEAATFTDISEVETFVKWIDEELSSLVDERAVLKHFPKWPERKVDSLREAACNYKRPK 255
Query: 614 KLESEVSSYKDDPRLPCDIALKKMVALSEKMERTVYTLLRTRDSLMRNCKEFQIPVEWML 673
L +E+ S+KD+P+ AL+++ +L +++E +V + RDS + K+FQIP EWML
Sbjct: 256 NLGNEILSFKDNPKDSLTQALQRIQSLQDRLEESVNNTEKMRDSTGKRYKDFQIPWEWML 315
Query: 674 DNGIIGKIKLGSVKLAKKYMKRVAIEVQTKSAFDKDPAMDYMVLQGVRFAFRIHQ 728
D G+IG++K S++LA++YMKR+A E+++ + + M LQGVRFA+ IHQ
Sbjct: 316 DTGLIGQLKYSSLRLAQEYMKRIAKELESNGSGKEGNLM----LQGVRFAYTIHQ 366
>At4g04980
Length = 681
Score = 80.9 bits (198), Expect = 2e-15
Identities = 62/256 (24%), Positives = 122/256 (47%), Gaps = 23/256 (8%)
Query: 437 RIPNPPPRPSCSISSKTKQEC--------SAQVQPPPPPPPPPPPMSFASRGNTAMVKRA 488
+ P PPP P S +S++ + C + P P PP PP + + T+ ++R+
Sbjct: 335 KAPAPPPPPPMSKASESGEFCQFSKTHSTNGDNAPSMPAPPAPPGSGRSLKKATSKLRRS 394
Query: 489 PQVVELYHSLMKR------DSRRDSSSGGLSDAPDVADV---RSSM---IGEIENRSSHL 536
Q+ LY +L + + + +S G + + + V RS M + E+ RSS+
Sbjct: 395 AQIANLYWALKGKLEGRGVEGKTKKASKGQNSVAEKSPVKVARSGMADALAEMTKRSSYF 454
Query: 537 LAIKADIETQGEFVNSLIREVNDAVYENIDDVVAFVKWLDDELGFLVDERAVLKHFD-WP 595
I+ D++ + + L ++ +++ +++ F ++ L L DE VL F+ +P
Sbjct: 455 QQIEEDVQKYAKSIEELKSSIHSFQTKDMKELLEFHSKVESILEKLTDETQVLARFEGFP 514
Query: 596 EKKADTLREAAFGYQDLKKLESEVSSYKDDPRLPCDIALKKMVALSEKMERTVYTLLRTR 655
EKK + +R A Y+ L + E+ ++K +P P + L K+ K + + T+ RT+
Sbjct: 515 EKKLEVIRTAGALYKKLDGILVELKNWKIEP--PLNDLLDKIERYFNKFKGEIETVERTK 572
Query: 656 DSLMRNCKEFQIPVEW 671
D + K + I +++
Sbjct: 573 DEDAKMFKRYNINIDF 588
>At1g52080 unknown protein
Length = 573
Score = 72.4 bits (176), Expect = 8e-13
Identities = 53/171 (30%), Positives = 91/171 (52%), Gaps = 18/171 (10%)
Query: 129 ELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQLVALSRS 188
ELD +S +Q K+ KLN + +++ +++ L ++VA L+EE+ + L L
Sbjct: 219 ELDMAKSQVQVLKK---KLN---INTQQHVAQILSLKQRVARLQEEE--IKAVLPDLEAD 270
Query: 189 CGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSSESDI 248
++R ++LE E+ EL N L + L+ +L S+ Q+ + E +
Sbjct: 271 KMMQR--------LRDLESEINELTDTNTRLQFENFELSEKLESV--QIIANSKLEEPEE 320
Query: 249 VAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELK 299
+ + + + LR NE+L K VE LQ R ++E+L YLRW+N+CLR EL+
Sbjct: 321 IETLREDCNRLRSENEELKKDVEQLQGDRCTDLEQLVYLRWINACLRYELR 371
>At1g61080 hypothetical protein
Length = 907
Score = 69.7 bits (169), Expect = 5e-12
Identities = 68/253 (26%), Positives = 108/253 (41%), Gaps = 41/253 (16%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPP-----------PPPPPPPMSFASRGNTAMVKR 487
P PPP P + +A PPPP PPPP S + +KR
Sbjct: 605 PPPPPPPMAMANG------AAGPPPPPPRMGMANGAAGPPPPPGAARSLRPKKAATKLKR 658
Query: 488 APQVVELYHSLMKRDSRRD------SSSG---GLSDAPDVADVRSSM---IGEIENRSSH 535
+ Q+ LY L + RD S SG G AP A + M + EI +S++
Sbjct: 659 STQLGNLYRILKGKVEGRDPNAKTGSGSGRKAGAGSAP--AGGKQGMADALAEITKKSAY 716
Query: 536 LLAIKADIETQGEFVNSLIREVNDAVYENIDDVVAFVKWLDDELGFLVDERAVLKHFD-W 594
L I+ADI +N L E+ +++ ++++F + ++ L L DE VL + +
Sbjct: 717 FLQIQADIAKYMTSINELKIEITKFQTKDMTELLSFHRRVESVLENLTDESQVLARCEGF 776
Query: 595 PEKKADTLREAAFGYQDLKKLESEVSSYKDDPRLPCDIALKKMVALSEKMERTVYTLLRT 654
P+KK + +R A Y L + +E+ + K +P L L +K+ER + T
Sbjct: 777 PQKKLEAMRMAVALYTKLHGMITELQNMKIEPPLN---------QLLDKVERYFTKIKET 827
Query: 655 RDSLMRNCKEFQI 667
+ NC E +
Sbjct: 828 MVDISSNCMELAL 840
Score = 45.8 bits (107), Expect = 8e-05
Identities = 48/201 (23%), Positives = 80/201 (38%), Gaps = 25/201 (12%)
Query: 307 SDKLSSPQSVVSSSGDSISSFSDQCGSANSFNLVKKPKKWPITSSDQLSQVECTNNSIID 366
S ++ SP S S S S + CGS + V P + +V NS
Sbjct: 249 SSRVLSPSESFSDSKSSFGSRNSFCGSPTTPRSVL-----PESMMGSPGRVGDFANSASH 303
Query: 367 KNW------IESISEGSNRRR--HSIS-------GSNSSEEEVSVLSKRRQSNCFDSFEC 411
W +E +S +R H +S ++ E+ +SV+ + +Q D E
Sbjct: 304 LLWNMRVQALEKLSPIDVKRLAIHILSQKEAQEPNESNDEDVISVVEEIKQKK--DEIES 361
Query: 412 LKEIEKESVPMPLFVQQCAL---EKRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPP 468
+ + + L + L + ++I + ++ K E S+Q+ PPPPPP
Sbjct: 362 IDVKMETEESVNLDEESVVLNGEQDTIMKISSLESTSESKLNHSEKYENSSQLFPPPPPP 421
Query: 469 PPPPPMSFASRGNTAMVKRAP 489
PPPPP+SF + + P
Sbjct: 422 PPPPPLSFIKTASLPLPSPPP 442
Score = 42.4 bits (98), Expect = 0.001
Identities = 41/147 (27%), Positives = 65/147 (43%), Gaps = 23/147 (15%)
Query: 339 LVKKPKKWPITSSDQ--LSQVECTNNSIIDKNWIESIS------EGSNRRRHSI--SGSN 388
L +K + P S+D+ +S VE K+ IESI E N S+ +G
Sbjct: 329 LSQKEAQEPNESNDEDVISVVEEIKQK---KDEIESIDVKMETEESVNLDEESVVLNGEQ 385
Query: 389 SSEEEVSVLSKRRQS--NCFDSFECLKEIEKESVPMPLFVQQCALEKRALRIPNPPPRPS 446
