

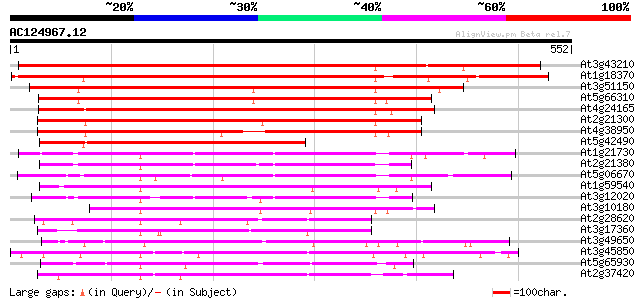
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124967.12 - phase: 0
(552 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g43210 kinesin -like protein 624 e-179
At1g18370 AtNACK1 kinesin-like protein (AtNACK1) 538 e-153
At3g51150 putative protein 414 e-116
At5g66310 kinesin heavy chain DNA binding protein-like 403 e-112
At4g24165 kinesin heavy chain like protein 401 e-112
At2g21300 putative kinesin heavy chain 377 e-104
At4g38950 kinesin like protein 370 e-102
At5g42490 kinesin heavy chain-like protein 280 1e-75
At1g21730 kinesin-related protein (MKRP1) 271 7e-73
At2g21380 putative kinesin heavy chain 265 5e-71
At5g06670 kinesin heavy chain-like protein 262 4e-70
At1g59540 zcf125 kinesin-like protein 258 6e-69
At3g12020 unknown protein 255 5e-68
At3g10180 putative kinesin-like centromere protein 249 2e-66
At2g28620 putative kinesin-like spindle protein 204 1e-52
At3g17360 kinesin-like protein 201 7e-52
At3g49650 kinesin-like protein 197 2e-50
At3g45850 kinesin-related protein - like 196 4e-50
At5g65930 kinesin-like calmodulin-binding protein 194 8e-50
At2g37420 putative kinesin heavy chain 194 8e-50
>At3g43210 kinesin -like protein
Length = 932
Score = 624 bits (1609), Expect = e-179
Identities = 332/535 (62%), Positives = 409/535 (76%), Gaps = 22/535 (4%)
Query: 9 TPSSKIRRKLADTPGGSKIREEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNPN 68
TP SKI + TP GSK+ EEKI VTVRMRPLN +E A YDLIAW+C DD+TIVFKNPN
Sbjct: 6 TPLSKIDKSNPYTPCGSKVTEEKILVTVRMRPLNWREHAKYDLIAWECPDDETIVFKNPN 65
Query: 69 QERPSTSYTFDRVFPPACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGI 128
++ T Y+FD+VF P C+TQ+VY+ G++DVALSAL+G NATIFAYGQTSSGKTFTMRG+
Sbjct: 66 PDKAPTKYSFDKVFEPTCATQEVYEGGSRDVALSALAGTNATIFAYGQTSSGKTFTMRGV 125
Query: 129 TENAIRDIYECIKNTPDRDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENL 188
TE+ ++DIYE I+ T +R FVLK+SALEIYNETV+DLLNR++GPLR+LDD EK T+VENL
Sbjct: 126 TESVVKDIYEHIRKTQERSFVLKVSALEIYNETVVDLLNRDTGPLRLLDDPEKGTIVENL 185
Query: 189 FEEVARDAQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRESPDHVKSYIA 248
EEV QHL+HLI ICE RQVGET LNDKSSRSHQIIRLT+ S RE V+S++A
Sbjct: 186 VEEVVESRQHLQHLISICEDQRQVGETALNDKSSRSHQIIRLTIHSSLREIAGCVQSFMA 245
Query: 249 SLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSG-KSGHISYRTSKLT 307
+LN VDLAGSERA QTN G RLKEGSHIN+SLL L VIR+LSSG K H+ YR SKLT
Sbjct: 246 TLNLVDLAGSERAFQTNADGLRLKEGSHINRSLLTLTTVIRKLSSGRKRDHVPYRDSKLT 305
Query: 308 RILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMV--------- 358
