

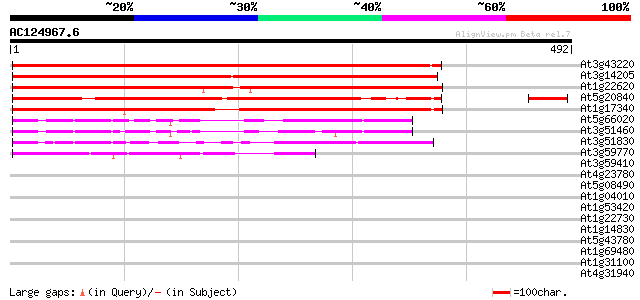
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124967.6 - phase: 0 /pseudo
(492 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g43220 putative protein 529 e-150
At3g14205 unknown protein 514 e-146
At1g22620 unknown protein (At1g22620) 421 e-118
At5g20840 unknown protein 410 e-115
At1g17340 unknown protein 367 e-102
At5g66020 SAC1-like protein AtSAC1b 97 2e-20
At3g51460 unknown protein 97 3e-20
At3g51830 transmembrane protein G5p 96 6e-20
At3g59770 hypothetical protein 86 5e-17
At3g59410 protein kinase like 33 0.36
At4g23780 putative protein 33 0.47
At5g08490 selenium-binding protein-like 32 0.80
At1g04010 unknown protein 32 0.80
At1g53420 hypothetical protein, 5' partial 32 1.0
At1g22730 putative topoisomerase 32 1.0
At1g14830 dynamin-like protein 31 1.4
At5g43780 ATP sulfurylase precursor (gb|AAD26634.1) 30 4.0
At1g69480 hypothetical protein 30 4.0
At1g31100 hypothetical protein 30 4.0
At4g31940 Cytochrome P450-like protein 29 5.2
>At3g43220 putative protein
Length = 794
Score = 529 bits (1362), Expect = e-150
Identities = 263/376 (69%), Positives = 303/376 (79%), Gaps = 1/376 (0%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
RYL+RGVN G VANDVETEQIV EDVPE P++ISSV+QNRGSIPLFWSQETSRLN+KP
Sbjct: 231 RYLKRGVNRNGDVANDVETEQIVSEDVPEDHPMQISSVVQNRGSIPLFWSQETSRLNLKP 290
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANAIDFINK 122
DI+LSKK+ +Y+AT+LHF+NLV+RYGNPIIILNLIKT E++PRE+ILR+EF NAIDFINK
Sbjct: 291 DIVLSKKEPNYEATRLHFDNLVERYGNPIIILNLIKTKERRPRESILREEFVNAIDFINK 350
Query: 123 DLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLRPEDCLKW 182
DL EENRLRFLHWDLHKHF+SK NVL LL KVA AL LT FY QV+ + ED +
Sbjct: 351 DLPEENRLRFLHWDLHKHFRSKTKNVLALLCKVATCALMLTDLFYYQVTPAMTIEDSMSL 410
Query: 183 PSTDDVDKGSFSPTVHVEYDNEDANDLERKPSDEINVSNENHSAKPPRLQNGVLRTNCID 242
S+ D D G SP + DN D + LE+K S N++ KPPRLQ+GVLRTNCID
Sbjct: 411 SSSSDADTGDISPHTSSDDDNGDHDSLEKKSSRSKNIAYGKCDVKPPRLQSGVLRTNCID 470
Query: 243 CLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFYERMGDTLAHQYGGS 302
CLDRTNVAQYAYG AA+G QLH LGI + P I+LDDP+A LM YERMGDTLAHQYGGS
Sbjct: 471 CLDRTNVAQYAYGWAALGQQLHVLGIRDVPAIELDDPLAISLMGLYERMGDTLAHQYGGS 530
Query: 303 AAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINVFLGHFQPQQDKPAL 362
AAHNK+FS RRGQWRAATQSQEF RTLQRYY+NA+MDA+KQDAIN+FLG FQP+Q PA+
Sbjct: 531 AAHNKVFSERRGQWRAATQSQEFLRTLQRYYNNAYMDADKQDAINIFLGTFQPEQGMPAI 590
Query: 363 WELGSDQHYDTGRIGD 378
WEL S+ GR G+
Sbjct: 591 WELRSNS-LSNGRNGE 605
Score = 34.7 bits (78), Expect = 0.12
Identities = 16/32 (50%), Positives = 20/32 (62%), Gaps = 1/32 (3%)