+ ++S L +S N + +E ++ P P ++ +L +P+PPP P
Sbjct: 386 DTIMKISSLESTSESKLNHSEKYENSSQLFPPPPPPPPPPPLSFIKTASLPLPSPPPTPP 445
Query: 447 CSISSKTKQECSAQVQPPPPPPPPPPP 473
+ + + PPPPPPPPPPP
Sbjct: 446 IADIAIS--------MPPPPPPPPPPP 464
Score = 35.4 bits (80), Expect = 0.11
Identities = 18/57 (31%), Positives = 24/57 (41%), Gaps = 8/57 (14%)
Query: 433 KRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPP--------PPPMSFASRGN 481
+ A+ P PPP P + + PPPPPPP PPPM + G+
Sbjct: 531 RAAVAPPPPPPPPGTAAAPPPPPPPPGTQAAPPPPPPPPMQNRAPSPPPMPMGNSGS 587
Score = 34.7 bits (78), Expect = 0.20
Identities = 13/33 (39%), Positives = 16/33 (48%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPP 471
P PPP P +++ PPPPPPPP
Sbjct: 524 PPPPPPPRAAVAPPPPPPPPGTAAAPPPPPPPP 556
Score = 31.6 bits (70), Expect = 1.7
Identities = 14/34 (41%), Positives = 19/34 (55%), Gaps = 1/34 (2%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPP 472
P PPP P+ +I++ + PPPPPPPP
Sbjct: 511 PPPPPLPT-TIAAPPPPPPPPRAAVAPPPPPPPP 543
Score = 30.0 bits (66), Expect = 4.8
Identities = 15/38 (39%), Positives = 17/38 (44%), Gaps = 8/38 (21%)
Query: 441 PPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFAS 478
PPP P K PPPPPPPP P + A+
Sbjct: 493 PPPTPPAFKPLKGSA--------PPPPPPPPLPTTIAA 522
>At1g11070 hypothetical protein
Length = 554
Score = 53.9 bits (128), Expect = 3e-07
Identities = 49/215 (22%), Positives = 89/215 (40%), Gaps = 17/215 (7%)
Query: 421 PMPLFVQQCALEKRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFASRG 480
P+P+ A+ K P PPP + + + PPPPPP ++
Sbjct: 237 PLPM-----AVRKGVAAPPLPPPGTAALPPPPPLPMAAGKGVAAPPPPPPGARGGLGAKK 291
Query: 481 NTAMVKRAPQVVELYHSLMKR------DSRRDSSSGGLSDAPDVADV-----RSSMIGEI 529
T+ +KR+ + L+ L + + R + GG A A + + EI
Sbjct: 292 VTSKLKRSTHLGALFRFLKGKLEGKNPEVRSRGAGGGSKGATGSAPASGKQGMADALAEI 351
Query: 530 ENRSSHLLAIKADIETQGEFVNSLIREVNDAVYENIDDVVAFVKWLDDELGFLVDERAVL 589
+S + I+ D+ +N L ++ ++I ++ F ++ L L DE VL
Sbjct: 352 TKKSPYFQKIEEDVRMYMTSINELKTDITKFKNKDITELQKFHHRIESVLEKLEDETQVL 411
Query: 590 KHFD-WPEKKADTLREAAFGYQDLKKLESEVSSYK 623
+ +P KK + +R AA Y L+ + E+ ++K
Sbjct: 412 ARCEGFPHKKLEAIRMAAALYSKLEGMIKELKNWK 446
>At1g77580 unknown protein
Length = 779
Score = 50.1 bits (118), Expect = 4e-06
Identities = 48/196 (24%), Positives = 82/196 (41%), Gaps = 26/196 (13%)
Query: 122 SSTDLFTELDHMRSLLQESKEREAKLNAEL------VECRKNQ------------SEVDE 163
S L +E++ + S ++E +E+ KL AE V+C + + S E
Sbjct: 337 SENSLASEIEVLTSRIKELEEKLEKLEAEKHELENEVKCNREEAVVHIENSEVLTSRTKE 396
Query: 164 LVKKVALLEEEKSGLSEQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQK 223
L +K+ LE EK L ++ C E+ + S L E+ L KEL Q
Sbjct: 397 LEEKLEKLEAEKEELKSEV-----KCNREKAVVHVENS---LAAEIEVLTSRTKELEEQL 448
Query: 224 RNLTCRLSSMESQLSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEE 283
L +ES++ C+ + + + E +L + L +++E L+ + E
Sbjct: 449 EKLEAEKVELESEVKCNREEAVAQVENSLATEIEVLTCRIKQLEEKLEKLEVEKDELKSE 508
Query: 284 LAYLRWVNSCLRTELK 299
+ R V S LR EL+
Sbjct: 509 VKCNREVESTLRFELE 524
>At1g72790 unknown protein
Length = 561
Score = 50.1 bits (118), Expect = 4e-06
Identities = 22/51 (43%), Positives = 29/51 (56%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFASRGNTAMVKRAP 489
P PPP SSK + PPPPPPPPPP + SR +T+ +++AP
Sbjct: 370 PPPPPFFQGLFSSKKGKSKKNNSNPPPPPPPPPPERRYESRASTSKLRKAP 420
Score = 33.5 bits (75), Expect = 0.44
Identities = 16/56 (28%), Positives = 28/56 (49%), Gaps = 1/56 (1%)
Query: 426 VQQCALEKRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFASRGN 481
+++ +E R + PNPP + + + + ++ PPPPPP P F RG+
Sbjct: 416 LRKAPVESRTSK-PNPPAKVTQYVGTGSESPLMPIPPPPPPPPFKMPAWKFVKRGD 470
Score = 32.3 bits (72), Expect = 0.97
Identities = 11/13 (84%), Positives = 11/13 (84%)
Query: 461 VQPPPPPPPPPPP 473
V PPPPPPPPPP
Sbjct: 362 VYSPPPPPPPPPP 374
Score = 29.6 bits (65), Expect = 6.3
Identities = 10/12 (83%), Positives = 10/12 (83%)
Query: 465 PPPPPPPPPMSF 476
PPPPPPPPP F
Sbjct: 365 PPPPPPPPPPFF 376
>At5g41790 myosin heavy chain-like protein
Length = 1305
Score = 49.7 bits (117), Expect = 6e-06
Identities = 63/220 (28%), Positives = 102/220 (45%), Gaps = 32/220 (14%)
Query: 117 THRRQSSTDLFTELDHMRSLLQE----------SKEREAKLNAELV-----ECRKNQSEV 161
TH+R+SST L +EL+ LL++ + E E K + ++ E ++ QS+V
Sbjct: 478 THQRESSTRL-SELETQLKLLEQRVVDLSASLNAAEEEKKSLSSMILEITDELKQAQSKV 536
Query: 162 DELVKKVALLEEEKSGLSEQLVALSRSCGL-ERQEEDKDGSTQNLELEVVELRRLNKELH 220
ELV ++A E K L+++ LS + E + D + LE V KEL+