RILQ+SLGGNARTAIICT+SP+LSHVEQT+ TLSFA +AKEV N A+VNMV
Sbjct: 306 RILQNSLGGNARTAIICTISPALSHVEQTKKTLSFAMSAKEVTNCAKVNMVVSEKKLLKH 365
Query: 359 --------EGELRNPEPEHAG-LRSLLAEKELKIQQMEKDMEDLRRQRDLAQCQLDLERR 409
E ELR+PEP + L+SLL EKE+KIQQME +M++L+RQRD+AQ +LDLER+
Sbjct: 366 LQQKVAKLESELRSPEPSSSTCLKSLLIEKEMKIQQMESEMKELKRQRDIAQSELDLERK 425
Query: 410 ANKVQKGSSDYGPSSQVVRCLSFAEENELAIGKHTP--ERTETVSRQAMLKNLLASPDPS 467
A K +KGSS+ P SQV RCLS+ + E K P RT R+ ++ L S DP+
Sbjct: 426 A-KERKGSSECEPFSQVARCLSYHTKEESIPSKSVPSSRRTARDRRKDNVRQSLTSADPT 484
Query: 468 ILVDEIQKLEHRQLQLCEDANRALEVLHKDFATHNLGNQETAETMSKVLSEIKDL 522
LV EI+ LE Q +L E+AN+AL+++HK+ +H LG+Q+ AE ++K+LSEI+D+
Sbjct: 485 ALVQEIRLLEKHQKKLGEEANQALDLIHKEVTSHKLGDQQAAEKVAKMLSEIRDM 539
>At1g18370 AtNACK1 kinesin-like protein (AtNACK1)
Length = 974
Score = 538 bits (1385), Expect = e-153
Identities = 313/560 (55%), Positives = 386/560 (68%), Gaps = 43/560 (7%)
Query: 2 VKTPMAATPSSKIRRKLADTPGGS-KIREEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDK 60
+KTP TP SK+ R A TPGGS + REEKI VTVR+RP+N++E D +AW+C++D
Sbjct: 3 IKTP--GTPVSKMDRTPAVTPGGSSRSREEKIVVTVRLRPMNKRELLAKDQVAWECVNDH 60
Query: 61 TIVFKNPNQER--PSTSYTFDRVFPPACSTQKVYDEGAKDVALSALSGINATIFAYGQTS 118
TIV K QER +S+TFD+VF P T+ VY++G K+VALSAL GINATIFAYGQTS
Sbjct: 61 TIVSKPQVQERLHHQSSFTFDKVFGPESLTENVYEDGVKNVALSALMGINATIFAYGQTS 120
Query: 119 SGKTFTMRGITENAIRDIYECIKNTPDRDFVLKISALEIYNETVIDLLNRESG-PLRILD 177
SGKT+TMRG+TE A+ DIY I TP+RDF +KIS LEIYNE V DLLN +SG L++LD
Sbjct: 121 SGKTYTMRGVTEKAVNDIYNHIIKTPERDFTIKISGLEIYNENVRDLLNSDSGRALKLLD 180
Query: 178 DTEKVTVVENLFEEVARDAQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHR 237
D EK TVVE L EE A + HLRHLI ICEA RQVGET LND SSRSHQIIRLT++S HR
Sbjct: 181 DPEKGTVVEKLVEETANNDNHLRHLISICEAQRQVGETALNDTSSRSHQIIRLTIQSTHR 240
Query: 238 ESPDHVKSYIASLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSGK-S 296
E+ D V+SY+ASLNFVDLAGSERASQ+ GTRL+EG HIN SL+ L VIR+LS GK S
Sbjct: 241 ENSDCVRSYMASLNFVDLAGSERASQSQADGTRLREGCHINLSLMTLTTVIRKLSVGKRS 300
Query: 297 GHISYRTSKLTRILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVN 356
GHI YR SKLTRILQ SLGGNARTAIICT+SP+L+HVEQ+RNTL FA AKEV N A VN
Sbjct: 301 GHIPYRDSKLTRILQHSLGGNARTAIICTLSPALAHVEQSRNTLYFANRAKEVTNNAHVN 360
Query: 357 MV-----------------EGELRNPEPEHAGLRSLLAEKELKIQQMEKDMEDLRRQRDL 399
MV E E R P P EK+ KIQQME ++ +LRRQRD
Sbjct: 361 MVVSDKQLVKHLQKEVARLEAERRTPGPS--------TEKDFKIQQMEMEIGELRRQRDD 412
Query: 400 AQCQLDLERRA----NKVQKGSSDY-GPSSQVVRCLSFAEENELAIGKHTPERTE----T 450