Query: 456 SHHIYYSEHGDSFSCSNFVDLDWLSSSANSCE 487
S +Y+ D S SNFVD++WLSS N CE
Sbjct: 691 SDQVYFGGEEDMSSVSNFVDIEWLSSE-NPCE 721
>At3g14205 unknown protein
Length = 808
Score = 514 bits (1325), Expect = e-146
Identities = 263/374 (70%), Positives = 298/374 (79%), Gaps = 2/374 (0%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
RYL+RGVNEKGRVANDVETEQIVFE+ +G P +ISSV+QNRGSIPLFWSQETSRLNIKP
Sbjct: 253 RYLKRGVNEKGRVANDVETEQIVFEEAQDGNPGRISSVVQNRGSIPLFWSQETSRLNIKP 312
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANAIDFINK 122
DIILS KD +++AT+LHFENL +RYGNPIIILNLIKT EK+PRE ILR EFANAI FINK
Sbjct: 313 DIILSPKDPNFEATRLHFENLGRRYGNPIIILNLIKTREKRPRETILRAEFANAIRFINK 372
Query: 123 DLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLRPEDCLKW 182
LS+E+RLR LHWDLHKH + K TNVL +LG++A YAL LT FYCQ++ LR E
Sbjct: 373 GLSKEDRLRPLHWDLHKHSRKKGTNVLAILGRLATYALNLTSIFYCQLTPDLRGEGFQNQ 432
Query: 183 -PSTDDVDKGSFSPTVHVEYDNEDANDLERKPSDEINVSNENHSAKPPRLQNGVLRTNCI 241
PST + D G S T +E A +L + ++ + E+ + LQ GVLRTNCI
Sbjct: 433 NPSTLENDDGECS-TYDPPSKDETAPNLVVENGNDSKDAKEDQQKEVTMLQKGVLRTNCI 491
Query: 242 DCLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFYERMGDTLAHQYGG 301
DCLDRTNVAQYAYGL A G QLH+LG+ E IDLD+P+A DLM YE MGDTLA QYGG
Sbjct: 492 DCLDRTNVAQYAYGLVAFGRQLHALGLTESTNIDLDNPLAEDLMGIYETMGDTLALQYGG 551
Query: 302 SAAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINVFLGHFQPQQDKPA 361
SAAHNKIF RRGQWRAATQSQEFFRTLQRYYSNA+MDAEKQDAINVFLG+FQPQ DKPA
Sbjct: 552 SAAHNKIFCERRGQWRAATQSQEFFRTLQRYYSNAYMDAEKQDAINVFLGYFQPQSDKPA 611
Query: 362 LWELGSDQHYDTGR 375
LWELGSDQHY+ R
Sbjct: 612 LWELGSDQHYNAAR 625
Score = 33.9 bits (76), Expect = 0.21
Identities = 13/26 (50%), Positives = 21/26 (80%), Gaps = 1/26 (3%)
Query: 464 HGDSFSCSNFVDLDWLSSSANSCEED 489
+G+ +C+ F D+DW+SSS NSCE++
Sbjct: 720 NGEQCTCAAF-DMDWVSSSGNSCEDE 744
>At1g22620 unknown protein (At1g22620)
Length = 912
Score = 421 bits (1083), Expect = e-118
Identities = 213/391 (54%), Positives = 276/391 (70%), Gaps = 19/391 (4%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
RYL+RGVN++GRVANDVETEQ+V +D K+SSV+Q RGSIPLFWSQE SR + KP
Sbjct: 268 RYLKRGVNDRGRVANDVETEQLVLDDEAGSCKGKMSSVVQMRGSIPLFWSQEASRFSPKP 327
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANAIDFINK 122
DI L + D +Y++TK+HFE+LV RYGNPII+LNLIKT EK+PRE +LR+EFANA+ ++N
Sbjct: 328 DIFLQRYDPTYESTKMHFEDLVNRYGNPIIVLNLIKTVEKRPREMVLRREFANAVGYLNS 387
Query: 123 DLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYC-------QVSSTLR 175
EEN L+F+HWD HK +SK+ NVL +LG VA+ AL LTG ++ + +S L
Sbjct: 388 IFREENHLKFIHWDFHKFAKSKSANVLAVLGAVASEALDLTGLYFSGKPKIVKKKASQLS 447