Sbjct: 537 QELVTELA---ESKDTLTQKENELSSFVEVHEAHKRDSSSQVKELEARVESAEEQVKELN 593
Query: 221 MQKRNLTCRLSSMESQLSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNE 280
+NL SS E + S SE I K +AE+++ ++LS + E L+ S +
Sbjct: 594 ---QNLN---SSEEEKKILSQQISEMSIKIK-RAESTI-----QELSSESERLKGSHAEK 641
Query: 281 VEELAYLRWVNSCLRTELKNTCSTLDSDKLSSPQSVVSSS 320
EL LR ++ + EL L++ SS V+ S
Sbjct: 642 DNELFSLRDIHETHQRELSTQLRGLEAQLESSEHRVLELS 681
Score = 42.0 bits (97), Expect = 0.001
Identities = 70/288 (24%), Positives = 113/288 (38%), Gaps = 42/288 (14%)
Query: 136 LLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQLVALSRSCGLERQE 195
L ++ E+E+KL + K+Q ++ EL VA LE E + +++ LE +
Sbjct: 722 LKEQLAEKESKLFLLTEKDSKSQVQIKELEATVATLELELESVRARII------DLETEI 775
Query: 196 EDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSSESDIVAKFKAE 255
K + LE + E+ EL LS++ +L +D S S I AE
Sbjct: 776 ASKTTVVEQLEAQNREMVARISELEKTMEERGTELSALTQKLEDNDKQSSSSI-ETLTAE 834
Query: 256 ASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVNSCLRTELKNTCSTLDSDKLSSPQS 315
LR + +S Q E ++ + + EE S K+
Sbjct: 835 IDGLRAELDSMSVQKEEVEKQMVCKSEEA----------------------SVKIKRLDD 872
Query: 316 VVSSSGDSISSFSDQCGSANSFNLVKKPKKWPITSSDQLSQVECTNNSIIDKNWI-ESIS 374
V+ ++S Q L KK ++ S+ LSQ+ II+K + ESI
Sbjct: 873 EVNGLRQQVASLDSQRAEL-EIQLEKKSEE----ISEYLSQITNLKEEIINKVKVHESIL 927
Query: 375 EGSNRRRHSISGSNSSEEEVSVLSKRRQSNCFDSFECLKEIEKESVPM 422
E N I G E E+ L K+R + E L+ ++E+V M
Sbjct: 928 EEINGLSEKIKG---RELELETLGKQRS----ELDEELRTKKEENVQM 968
Score = 30.0 bits (66), Expect = 4.8
Identities = 37/178 (20%), Positives = 76/178 (41%), Gaps = 20/178 (11%)
Query: 124 TDLFTEL-DHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQL 182
T+L L + + SL + E EA+L E E + +++ ++ K + E + L E+
Sbjct: 983 TELINNLKNELDSLQVQKSETEAELEREKQEKSELSNQITDVQKALVEQEAAYNTLEEEH 1042
Query: 183 VALSRSCGLERQEEDKDGSTQNLELEVVELRRL----NKELHMQKRNLTCRLSSMES--- 235
++ ++ + + + ++ E +RL KE+ + + +MES
Sbjct: 1043 KQINELF------KETEATLNKVTVDYKEAQRLLEERGKEVTSRDSTIGVHEETMESLRN 1096
Query: 236 QLSCSDNSSE------SDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYL 287
+L + E S+I K + LR+T + L+++ E + +EE A L
Sbjct: 1097 ELEMKGDEIETLMEKISNIEVKLRLSNQKLRVTEQVLTEKEEAFRKEEAKHLEEQALL 1154
>At1g31810 hypothetical protein
Length = 1201
Score = 47.0 bits (110), Expect = 4e-05
Identities = 26/65 (40%), Positives = 28/65 (43%), Gaps = 14/65 (21%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPP--------------PPPPMSFASRGNTAM 484
P PPP PS SI Q + PPPPPPP PPPP SF S GN
Sbjct: 577 PPPPPLPSRSIPPPLAQPPPPRPPPPPPPPPSSRSIPSPSAPPPPPPPPPSFGSTGNKRQ 636
Query: 485 VKRAP 489
+ P
Sbjct: 637 AQPPP 641
Score = 45.1 bits (105), Expect = 1e-04
Identities = 24/61 (39%), Positives = 31/61 (50%), Gaps = 3/61 (4%)
Query: 421 PMPLFVQQ---CALEKRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFA 477
P+P F + L + + P PPP P + S++ AQ PP PPPPPPPP S
Sbjct: 551 PLPSFSNRDPLTTLHQPINKTPPPPPPPPPPLPSRSIPPPLAQPPPPRPPPPPPPPPSSR 610
Query: 478 S 478
S
Sbjct: 611 S 611
Score = 42.7 bits (99), Expect = 7e-04
Identities = 19/34 (55%), Positives = 20/34 (57%), Gaps = 3/34 (8%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPP 472
P PPP P S S + Q QPPPPPPPPPP
Sbjct: 618 PPPPPPPPPSFGSTGNKR---QAQPPPPPPPPPP 648
Score = 40.8 bits (94), Expect = 0.003
Identities = 44/183 (24%), Positives = 67/183 (36%), Gaps = 29/183 (15%)
Query: 311 SSPQSVVSSSGDSISSFSDQCGSANSFNLVKKPKKWPITSSDQLSQVECTNNSIIDKNWI 370
+SP S ++ S + SD+ F +++P+ +D + + +
Sbjct: 391 NSPDSEEETNTSSAADSSDE-----GFEAIQRPRIHIPFDNDDTDDITLS----VAHESS 441
Query: 371 ESISEGSNRRRHSISGSNSSEEEVSVLSKRRQSNCFDSFECLKEIEKESVPMPLFVQQCA 430
E E S+ H I +S + +++ S S L P PLF +
Sbjct: 442 EEPHEFSHHHHHEIPAKDSVDNPLNLPSDPPSSG---DHVTLLPPPPPPPPPPLFTSTTS 498
Query: 431 LEKRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPP-------------PPPPPPM-SF 476
P PPP P S T S PPPPP PPPPPP+ SF
Sbjct: 499 FSPSQ---PPPPPPPPPLFMSTTSFSPSQPPPPPPPPPLFTSTTSFSPSQPPPPPPLPSF 555
Query: 477 ASR 479
++R
Sbjct: 556 SNR 558
Score = 39.3 bits (90), Expect = 0.008
Identities = 18/42 (42%), Positives = 22/42 (51%), Gaps = 5/42 (11%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPP-----PPPPPPPPMS 475
P PPP P +IS+ K + P PPPPPPPP+S
Sbjct: 682 PPPPPPPKANISNAPKPPAPPPLPPSSTRLGAPPPPPPPPLS 723
Score = 38.5 bits (88), Expect = 0.014
Identities = 22/55 (40%), Positives = 26/55 (47%), Gaps = 4/55 (7%)
Query: 421 PMPLFVQQCALEKRALRIPNPPPRPSCSISSK--TKQECSAQVQPPPPPPPPPPP 473