AQ QL+ R+ + KG + + P V +CLS++ + T R E T
Sbjct: 413 AQIQLEELRQKLQGDQQQNKGLNPFESPDPPVRKCLSYSVAVTPSSENKTLNRNERARKT 472
Query: 451 VSRQAMLKNLLASPDPSILVDEIQKLEHRQLQLCEDANRALEVLHKDFATHNLGNQETAE 510
RQ+M++ +S P L+ EI+KLEH Q QL E+A +ALEVL K+ A H LGNQ+ A+
Sbjct: 473 TMRQSMIRQ--SSTAPFTLMHEIRKLEHLQEQLGEEATKALEVLQKEVACHRLGNQDAAQ 530
Query: 511 TMSKVLSEIKDLVAASSTAL 530
T++K+ +EI+++ +A+
Sbjct: 531 TIAKLQAEIREMRTVKPSAM 550
>At3g51150 putative protein
Length = 968
Score = 414 bits (1064), Expect = e-116
Identities = 224/445 (50%), Positives = 307/445 (68%), Gaps = 18/445 (4%)
Query: 20 DTPGGSKIREEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKN----PNQERPSTS 75
D GS REEKI V+VR+RPLN +E+A D+ W+C++D+T+++++ + T+
Sbjct: 6 DQMQGSSGREEKIFVSVRLRPLNVRERARNDVADWECINDETVIYRSHLSISERSMYPTA 65
Query: 76 YTFDRVFPPACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGITENAIRD 135
YTFDRVF P CST++VYD+GAK+VALS +SG++A++FAYGQTSSGKT+TM GIT+ A+ D
Sbjct: 66 YTFDRVFGPECSTREVYDQGAKEVALSVVSGVHASVFAYGQTSSGKTYTMIGITDYALAD 125
Query: 136 IYECIKNTPDRDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENLFEEVARD 195
IY+ I+ +R+F+LK SA+EIYNE+V DLL+ + PLR+LDD EK TVVE L EE RD
Sbjct: 126 IYDYIEKHNEREFILKFSAMEIYNESVRDLLSTDISPLRVLDDPEKGTVVEKLTEETLRD 185
Query: 196 AQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRE--SPDHVKSYIASLNFV 253
H + L+ IC A RQ+GET LN+ SSRSHQI+RLTVES RE + D + A++NF+
Sbjct: 186 WNHFKELLSICIAQRQIGETALNEVSSRSHQILRLTVESTAREYLAKDKFSTLTATVNFI 245
Query: 254 DLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSS 313
DLAGSERASQ+ + GTRLKEG HIN+SLL L VIR+LS GK+GHI +R SKLTRILQ+S
Sbjct: 246 DLAGSERASQSLSAGTRLKEGGHINRSLLTLGTVIRKLSKGKNGHIPFRDSKLTRILQTS 305
Query: 314 LGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMV----------EGELR 363
LGGNART+IICT+SP+ HVEQ+RNTL FA+ AKEV A+VN+V + EL
Sbjct: 306 LGGNARTSIICTLSPARVHVEQSRNTLLFASCAKEVTTNAQVNVVMSDKALVRHLQRELA 365
Query: 364 NPEPEHAGLRSLLAEKELKIQQMEKDMEDLRRQRDLAQCQLDLERRANKVQKGSSDYG-- 421
E E + R L + EKD++ + +++ Q +LER ++++ G
Sbjct: 366 KLESELSSPRQALVVSDTTALLKEKDLQIEKLNKEVFQLAQELERAYSRIEDLQQIIGEA 425
Query: 422 PSSQVVRCLSFAEENELAIGKHTPE 446
P +++ S + +G+ P+
Sbjct: 426 PQQEILSTDSEQTNTNVVLGRQYPK 450
>At5g66310 kinesin heavy chain DNA binding protein-like
Length = 1037
Score = 403 bits (1035), Expect = e-112
Identities = 216/412 (52%), Positives = 295/412 (71%), Gaps = 25/412 (6%)
Query: 29 EEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKN----PNQERPSTSYTFDRVFPP 84
+EKI V+VRMRPLN KE+ D+ W+C+++ TI++++ + ++YTFDRVF P