Query: 176 PEDCLKWPSTDDVDKGSFSPTVHVEYDNEDANDL-------ERKPSDEINVSNENHSAKP 228
+ + PS D+ S + E AND+ E++ +E ++
Sbjct: 448 HANTAREPSLRDLRAYSAELS-----RGESANDILSALANREKEMKLTQQKKDEGTNSSA 502
Query: 229 PRLQNGVLRTNCIDCLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFY 288
PR Q+GVLRTNCIDCLDRTNVAQYAYGLAA+G QLH++G+ + PKID D +A LM Y
Sbjct: 503 PRYQSGVLRTNCIDCLDRTNVAQYAYGLAALGRQLHAMGLSDTPKIDPDSSIAAALMDMY 562
Query: 289 ERMGDTLAHQYGGSAAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINV 348
+ MGD LA QYGGSAAHN +F R+G+W+A TQS+EF ++++RYYSN + D EKQDAIN+
Sbjct: 563 QSMGDALAQQYGGSAAHNTVFPERQGKWKATTQSREFLKSIKRYYSNTYTDGEKQDAINL 622
Query: 349 FLGHFQPQQDKPALWELGSDQHYDTGRIGDD 379
FLG+FQPQ+ KPALWEL SD + IGDD
Sbjct: 623 FLGYFQPQEGKPALWELDSDYYLHVSGIGDD 653
>At5g20840 unknown protein
Length = 774
Score = 410 bits (1055), Expect = e-115
Identities = 223/376 (59%), Positives = 264/376 (69%), Gaps = 37/376 (9%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
RYL+RG+NE G VANDVETEQIV EDVP P++ISSV+QNRGSIPLFWSQETSR+ +KP
Sbjct: 232 RYLKRGINESGNVANDVETEQIVSEDVPVDRPMQISSVVQNRGSIPLFWSQETSRMKVKP 291
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANAIDFINK 122
DI AT++HFENLV+RYG PIIILNLIKT+E+KPRE+ILR EFANAIDFINK
Sbjct: 292 DI----------ATRVHFENLVERYGVPIIILNLIKTNERKPRESILRAEFANAIDFINK 341
Query: 123 DLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLRPEDCLKW 182
DL EENRLRFLHWDLHKHF SK NVL LLGKVAA AL LTGFFY Q++ ++ E +
Sbjct: 342 DLPEENRLRFLHWDLHKHFHSKTENVLALLGKVAACALMLTGFFYYQLTPAMKLEGYMSL 401
Query: 183 PSTDDVDKGSFSPTVHVEYDNEDANDLERKPSDEINVSNENHSAKPPRLQNGVLRTNCID 242
S+D SP + D+ D + LE+ NV+N ++ KP RLQ+GVLRTNCID
Sbjct: 402 SSSD----ADTSPHNSSDDDSRDYDSLEKNCRPSKNVANGDYDVKPSRLQSGVLRTNCID 457
Query: 243 CLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFYERMGDTLAHQYGGS 302
CLDRTNVAQYAYG AA+G QLH+LGI + P I+LDDP+++ LM YERMGDTLA+QYGGS
Sbjct: 458 CLDRTNVAQYAYGWAALGQQLHALGIRDAPTIELDDPLSSTLMGLYERMGDTLAYQYGGS 517
Query: 303 AAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINVFLGHFQPQQDKPAL 362
AAHNK R S R L+ D+ FLG F+P+Q A+
Sbjct: 518 AAHNKA--------RPVESSNPVTRVLE--------DSA------TFLGTFRPEQGSQAV 555
Query: 363 WELGSDQHYDTGRIGD 378
WEL SD H GR G+
Sbjct: 556 WELRSDSH-SNGRSGE 570
Score = 45.1 bits (105), Expect = 9e-05
Identities = 21/34 (61%), Positives = 23/34 (66%)
Query: 456 SHHIYYSEHGDSFSCSNFVDLDWLSSSANSCEED 489
S IY SE D S SNFVD++WLSSS N CE D
Sbjct: 664 SSSIYLSEQEDMSSVSNFVDIEWLSSSENLCEND 697
>At1g17340 unknown protein
Length = 785
Score = 367 bits (943), Expect = e-102