P PLF + + P PPP PS S T + + PPPPPPPPP P
Sbjct: 531 PPPLFTSTTSFSPS--QPPPPPPLPSFSNRDPLTTLHQPINKTPPPPPPPPPPLP 583
Score = 37.7 bits (86), Expect = 0.023
Identities = 20/43 (46%), Positives = 22/43 (50%), Gaps = 6/43 (13%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFASRGN 481
P PPP P S S S +V PP PPPPPPP A+ N
Sbjct: 658 PPPPPPPPTSHSG------SIRVGPPSTPPPPPPPPPKANISN 694
Score = 36.6 bits (83), Expect = 0.051
Identities = 25/79 (31%), Positives = 28/79 (34%), Gaps = 23/79 (29%)
Query: 421 PMPLFVQQCALEKRALRIPNPPPRPSCSISSKTKQECSAQVQPPP--------------- 465
P PLF+ + P PPP P +S T S PPP
Sbjct: 510 PPPLFMSTTSFSPSQ---PPPPPPPPPLFTSTTSFSPSQPPPPPPLPSFSNRDPLTTLHQ 566
Query: 466 -----PPPPPPPPMSFASR 479
PPPPPPPP SR
Sbjct: 567 PINKTPPPPPPPPPPLPSR 585
Score = 33.1 bits (74), Expect = 0.57
Identities = 21/71 (29%), Positives = 27/71 (37%), Gaps = 19/71 (26%)
Query: 433 KRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPP--------------PPPPMSFAS 478
KR + P PPP P T+ + PPPPPPP PPPP
Sbjct: 634 KRQAQPPPPPPPPP-----PTRIPAAKCAPPPPPPPPTSHSGSIRVGPPSTPPPPPPPPP 688
Query: 479 RGNTAMVKRAP 489
+ N + + P
Sbjct: 689 KANISNAPKPP 699
Score = 33.1 bits (74), Expect = 0.57
Identities = 16/39 (41%), Positives = 21/39 (53%), Gaps = 7/39 (17%)
Query: 432 EKRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPP 470
+++A P PPP P I + A+ PPPPPPPP
Sbjct: 634 KRQAQPPPPPPPPPPTRIPA-------AKCAPPPPPPPP 665
>At3g51290 putative protein
Length = 602
Score = 44.7 bits (104), Expect = 2e-04
Identities = 19/39 (48%), Positives = 23/39 (58%)
Query: 437 RIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMS 475
R P PP P ++ T S+ + PPPPPPPPPPP S
Sbjct: 80 RPPPPPLSPGSETTTWTTTTTSSVLPPPPPPPPPPPPPS 118
Score = 41.6 bits (96), Expect = 0.002
Identities = 21/45 (46%), Positives = 23/45 (50%), Gaps = 8/45 (17%)
Query: 439 PNPPPRP--------SCSISSKTKQECSAQVQPPPPPPPPPPPMS 475
P PPPRP S + + T S PPPPPPPPPPP S
Sbjct: 75 PPPPPRPPPPPLSPGSETTTWTTTTTSSVLPPPPPPPPPPPPPSS 119
Score = 34.3 bits (77), Expect = 0.26
Identities = 13/20 (65%), Positives = 14/20 (70%)
Query: 463 PPPPPPPPPPPMSFASRGNT 482
PPPPP PPPPP+S S T
Sbjct: 75 PPPPPRPPPPPLSPGSETTT 94
>At4g33390 putative protein
Length = 779
Score = 44.3 bits (103), Expect = 2e-04
Identities = 43/216 (19%), Positives = 98/216 (44%), Gaps = 10/216 (4%)
Query: 62 KQLPTKKQQNTTTITTSDVVTNQKNVNPFVPPHSRVKRSLMGDLSCSQVHPHAFPTHRRQ 121
K+ K++ + T+ T+ ++ Q+ + K+ L +++ + + +
Sbjct: 422 KESSKVKEETSETVVTNIEISLQEKTTDIQKAVASAKKELE-EVNANVEKATSEVNCLKV 480
Query: 122 SSTDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQ 181
+S+ L E+D +S L K+RE + + ++E+D ++AL++ ++ E+
Sbjct: 481 ASSSLRLEIDKEKSALDSLKQREGMASVTVASL---EAEIDITRCEIALVKSKEKETREE 537
Query: 182 LVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSD 241
+V L + L++ ++ D + EL ELR+ +E K S+MES+L +
Sbjct: 538 MVELPKQ--LQQASQEADEAKSFAELAREELRKSQEEAEQAKAG----ASTMESRLFAAQ 591
Query: 242 NSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSR 277
E+ ++ A A++ L + S + + + R
Sbjct: 592 KEIEAIKASERLALAAIKALQESESSSKENAVDSPR 627
Score = 29.3 bits (64), Expect = 8.2
Identities = 14/50 (28%), Positives = 24/50 (48%), Gaps = 3/50 (6%)
Query: 593 DWPEKKADTLREAAFGYQDLKKLESEVSSYKDDPRLPCDIALKKMVALSE 642
DW + L F Q+L K++ E+ YK + + + KM+A+ E
Sbjct: 177 DWKAHRMKVLERRNFVEQELDKIQEEIPEYKKKSEM---VEMSKMLAVEE 223
>At4g11430 putative proline-rich protein
Length = 219
Score = 44.3 bits (103), Expect = 2e-04
Identities = 18/35 (51%), Positives = 20/35 (56%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P S + T + PPPPPPPPPPP
Sbjct: 97 PPPPPPPPLSAITTTGHHHHRRSPPPPPPPPPPPP 131
Score = 43.1 bits (100), Expect = 5e-04
Identities = 18/40 (45%), Positives = 23/40 (57%)
Query: 436 LRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMS 475
LR P PPP P +I++ PPPPPPPPPP ++
Sbjct: 95 LRPPPPPPPPLSAITTTGHHHHRRSPPPPPPPPPPPPTIT 134
Score = 40.8 bits (94), Expect = 0.003
Identities = 16/35 (45%), Positives = 21/35 (59%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP + +++ T + PPPPPPPPPPP
Sbjct: 127 PPPPPTITPPVTTTTAGHHHHRRSPPPPPPPPPPP 161
Score = 38.1 bits (87), Expect = 0.018
Identities = 17/40 (42%), Positives = 21/40 (52%), Gaps = 5/40 (12%)
Query: 439 PNPPPRPSCSISSKTKQECSA-----QVQPPPPPPPPPPP 473
P PPP P +I+ + + PPPPPPPPPPP
Sbjct: 123 PPPPPPPPPTITPPVTTTTAGHHHHRRSPPPPPPPPPPPP 162
Score = 30.4 bits (67), Expect = 3.7
Identities = 14/40 (35%), Positives = 19/40 (47%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFAS 478