Identities = 196/379 (51%), Positives = 256/379 (66%), Gaps = 23/379 (6%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
RYLRRGVN+ GRVANDVETEQIV + VP G I I+SV+Q RGSIPLFWSQE S N +P
Sbjct: 253 RYLRRGVNDIGRVANDVETEQIVSKVVPAGQKIPITSVVQVRGSIPLFWSQEASVFNPQP 312
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKT--HEKKPREAILRQEFANAIDFI 120
+IIL+KKD +Y+AT+ HF+NL +RYGN IIILNL+KT EKK RE ILR EFA I FI
Sbjct: 313 EIILNKKDANYEATQHHFQNLRQRYGNRIIILNLLKTVTGEKKHRETILRGEFAKTIRFI 372
Query: 121 NKDLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLRPEDCL 180
NK + E+RL+ +H+DL KH++ A L + +L LT FYC+ S + E+ +
Sbjct: 373 NKGMDREHRLKAIHFDLSKHYKKGADGAFNHLCIFSRKSLELTDLFYCKAPSGVGAEEVI 432
Query: 181 KWPSTDDVDKGSFSPTVHVEYDNEDANDLERKPSDEINVSNENHSAKPPRLQNGVLRTNC 240
YD+ N + + + + E+ A LQNGVLRTNC
Sbjct: 433 --------------------YDSFFNNPIPSQDEEASSPEKEDMKADIFLLQNGVLRTNC 472
Query: 241 IDCLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFYERMGDTLAHQYG 300
IDCLDRTN AQYA+GL ++GHQL +LGI P +DL++P+A +LM Y++MG+TLA QYG
Sbjct: 473 IDCLDRTNFAQYAHGLVSLGHQLRTLGISGPPVVDLNNPLAIELMDAYQKMGNTLAMQYG 532
Query: 301 GSAAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINVFLGHFQPQQDKP 360
GS AH+K+F RG W + ++ F ++RYYSNA+ D++KQ+AINVFLGHF+P+ +P
Sbjct: 533 GSEAHSKMFCDLRGNWNMVMRQRDIFTAVRRYYSNAYQDSDKQNAINVFLGHFRPRLGRP 592
Query: 361 ALWELGSDQHYDTGRIGDD 379
ALWEL SDQH + GR G +
Sbjct: 593 ALWELDSDQH-NIGRSGSN 610
>At5g66020 SAC1-like protein AtSAC1b
Length = 593
Score = 97.1 bits (240), Expect = 2e-20
Identities = 96/357 (26%), Positives = 147/357 (40%), Gaps = 82/357 (22%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
R RRG + G VAN VETEQIV + SS +Q RGS+P W Q L KP
Sbjct: 227 RMWRRGADPDGYVANFVETEQIV------RMNGYTSSFVQIRGSMPFMWEQIVD-LTYKP 279
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANAIDFINK 122
+ + +++ + + HF +L K+YG+ ++ ++L+ H + R L + FA A+ I
Sbjct: 280 KFEIVQPEEAARIAERHFLDLRKKYGS-VLAVDLVNKHGGEGR---LSERFAGAMQHITG 335
Query: 123 DLSEENRLRFLHWDLHK-----HFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLRPE 177
D +R+LH+D H HF+ L +L + L G+F
Sbjct: 336 D-----DVRYLHFDFHHICGHIHFER-----LAILYEQMEDFLEKNGYFL---------- 375
Query: 178 DCLKWPSTDDVDKGSFSPTVHVEYDNEDANDLERKPSDEINVSNENHSAKPPRLQNGVLR 237
N+ K +++ + N
Sbjct: 376 ----------------------------LNEKGEKMKEQLGIVRTN-------------- 393
Query: 238 TNCIDCLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQ-FYERMGDTLA 296
CIDCLDRTNV Q G + QL +G+ + ++ + + GD ++
Sbjct: 394 --CIDCLDRTNVTQSMIGRKLLELQLKRIGVFGAEETIRSHQNFDECYKILWANHGDDIS 451
Query: 297 HQYGGSAAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINVFLGHF 353
QY G+ A F R GQ Q+ + L RYY N + D KQDAI++ GH+
Sbjct: 452 IQYSGTPALKGDF-VRYGQRTIQGVLQDGWNALARYYLNNFADGTKQDAIDLVQGHY 507
>At3g51460 unknown protein
Length = 597
Score = 96.7 bits (239), Expect = 3e-20
Identities = 95/362 (26%), Positives = 157/362 (43%), Gaps = 92/362 (25%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
R RRG + G VAN VETEQIV + SS +Q RGS+P W Q L KP
Sbjct: 229 RMWRRGADLDGYVANFVETEQIV------QMNGYTSSFVQVRGSMPFMWEQVVD-LTYKP 281
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANAIDFINK 122
+ + +++ + + HF +L K+YG+ ++ ++L+ K+ E L +++A + I
Sbjct: 282 KFEIVQPEEAKRIAERHFLDLRKKYGS-VLAVDLV---NKQGGEGRLCEKYATVMQHITG 337
Query: 123 DLSEENRLRFLHWDLHK-----HFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLRPE 177
D +R+LH+D H+ HF+ + +L ++ + L G+F
Sbjct: 338 D-----DIRYLHFDFHQICGHIHFE----RLSILYEQIEGF-LEKNGYFL---------- 377
Query: 178 DCLKWPSTDDVDKGSFSPTVHVEYDNEDANDLERKPSDEINVSNENHSAKPPRLQNGVLR 237
N+ K +++ V +R
Sbjct: 378 ----------------------------LNEKGEKMKEQLGV----------------VR 393
Query: 238 TNCIDCLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDL------MQFYERM 291
+NCIDCLDRTNV Q G + QL +G+ ++ +++ L +
Sbjct: 394 SNCIDCLDRTNVTQSMIGRKMLEVQLKRIGV-----FGAEETISSHLNFDEHYKILWANH 448
Query: 292 GDTLAHQYGGSAAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINVFLG 351
GD ++ QY G+ A F R G A ++ + +L+RYY N + D KQDAI++ G
Sbjct: 449 GDEISIQYSGTPALKGDF-VRYGHRTAHGVLKDGWSSLRRYYLNNFADGTKQDAIDLLQG 507
Query: 352 HF 353
H+
Sbjct: 508 HY 509
>At3g51830 transmembrane protein G5p
Length = 588
Score = 95.5 bits (236), Expect = 6e-20
Identities = 98/371 (26%), Positives = 159/371 (42%), Gaps = 75/371 (20%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
R RRG N +G AN VE+EQIV E K S ++Q RGSIPL W Q L+ KP
Sbjct: 228 RMWRRGANLEGDAANFVESEQIV-----EINGFKFS-LLQVRGSIPLLWEQIVD-LSYKP 280
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANAIDFINK 122
+ ++K +++ + + HF +L +RYG I+ ++L H E L + +A ++ +
Sbjct: 281 RLKINKHEETPKVVQRHFHDLCQRYGE-IMAVDLTDQHGD---EGALSKAYATEMEKLPD 336
Query: 123 DLSEENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLRPEDCLKW 182
+R++ +D H+ + + L G Y Q+
Sbjct: 337 -------VRYVSFDFHQVCGTTNFDNL--------------GVLYEQIG----------- 364
Query: 183 PSTDDVDKGSFSPTVHVEYDNEDANDLERKPSDEINVSNENHSAKPPRLQNGVLRTNCID 242
D+ +K + + D N LE Q GV+R+NCID
Sbjct: 365 ---DEFEKQGYFLV------DADENILEE--------------------QKGVIRSNCID 395
Query: 243 CLDRTNVAQYAYGLAAIGHQLHSLGIIEHPK-IDLDDPVANDLMQFYERMGDTLAHQYGG 301
CLDRTNV Q G ++ QL +G+ + + I + + GD ++ QY G
Sbjct: 396 CLDRTNVTQSFMGQKSLNLQLQRIGVCDSTECISTFEDDYTKFRTIWAEQGDEVSLQYAG 455
Query: 302 SAAHNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMDAEKQDAINVFLGHFQPQQDKPA 361