P PPP P+ + T +PPPPPP P++ S
Sbjct: 156 PPPPPPPTITPPVTTTTTGHHHHRPPPPPPATTTPITNTS 195
Score = 29.6 bits (65), Expect = 6.3
Identities = 18/53 (33%), Positives = 23/53 (42%), Gaps = 5/53 (9%)
Query: 434 RALRIPNPPPRPS-----CSISSKTKQECSAQVQPPPPPPPPPPPMSFASRGN 481
R LR P+ P + C S S Q PPPPPPPP + + G+
Sbjct: 61 RQLRPPSIPVTTNTGHRHCRPPSNPATTNSGHHQLRPPPPPPPPLSAITTTGH 113
Score = 29.6 bits (65), Expect = 6.3
Identities = 13/30 (43%), Positives = 17/30 (56%), Gaps = 6/30 (20%)
Query: 441 PPPRPSCSISSKTKQECSAQVQPPPPPPPP 470
PP P+ + S Q++PPPPPPPP
Sbjct: 81 PPSNPATTNSGHH------QLRPPPPPPPP 104
>At5g07750 putative protein
Length = 1289
Score = 43.9 bits (102), Expect = 3e-04
Identities = 21/63 (33%), Positives = 29/63 (45%), Gaps = 2/63 (3%)
Query: 429 CALEKRALRIPNPPPRPSC--SISSKTKQECSAQVQPPPPPPPPPPPMSFASRGNTAMVK 486
C+ + P PPP P S+ K+ ++Q+ PPPPPPPPP S T +
Sbjct: 752 CSTSQAPTSSPTPPPPPPAYYSVGQKSSDLQTSQLPSPPPPPPPPPFASVRRNSETLLPP 811
Query: 487 RAP 489
P
Sbjct: 812 PPP 814
Score = 43.5 bits (101), Expect = 4e-04
Identities = 18/32 (56%), Positives = 20/32 (62%)
Query: 441 PPPRPSCSISSKTKQECSAQVQPPPPPPPPPP 472
PPP PS + +KT S V PPPPPPPPP
Sbjct: 624 PPPLPSLTSEAKTVLHSSQAVASPPPPPPPPP 655
Score = 43.1 bits (100), Expect = 5e-04
Identities = 27/81 (33%), Positives = 38/81 (46%), Gaps = 6/81 (7%)
Query: 439 PNPPPRPSCSISS---KTKQECSAQVQPPPPPPPPPPPMSFASRGNTAMVKRAPQVVELY 495
P PPP P S+ + +T + CS PPPPPPPPP ++ N ++ P Y
Sbjct: 833 PPPPPPPWKSLYASTFETHEACSTSSSPPPPPPPPPFSPLNTTKANDYILPPPPLP---Y 889
Query: 496 HSLMKRDSRRDSSSGGLSDAP 516
S+ S + G+S AP
Sbjct: 890 TSIAPSPSVKILPLHGISSAP 910
Score = 42.7 bits (99), Expect = 7e-04
Identities = 21/71 (29%), Positives = 38/71 (52%), Gaps = 4/71 (5%)
Query: 421 PMPLF--VQQCALEKRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFAS 478
P P + V Q + + + ++P+PPP P + ++ +++ PPPPPPPPPP +
Sbjct: 767 PPPAYYSVGQKSSDLQTSQLPSPPPPPPPPPFASVRR--NSETLLPPPPPPPPPPFASVR 824
Query: 479 RGNTAMVKRAP 489
R + ++ P
Sbjct: 825 RNSETLLPPPP 835
Score = 37.7 bits (86), Expect = 0.023
Identities = 25/71 (35%), Positives = 31/71 (43%), Gaps = 21/71 (29%)
Query: 437 RIPNPPPRPSCSISSKTKQECSAQVQPPPP-----------------PPPPPPPMSFAS- 478
++P PPP P S + S V PPPP PPPPPPP+ F+S
Sbjct: 666 QLPPPPPPPPPFSSERPN---SGTVLPPPPPPPPPFSSERPNSGTVLPPPPPPPLPFSSE 722
Query: 479 RGNTAMVKRAP 489
R N+ V P
Sbjct: 723 RPNSGTVLPPP 733
Score = 37.0 bits (84), Expect = 0.039
Identities = 19/50 (38%), Positives = 22/50 (44%), Gaps = 10/50 (20%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPP----------PPPPPPPMSFAS 478
P PPP P + S + PPPP PPPPPPP S+ S
Sbjct: 919 PPPPPPPFSNAHSVLSPPPPSYGSPPPPPPPPPSYGSPPPPPPPPPSYGS 968
Score = 36.6 bits (83), Expect = 0.051
Identities = 20/44 (45%), Positives = 21/44 (47%), Gaps = 9/44 (20%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPP----PPPPPPPPMSFAS 478
P PPP P S S PPP PPPPPPPP S+ S
Sbjct: 956 PPPPPPPPPSYGSPPPPP-----PPPPGYGSPPPPPPPPPSYGS 994
Score = 35.4 bits (80), Expect = 0.11
Identities = 19/41 (46%), Positives = 19/41 (46%), Gaps = 14/41 (34%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPP-----PPPPPPPM 474
P PPP P AQ PPPP PPPPPPPM
Sbjct: 1062 PPPPPPPMFG---------GAQPPPPPPMRGGAPPPPPPPM 1093
Score = 35.4 bits (80), Expect = 0.11
Identities = 15/34 (44%), Positives = 16/34 (46%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPP 472
P PPP P S + PPPPPPPPP
Sbjct: 918 PPPPPPPPFSNAHSVLSPPPPSYGSPPPPPPPPP 951
Score = 35.0 bits (79), Expect = 0.15
Identities = 19/50 (38%), Positives = 19/50 (38%), Gaps = 10/50 (20%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPP----------PPMSFAS 478
P PPP PS PPPPPPPPP PP S S
Sbjct: 958 PPPPPPPSYGSPPPPPPPPPGYGSPPPPPPPPPSYGSPPPPPPPPFSHVS 1007
Score = 35.0 bits (79), Expect = 0.15
Identities = 20/49 (40%), Positives = 21/49 (42%), Gaps = 13/49 (26%)
Query: 439 PNPPPRPSCSISS------KTKQECSAQVQPPPPP-------PPPPPPM 474
P PPP P +SS A PPPPP PPPPPPM
Sbjct: 996 PPPPPPPFSHVSSIPPPPPPPPMHGGAPPPPPPPPMHGGAPPPPPPPPM 1044
Score = 34.3 bits (77), Expect = 0.26
Identities = 24/79 (30%), Positives = 31/79 (38%), Gaps = 5/79 (6%)
Query: 421 PMPLFVQQCALEKRALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFASRG 480
P P F + L P PPP P S ++ S V PPPPPPP P + G
Sbjct: 673 PPPPFSSERPNSGTVLPPPPPPPPPFSS-----ERPNSGTVLPPPPPPPLPFSSERPNSG 727
Query: 481 NTAMVKRAPQVVELYHSLM 499
+P +Y S +
Sbjct: 728 TVLPPPPSPPWKSVYASAL 746
Score = 34.3 bits (77), Expect = 0.26