+ A K R G+ ++ + RYY N + D +QDA+++ G + P+
Sbjct: 456 TYA-LKGDLVRYGKQTMTGAIKDGLSAMSRYYLNNFQDGVRQDALDLISGRYTVGTHSPS 514
Query: 362 -LWELGSDQHY 371
L +GS +
Sbjct: 515 QLQPIGSQPSF 525
>At3g59770 hypothetical protein
Length = 1616
Score = 85.9 bits (211), Expect = 5e-17
Identities = 78/288 (27%), Positives = 120/288 (41%), Gaps = 59/288 (20%)
Query: 3 RYLRRGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRLNIKP 62
RYL RG+N N+VE EQ+V+ G I +S I RG+IP++W E +
Sbjct: 237 RYLARGINSCSGTGNEVECEQLVWIPKRNGQSIAFNSYIWRRGTIPIWWGAELKMTAAEA 296
Query: 63 DIILSKKDQSYQATKLHFENLVKRYGN-----------------PIIILNLIKTHEKKPR 105
+I ++ +D Y+ + +++ L KRY PI+ +NL+++ E K
Sbjct: 297 EIYVADRD-PYKGSTEYYQRLSKRYDTRNLDAPVGENQKKKAFVPIVCVNLLRSGEGK-S 354
Query: 106 EAILRQEFANAIDFI-NKDLSEENRLRFLHWDLHKHFQSKATNV----LLLLGKVAAYAL 160
E IL Q F +++FI + R+ +++D H + K L + K A+
Sbjct: 355 ECILVQHFEESMNFIKSSGKLPYTRVHLINYDWHASVKLKGEQQTIEGLWMYLKSPTMAI 414
Query: 161 TLTGFFYCQVSSTLRPEDCLKWPSTDDVDKGSFSPTVHVEYDNEDANDLERKPSDEINVS 220
++ Y + S R +DC D +G+F H
Sbjct: 415 GISEGDY--LPSRQRLKDCRGEVICIDDIEGAFCLRSH---------------------- 450
Query: 221 NENHSAKPPRLQNGVLRTNCIDCLDRTNVAQYAYGLAAIGHQLHSLGI 268
QNGV+R NC D LDRTN A + GL Q LGI
Sbjct: 451 -----------QNGVIRFNCADSLDRTNAASFFGGLQVFVEQCRRLGI 487
Score = 40.4 bits (93), Expect = 0.002
Identities = 25/75 (33%), Positives = 41/75 (54%), Gaps = 5/75 (6%)
Query: 282 NDLMQFYERMGDTLAHQYGGSAAHNK----IFSARRGQWRAATQSQEFFR-TLQRYYSNA 336
++L + + GD A Y GS A + IFS G ++ + +Q+ + TLQR Y NA
Sbjct: 566 SELADLFLQQGDIHATLYTGSKAMHSQILNIFSEESGAFKQFSAAQKNMKITLQRRYKNA 625
Query: 337 WMDAEKQDAINVFLG 351
+D+ +Q + +FLG
Sbjct: 626 MVDSSRQKQLEMFLG 640
>At3g59410 protein kinase like
Length = 1271
Score = 33.1 bits (74), Expect = 0.36
Identities = 30/116 (25%), Positives = 44/116 (37%), Gaps = 16/116 (13%)
Query: 184 STDDVDKGSFS----------PTVHVEYDNEDANDLERKPSDEINVSNENHSAKPPRLQN 233
STDDV+ G F T + N ++ L SD + +K L
Sbjct: 289 STDDVESGLFQNEKKESNLQDDTAEDDSTNSESESLGSWSSDSLAQDQVPQISKKDLLMV 348
Query: 234 GVLRTNCIDCLDRTNVAQYAYGLAAIGHQLHSLGIIEHPKIDLDDPVANDLMQFYE 289
+LR C T+ A L I +LH LGI+ +DL + D + +E
Sbjct: 349 HLLRVAC------TSRGPLADALPQITDELHELGILSEEVLDLASKSSPDFNRTFE 398
>At4g23780 putative protein
Length = 148
Score = 32.7 bits (73), Expect = 0.47
Identities = 24/91 (26%), Positives = 46/91 (50%), Gaps = 14/91 (15%)
Query: 52 SQETSRLNIKPDIILSKKD---QSYQATKLHFENLVKRYGNPIIILNLIKTHE------- 101
S++ S ++ L++ + + Y++ ENLVK + I L LI+ HE
Sbjct: 8 SEKNSNNKFSDNVPLAQHEIWKEKYESLVTEHENLVKEHETLIKTLELIEKHEITLEDLV 67
Query: 102 -KKPREAILRQEFANAIDFINKDLSEENRLR 131
KK REA++ +E I+ +K++ E +++
Sbjct: 68 KKKQREALVGKE---EIEKFDKEIGERKKMQ 95
>At5g08490 selenium-binding protein-like
Length = 849
Score = 32.0 bits (71), Expect = 0.80
Identities = 19/50 (38%), Positives = 25/50 (50%), Gaps = 1/50 (2%)
Query: 258 AIGHQLHSLGIIEHPKIDLDDPVANDLMQFYERMGDTLAHQYGGSAAHNK 307
A G ++HS I+ H + D V N L+ FY R GDT A + S K
Sbjct: 347 ASGKEIHSY-ILRHSYLLEDTSVGNALISFYARFGDTSAAYWAFSLMSTK 395
>At1g04010 unknown protein
Length = 629
Score = 32.0 bits (71), Expect = 0.80
Identities = 24/99 (24%), Positives = 45/99 (45%), Gaps = 14/99 (14%)
Query: 66 LSKKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANAIDFINKDLS 125
L ++D + KL FE +K G P I+ ++ F ++++ +++
Sbjct: 164 LEERDLYFHKLKLTFETALKLRGGPSIVF----------AHSMGNNVFRYFLEWLRLEIA 213
Query: 126 EENRLRFLHWDLHKHFQSKATNVLLLLGKVAAYALTLTG 164
++ L++L +H +F A LLG V A TL+G
Sbjct: 214 PKHYLKWLDQHIHAYFAVGAP----LLGSVEAIKSTLSG 248
>At1g53420 hypothetical protein, 5' partial
Length = 856
Score = 31.6 bits (70), Expect = 1.0
Identities = 43/166 (25%), Positives = 69/166 (40%), Gaps = 35/166 (21%)
Query: 78 LHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANAIDFINKD-------------- 123
LH NLVK YG + L+ +E ++ R F + D
Sbjct: 578 LHHPNLVKLYGCCVEGGQLLLVYEFVENNSLARALFGPQETQLRLDWPTRRKICIGVARG 637
Query: 124 ---LSEENRLRFLHWDLHKHFQSKATNVLL---LLGKVAAYALTL-----TGFFYCQVSS 172
L EE+RL+ +H D+ KATNVLL L K++ + L + +++
Sbjct: 638 LAYLHEESRLKIVHRDI------KATNVLLDKQLNPKISDFGLAKLDEEDSTHISTRIAG 691
Query: 173 T---LRPEDCLKWPSTDDVDKGSFSPTVHVEYDNEDANDLERKPSD 215
T + PE ++ TD D SF V +E + +N +ER ++
Sbjct: 692 TFGYMAPEYAMRGHLTDKADVYSFG-IVALEIVHGRSNKIERSKNN 736
>At1g22730 putative topoisomerase
Length = 693
Score = 31.6 bits (70), Expect = 1.0
Identities = 25/84 (29%), Positives = 41/84 (48%), Gaps = 5/84 (5%)
Query: 7 RGVNEKGRVANDVETEQIVFEDVPEGLPIKISSVIQNRGSIPLFWSQETSRL--NIKPDI 64
RG N+ A+D+ + D + L + ++ I + P F ++ L N K
Sbjct: 165 RGFNKLVASADDLSVD---IPDAVDVLAVFVARAIVDDILPPAFLKKQMKLLPDNSKGVE 221
Query: 65 ILSKKDQSYQATKLHFENLVKRYG 88
+L K ++SY AT LH E + KR+G
Sbjct: 222 VLRKAEKSYLATPLHAEVVEKRWG 245
>At1g14830 dynamin-like protein
Length = 614
Score = 31.2 bits (69), Expect = 1.4
Identities = 20/72 (27%), Positives = 37/72 (50%), Gaps = 6/72 (8%)
Query: 58 LNIKPDIILSK-KDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAILRQEFANA 116
+N + D+I ++ K+Q Y T + +L R G+ + L+ H E ++RQ+ +
Sbjct: 249 INKRVDMIAARRKEQEYFETSPEYGHLASRMGSEYLA-KLLSQH----LETVIRQKIPSI 303
Query: 117 IDFINKDLSEEN 128
+ INK + E N
Sbjct: 304 VALINKSIDEIN 315
>At5g43780 ATP sulfurylase precursor (gb|AAD26634.1)
Length = 469
Score = 29.6 bits (65), Expect = 4.0