Identities = 17/45 (37%), Positives = 24/45 (52%), Gaps = 3/45 (6%)
Query: 438 IPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFASRGNT 482
+P PPP P +S + +++ PPPPPPPP +AS T
Sbjct: 809 LPPPPPPPPPPFASVRR---NSETLLPPPPPPPPWKSLYASTFET 850
Score = 34.3 bits (77), Expect = 0.26
Identities = 19/50 (38%), Positives = 21/50 (42%), Gaps = 7/50 (14%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPP-------PPPPPMSFASRGN 481
P PPP PS + PPPPPP PPPPP S G+
Sbjct: 945 PPPPPPPSYGSPPPPPPPPPSYGSPPPPPPPPPGYGSPPPPPPPPPSYGS 994
Score = 33.9 bits (76), Expect = 0.33
Identities = 19/43 (44%), Positives = 19/43 (44%), Gaps = 13/43 (30%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPP-------PPPPPPPPM 474
P PPP P S S PPP PPPPPPPPM
Sbjct: 982 PPPPPPPPPSYGSPPPPP------PPPFSHVSSIPPPPPPPPM 1018
Score = 33.5 bits (75), Expect = 0.44
Identities = 16/38 (42%), Positives = 18/38 (47%), Gaps = 3/38 (7%)
Query: 434 RALRIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPP 471
+A+ P PPP P T PPPPPPPPP
Sbjct: 642 QAVASPPPPPPPP---PLPTYSHYQTSQLPPPPPPPPP 676
Score = 33.1 bits (74), Expect = 0.57
Identities = 18/41 (43%), Positives = 18/41 (43%), Gaps = 14/41 (34%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPP-----PPPPPPPM 474
P PPP P A PPPP PPPPPPPM
Sbjct: 1098 PPPPPPP---------MRGGAPPPPPPPMHGGAPPPPPPPM 1129
Score = 32.7 bits (73), Expect = 0.74
Identities = 19/46 (41%), Positives = 22/46 (47%), Gaps = 9/46 (19%)
Query: 441 PPPRPSCSISSKTKQEC------SAQVQPP---PPPPPPPPPMSFA 477
PPP P SI+ + S+ PP PPPPPPPP S A
Sbjct: 884 PPPLPYTSIAPSPSVKILPLHGISSAPSPPVKTAPPPPPPPPFSNA 929
Score = 32.7 bits (73), Expect = 0.74
Identities = 18/42 (42%), Positives = 18/42 (42%), Gaps = 15/42 (35%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPP------PPPPPPM 474
P PPP P A PPPPP PPPPPPM
Sbjct: 1037 PPPPPPP---------MHGGAPPPPPPPPMHGGAPPPPPPPM 1069
Score = 32.3 bits (72), Expect = 0.97
Identities = 18/43 (41%), Positives = 18/43 (41%), Gaps = 16/43 (37%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPP-------PPPPPPM 474
P PPP P A PPPPP PPPPPPM
Sbjct: 1024 PPPPPPP---------MHGGAPPPPPPPPMHGGAPPPPPPPPM 1057
Score = 30.4 bits (67), Expect = 3.7
Identities = 17/42 (40%), Positives = 17/42 (40%), Gaps = 16/42 (38%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPP-------PPPPPPP 473
P PPP P A PPPP PPPPPPP
Sbjct: 1122 PPPPPPP---------MRGGAPPPPPPPGGRGPGAPPPPPPP 1154
>At1g65010 hypothetical protein
Length = 1318
Score = 43.5 bits (101), Expect = 4e-04
Identities = 51/201 (25%), Positives = 95/201 (46%), Gaps = 20/201 (9%)
Query: 126 LFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDEL---VKKVALLE---EEKSGLS 179
L +EL +++LL +E+EA E+V K +SE++ L ++KV++LE +E+ GL
Sbjct: 204 LASELGRLKALLGSKEEKEAIEGNEIVS--KLKSEIELLRGELEKVSILESSLKEQEGLV 261
Query: 180 EQL------VALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSM 233
EQL ++ SC EE K+ LE EV E R ++ +L+ +
Sbjct: 262 EQLKVDLEAAKMAESCTNSSVEEWKN-KVHELEKEVEESNRSKSSASESMESVMKQLAEL 320
Query: 234 ESQL--SCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRWVN 291
L + SDN+++ + + + R E+ +QV + +++E L + +
Sbjct: 321 NHVLHETKSDNAAQKEKIELLEKTIEAQRTDLEEYGRQV-CIAKEEASKLENL--VESIK 377
Query: 292 SCLRTELKNTCSTLDSDKLSS 312
S L + LD++K ++
Sbjct: 378 SELEISQEEKTRALDNEKAAT 398
Score = 37.7 bits (86), Expect = 0.023
Identities = 44/198 (22%), Positives = 92/198 (46%), Gaps = 11/198 (5%)
Query: 120 RQSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLS 179
R+ T L + + + L + ++ +KL + E + + +KK+ L + LS
Sbjct: 837 RERETTLLKKAEELSELNESLVDKASKLQTVVQENEELRERETAYLKKIEELSKLHEILS 896
Query: 180 EQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSC 239
+Q L S +EE K+ T L+ ++ EL ++ ++L ++ L + +E L
Sbjct: 897 DQETKLQIS--NHEKEELKERETAYLK-KIEELSKVQEDLLNKENELHGMVVEIE-DLRS 952
Query: 240 SDNSSESDIVAKFKAEASLLRLTNE-----DLSKQVEGLQTSRLNEVEELAYLRWVNSCL 294
D+ ++ I ASLL NE +++++ Q S L ++EL+ L+ S +
Sbjct: 953 KDSLAQKKIEELSNFNASLLIKENELQAVVCENEELKSKQVSTLKTIDELSDLK--QSLI 1010
Query: 295 RTELKNTCSTLDSDKLSS 312
E + + ++++KL +
Sbjct: 1011 HKEKELQAAIVENEKLKA 1028
Score = 37.4 bits (85), Expect = 0.030
Identities = 67/288 (23%), Positives = 121/288 (41%), Gaps = 42/288 (14%)
Query: 121 QSSTDLFTELDHMRSLLQESKEREAKLNAELVECRK-NQSEVDELVKKVALLEEEKSGLS 179
+S D T+L H+ +E + REA ++ E K N++ VD + + EE K
Sbjct: 741 ESLVDKETKLQHIDQEAEELRGREASHLKKIEELSKENENLVDNVANMQNIAEESKDLRE 800