Identities = 20/62 (32%), Positives = 32/62 (51%), Gaps = 1/62 (1%)
Query: 50 FWSQETSRLNIKPDIILS-KKDQSYQATKLHFENLVKRYGNPIIILNLIKTHEKKPREAI 108
F ++ S +N+ I+L+ DQ ++ + LV GNPI ILN I+ ++ E I
Sbjct: 124 FRLEDGSVVNMSVPIVLAIDDDQKFRIGDSNQVTLVDSVGNPIAILNDIEIYKHPKEERI 183
Query: 109 LR 110
R
Sbjct: 184 AR 185
>At1g69480 hypothetical protein
Length = 777
Score = 29.6 bits (65), Expect = 4.0
Identities = 30/155 (19%), Positives = 64/155 (40%), Gaps = 21/155 (13%)
Query: 197 VHVEYDNEDANDLERKPSDEINVSNENHSAKPPRLQNGVLRTNCIDCLDRTNVAQYAYGL 256
V ++ + D +LE+ PSD++ V +++ + G T+ + ++RTN+ +
Sbjct: 152 VKMQKPDVDNLNLEKHPSDKVVVDTSDNTMR----TQGTANTDMVHGIERTNIPE----- 202
Query: 257 AAIGHQLHSLGIIEHPKIDLDDPVANDLMQFYE-----RMGD-------TLAHQYGGSAA 304
H + + + H D ++ D E +M D TL +G S
Sbjct: 203 EEASHIMADIVPVSHTNGDEEEASIGDKQDLREILERVKMNDVLESPITTLKGVFGDSNE 262
Query: 305 HNKIFSARRGQWRAATQSQEFFRTLQRYYSNAWMD 339
++G+ + EF++ L+R ++M+
Sbjct: 263 PISKKGLKKGEEQLRLVFSEFYQKLRRLKEYSFMN 297
>At1g31100 hypothetical protein
Length = 1090
Score = 29.6 bits (65), Expect = 4.0
Identities = 18/66 (27%), Positives = 29/66 (43%), Gaps = 10/66 (15%)
Query: 137 LHKHFQSKATNVLLLLGKVAAYALTLTGFFYCQVSSTLRPEDCLK----------WPSTD 186
L K F+S + L G+V A ++G +S+ + P+ C+K W
Sbjct: 681 LAKRFRSWSVKCLSFAGRVQLIASVISGIINFWISTFILPKGCVKRIEALCARFLWSGNI 740
Query: 187 DVDKGS 192
DV KG+
Sbjct: 741 DVKKGA 746
>At4g31940 Cytochrome P450-like protein
Length = 524
Score = 29.3 bits (64), Expect = 5.2
Identities = 31/121 (25%), Positives = 61/121 (49%), Gaps = 16/121 (13%)
Query: 91 IIILNLIKTHEKKPREAILRQEFANAIDFINKDLSEENRLRFLHWDLHKHFQSKATNVLL 150
+I+ I+ H ++ + + ++ ++ ID + L+E+ +L L +D + K+T + L
Sbjct: 262 VILERWIENHRQQRKFSGTKENDSDFID-VMMSLAEQGKLSHLQYDANTSI--KSTCLAL 318
Query: 151 LLGKVAAYALTLTGFFYCQVSSTLRPEDCLKWPSTDDVDKGSFSPTVHVEYD-NEDANDL 209
+LG A TLT +S L ++ LK + D++D +HV D N + +D+
Sbjct: 319 ILGGSDTSASTLT----WAISLLLNNKEMLK-KAQDEID-------IHVGRDRNVEDSDI 366
Query: 210 E 210
E
Sbjct: 367 E 367
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.328 0.141 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,357,753
Number of Sequences: 26719
Number of extensions: 489540
Number of successful extensions: 1592
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 1550
Number of HSP's gapped (non-prelim): 42
length of query: 492
length of database: 11,318,596
effective HSP length: 103
effective length of query: 389
effective length of database: 8,566,539
effective search space: 3332383671
effective search space used: 3332383671
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC124967.6