Query: 180 EQLVALSRSCGLERQE---EDKDGSTQNLELEVVELR-------RLNKELHMQKRNLTCR 229
++ L + L D + QN+ E ELR + +EL +L +
Sbjct: 801 REVAYLKKIDELSTANGTLADNVTNLQNISEENKELRERETTLLKKAEELSELNESLVDK 860
Query: 230 LSSMESQLSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVE----ELA 285
S +++ + ++ E + K E L +E LS Q LQ S + E E A
Sbjct: 861 ASKLQTVVQENEELRERETAYLKKIEE--LSKLHEILSDQETKLQISNHEKEELKERETA 918
Query: 286 YLRWVN--SCLRTELKNTCSTL-----DSDKLSSPQSVVSSSGDSISSFSDQCGSANSFN 338
YL+ + S ++ +L N + L + + L S S+ + +S+F N+
Sbjct: 919 YLKKIEELSKVQEDLLNKENELHGMVVEIEDLRSKDSLAQKKIEELSNF-------NASL 971
Query: 339 LVKKPKKWPITSSDQLSQVECTNNSIIDK--NWIESISEGSNRRRHSI 384
L+K+ ++L V C N + K + +++I E S+ ++ I
Sbjct: 972 LIKE---------NELQAVVCENEELKSKQVSTLKTIDELSDLKQSLI 1010
Score = 35.0 bits (79), Expect = 0.15
Identities = 70/312 (22%), Positives = 135/312 (42%), Gaps = 41/312 (13%)
Query: 129 ELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQLVALSRS 188
+++ + LQE+ ++ A L+ C++ + V + L +E + E+++ +R+
Sbjct: 428 DMESLTLALQEASTESSEAKATLLVCQEELKNCESQVDSLKLASKETNEKYEKMLEDARN 487
Query: 189 CGLERQEEDKDGSTQNLELEVVELRRLNKELHMQKRNLTCRLSSMESQLSCSDNSSESDI 248
++ + D S QN E E + KELH+ + C S E +NSS +
Sbjct: 488 -EIDSLKSTVD-SIQN-EFENSKAGWEQKELHL----MGCVKKSEE------ENSSSQEE 534
Query: 249 VAKFKAEASLLRLTNED--LSKQVEGLQTSRLNEVE-ELAYLRWVNSCLRTE-LKNTCST 304
V++ +LL+ + ED K+ E + L E E+ YL+ + E +K S
Sbjct: 535 VSRL---VNLLKESEEDACARKEEEASLKNNLKVAEGEVKYLQETLGEAKAESMKLKESL 591
Query: 305 LDSDK-LSSPQSVVSSSGD---SISSFSDQCGSANSFNLVKKPKKWPITSS--------- 351
LD ++ L + + +SS + S+ ++ + K+ K IT
Sbjct: 592 LDKEEDLKNVTAEISSLREWEGSVLEKIEELSKVKESLVDKETKLQSITQEAEELKGREA 651
Query: 352 ---DQLSQVECTNNSIIDK-NWIESISEGSNRRRHSISGSNSSEEEVSVLSKRRQSNCFD 407
Q+ ++ N S++D+ ++SI + S + +G EE+SV ++ N D
Sbjct: 652 AHMKQIEELSTANASLVDEATKLQSIVQESEDLKEKEAGYLKKIEELSVANESLADNVTD 711
Query: 408 SFECLKEIEKES 419
L+ I +ES
Sbjct: 712 ----LQSIVQES 719
Score = 34.3 bits (77), Expect = 0.26
Identities = 77/388 (19%), Positives = 137/388 (34%), Gaps = 91/388 (23%)
Query: 119 RRQSSTDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGL 178
R T+L T+L+ ++ L+++ E+ EL++ K ++ +D+L + L+EE L
Sbjct: 49 RLVKGTELQTQLNQIQEDLKKADEQ-----IELLKKDKAKA-IDDLKESEKLVEEANEKL 102
Query: 179 SEQLVALSRS----------------CGLERQEEDKDGSTQNLE-------LEVVELRRL 215
E L A R+ GLE ++ S LE L++ L
Sbjct: 103 KEALAAQKRAEESFEVEKFRAVELEQAGLEAVQKKDVTSKNELESIRSQHALDISALLST 162
Query: 216 NKELHMQKRNLTCRLSSMESQLSCSDNSSE------------------------------ 245
+EL K L+ + LS ++ +++
Sbjct: 163 TEELQRVKHELSMTADAKNKALSHAEEATKIAEIHAEKAEILASELGRLKALLGSKEEKE 222
Query: 246 ----SDIVAKFKAEASLLRLTNEDLS------KQVEGLQTSRLNEVEELAYLRWVNSCLR 295
++IV+K K+E LLR E +S K+ EGL ++E +
Sbjct: 223 AIEGNEIVSKLKSEIELLRGELEKVSILESSLKEQEGLVEQLKVDLEAAKMAESCTNSSV 282
Query: 296 TELKNTCSTLDSDKLSSPQSVVSSSGDSISSF---------------SDQCGSANSFNLV 340
E KN L+ + S +S SS+ +S+ S SD L+
Sbjct: 283 EEWKNKVHELEKEVEESNRS-KSSASESMESVMKQLAELNHVLHETKSDNAAQKEKIELL 341
Query: 341 KKPKKWPITSSDQLSQVECTNNSIIDK--NWIESISE----GSNRRRHSISGSNSSEEEV 394
+K + T ++ + C K N +ESI + ++ ++ +
Sbjct: 342 EKTIEAQRTDLEEYGRQVCIAKEEASKLENLVESIKSELEISQEEKTRALDNEKAATSNI 401
Query: 395 SVLSKRRQSNCFDSFECLKEIEKESVPM 422
L +R + C E EK M
Sbjct: 402 QNLLDQRTELSIELERCKVEEEKSKKDM 429
>At3g10880 hypothetical protein
Length = 278
Score = 43.1 bits (100), Expect = 5e-04
Identities = 48/188 (25%), Positives = 89/188 (46%), Gaps = 32/188 (17%)
Query: 101 LMGDLSCSQVHPHAFPTHRRQSSTDLFTELDHMRSLLQESKER-----EAKLNAELVECR 155
L+ D+ C+++ T RR+ R++ E+++R E+KL+ + +
Sbjct: 100 LLADMFCAELE-----TARRELEA---------RNIAIETEKRYVVDLESKLSDSVYKIE 145
Query: 156 KNQSEVDELVKKVALLEEEKSGLSEQLVALSRSCGLERQEEDKDGSTQNLELEVVELRRL 215
K +SE+DE+ + + + E E S L E L C E+ + D + L+ ELR
Sbjct: 146 KLESELDEVKECLGVSEAEVSKLMEML----SECKNEKSKLQTDNADDLLDSLRAELRSR 201
Query: 216 NKELHMQKRNLTCRLSSMESQLSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQT 275
++ + L L E+++ S++ ++ +IV + +A+ E L KQVE LQ
Sbjct: 202 EIQIEQMEEYLNQVLCLNETEIK-SESETDKNIVEELRAKV-------EVLEKQVE-LQR 252
Query: 276 SRLNEVEE 283
+ + E EE
Sbjct: 253 NVITEREE 260
>At3g10170 hypothetical protein
Length = 634
Score = 43.1 bits (100), Expect = 5e-04
Identities = 45/198 (22%), Positives = 91/198 (45%), Gaps = 29/198 (14%)
Query: 124 TDLFTELDHMRSLLQESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQLV 183
TDL +L+H + L+ + +L L +Q +++ VK+ LLEE+ E+L
Sbjct: 343 TDLTEKLEHSNTKLEHLQNDVTELKTRLEVSSSDQQQLETNVKQ--LLEEK-----EELA 395
Query: 184 ALSRSCGLERQEEDKDGSTQNLEL-----------EVVELRRLNKELHMQKRNL-TCRLS 231
+ LE +EE S++ L + +++ L+KE+ +K+ L +CRL
Sbjct: 396 MHLANSLLEMEEEKAIWSSKEKALTEAVEEKIRLYKNIQIESLSKEMSEEKKELESCRLE 455
Query: 232 --SMESQLSCSDNSSESDIVAKFKAEASLLRLTNEDLSKQVEGLQTSRLNEVEELAYLRW 289
++ +L CS+ +++ D + + + RL +E S Q+ + L+
Sbjct: 456 CVTLADRLRCSEENAKQDKESSLEKSLEIDRLGDELRSADAVSKQSQEV--------LKS 507
Query: 290 VNSCLRTELKNTCSTLDS 307
L++E+++ C D+
Sbjct: 508 DIDILKSEVQHACKMSDT 525
Score = 31.2 bits (69), Expect = 2.2
Identities = 70/377 (18%), Positives = 140/377 (36%), Gaps = 46/377 (12%)
Query: 127 FTELDHMRSLL--QESKEREAKLNAELVECRKNQSEVDELVKKVALLEEEKSGLSEQLVA 184
F+ L H +L Q S++ ++ +N+ + +V ++ + LL+E+ +GL Q+
Sbjct: 108 FSPLIHDFTLFVRQSSEQHDSLINSYQTVQSSLKKKVLDVENEKLLLQEQCAGLQSQIEE 167
Query: 185 LSRSCG---------LERQEEDKDGSTQNLELEVVELRRL-NKELHMQKRNL-------T 227
L++ E E ++ ++E ++ L + L +K NL
Sbjct: 168 LNQEAQKHETSLKMLSEHHESERSDLLSHIECLEKDIGSLSSSSLAKEKENLRKDFEKTK 227
Query: 228 CRLSSMESQLSCSDNSSESDIVAKFKAEASLLRLTNE------DLSKQVEGLQTSRLNEV 281
+L ES+L S K AE L RL ++ D+SKQ R
Sbjct: 228 TKLKDTESKLKNSMQDKTKLEAEKASAERELKRLHSQKALLERDISKQESFAGKRR---- 283
Query: 282 EELAYLRWVNSCLRTELKN---TCSTLDSDKLSSPQSVVSSSGDSISSFSDQCGSANSFN 338
+ L R N L+ E K +++ S + + + G+ + G +
Sbjct: 284 DSLLVERSANQSLQEEFKQLEVLAFEMETTIASLEEELAAERGEKEEALCRNDGLGSEIT 343
Query: 339 LVKKPKKWPITSSDQLS----------QVECTNNSIIDKNWIESISEGSNRRRHSISGSN 388
+ + + T + L +V ++ ++ N + + E H +
Sbjct: 344 DLTEKLEHSNTKLEHLQNDVTELKTRLEVSSSDQQQLETNVKQLLEEKEELAMHLANSLL 403
Query: 389 SSEEEVSVLSKRRQS---NCFDSFECLKEIEKESVPMPLFVQQCALEKRALRIPNPPPRP 445
EEE ++ S + ++ + K I+ ES+ + ++ LE L R
Sbjct: 404 EMEEEKAIWSSKEKALTEAVEEKIRLYKNIQIESLSKEMSEEKKELESCRLECVTLADRL 463
Query: 446 SCSISSKTKQECSAQVQ 462
CS KQ+ + ++
Sbjct: 464 RCS-EENAKQDKESSLE 479
>At5g58160 strong similarity to unknown protein (gb|AAD23008.1)
Length = 1307
Score = 42.4 bits (98), Expect = 0.001
Identities = 20/49 (40%), Positives = 25/49 (50%)
Query: 441 PPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFASRGNTAMVKRAP 489
PPP P + T ++ PPPPPPPPP P + S G +AM P
Sbjct: 707 PPPPPPAPPAPPTPIVHTSSPPPPPPPPPPPAPPTPQSNGISAMKSSPP 755
Score = 36.2 bits (82), Expect = 0.067
Identities = 18/37 (48%), Positives = 19/37 (50%), Gaps = 7/37 (18%)
Query: 437 RIPNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
R P PPP P S+ TK PPPPPP PP P
Sbjct: 688 RPPPPPPPPPMQHSTVTKV-------PPPPPPAPPAP 717
Score = 33.9 bits (76), Expect = 0.33
Identities = 17/39 (43%), Positives = 22/39 (55%), Gaps = 9/39 (23%)
Query: 440 NPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFAS 478
+PPP IS+ K+ P PPPPPPPPPM ++
Sbjct: 673 SPPP-----ISNSDKKPAL----PRPPPPPPPPPMQHST 702
Score = 30.8 bits (68), Expect = 2.8
Identities = 18/46 (39%), Positives = 20/46 (43%), Gaps = 10/46 (21%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPP--------PPPP--PPPPPM 474
P PP P + S K A PP PPPP PPPPP+
Sbjct: 736 PPAPPTPQSNGISAMKSSPPAPPAPPRLPTHSASPPPPTAPPPPPL 781
Score = 30.4 bits (67), Expect = 3.7
Identities = 14/40 (35%), Positives = 19/40 (47%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPPMSFAS 478
P PPP P + ++ + + PP PP PP P AS
Sbjct: 730 PPPPPPPPAPPTPQSNGISAMKSSPPAPPAPPRLPTHSAS 769
Score = 29.6 bits (65), Expect = 6.3
Identities = 13/33 (39%), Positives = 15/33 (45%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPP 471
P PPP P + + SA PP PP PP
Sbjct: 729 PPPPPPPPPAPPTPQSNGISAMKSSPPAPPAPP 761
Score = 29.3 bits (64), Expect = 8.2
Identities = 13/35 (37%), Positives = 14/35 (39%)
Query: 439 PNPPPRPSCSISSKTKQECSAQVQPPPPPPPPPPP 473
P PPP P + T Q PP PP PP
Sbjct: 727 PPPPPPPPPPPAPPTPQSNGISAMKSSPPAPPAPP 761
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.128 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,974,273
Number of Sequences: 26719
Number of extensions: 801476
Number of successful extensions: 10738
Number of sequences better than 10.0: 374
Number of HSP's better than 10.0 without gapping: 169
Number of HSP's successfully gapped in prelim test: 219
Number of HSP's that attempted gapping in prelim test: 5671
Number of HSP's gapped (non-prelim): 1922
length of query: 732
length of database: 11,318,596
effective HSP length: 107
effective length of query: 625
effective length of database: 8,459,663
effective search space: 5287289375
effective search space used: 5287289375
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC124968.5