Sbjct: 16 QEKIYVSVRMRPLNDKEKFRNDVPDWECINNTTIIYRSHLSISERSMYPSAYTFDRVFSP 75
Query: 85 ACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGITENAIRDIYECIKNTP 144
C T++VY++GAK+VA S +SG+NA++FAYGQTSSGKT+TM GIT+ A+ DIY I
Sbjct: 76 ECCTRQVYEQGAKEVAFSVVSGVNASVFAYGQTSSGKTYTMSGITDCALVDIYGYIDKHK 135
Query: 145 DRDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENLFEEVARDAQHLRHLIG 204
+R+F+LK SA+EIYNE+V DLL+ ++ PLR+LDD EK TVVE L EE RD H + L+
Sbjct: 136 EREFILKFSAMEIYNESVRDLLSTDTSPLRLLDDPEKGTVVEKLTEETLRDWNHFKELLS 195
Query: 205 ICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRE--SPDHVKSYIASLNFVDLAGSERAS 262
+C+A RQ+GET LN+ SSRSHQI+RLTVES RE + D + A++NF+DLAGSERAS
Sbjct: 196 VCKAQRQIGETALNEVSSRSHQILRLTVESIAREFSTNDKFSTLTATVNFIDLAGSERAS 255
Query: 263 QTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSSLGGNARTAI 322
Q+ + GTRLKEG HIN+SLL L VIR+LS K+GHI +R SKLTRILQSSLGGNARTAI
Sbjct: 256 QSLSAGTRLKEGCHINRSLLTLGTVIRKLSKEKTGHIPFRDSKLTRILQSSLGGNARTAI 315
Query: 323 ICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMV-----------------EGELRNP 365
ICT+SP+ HVEQ+RNTL FA+ AKEV A+VN+V E ELR+P
Sbjct: 316 ICTMSPARIHVEQSRNTLLFASCAKEVTTNAQVNVVMSDKALVKHLQRELAKLESELRSP 375
Query: 366 EPEH--AGLRSLLAEKELKIQQMEKDMEDLRRQRDLAQCQLDLERRANKVQK 415
+ +LL EK+L++++++K++ L +Q + A+ ++ RR + +K
Sbjct: 376 SQASIVSDTTALLTEKDLEVEKLKKEVFQLAQQLEQARSEIKDLRRMVEEEK 427
>At4g24165 kinesin heavy chain like protein
Length = 1004
Score = 401 bits (1031), Expect = e-112
Identities = 229/415 (55%), Positives = 292/415 (70%), Gaps = 26/415 (6%)
Query: 29 EEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNPNQERPSTSYTFDRVFPPACST 88
EEKI V+VR+RPLN KE+ D W+C++D TI+ K N S SYTFD+VF C T
Sbjct: 4 EEKILVSVRVRPLNEKEKTRNDRCDWECINDTTIICKFHNLPDKS-SYTFDKVFGFECPT 62
Query: 89 QKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGITENAIRDIYECI-KNTPDRD 147
++VYD+GAK+VAL LSGIN++IFAYGQTSSGKT+TM GITE A+ DI+ I K+ +R
Sbjct: 63 KQVYDDGAKEVALCVLSGINSSIFAYGQTSSGKTYTMSGITEFAMDDIFAYIDKHKQERK 122
Query: 148 FVLKISALEIYNETVIDLLNRESG-PLRILDDTEKVTVVENLFEEVARDAQHLRHLIGIC 206
F LK SA+EIYNE V DLL +S PLR+LDD E+ TVVE L EE RD HL L+ IC
Sbjct: 123 FTLKFSAMEIYNEAVRDLLCEDSSTPLRLLDDPERGTVVEKLREETLRDRSHLEELLSIC 182
Query: 207 EAHRQVGETTLNDKSSRSHQIIRLTVESFHRE-SPDHVKSYIASLNFVDLAGSERASQTN 265
E R++GET+LN+ SSRSHQI+RLT+ES ++ SP+ + AS+ FVDLAGSERASQT
Sbjct: 183 ETQRKIGETSLNEISSRSHQILRLTIESSSQQFSPESSATLAASVCFVDLAGSERASQTL 242
Query: 266 TCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSSLGGNARTAIICT 325
+ G+RLKEG HIN+SLL L VIR+LS GK+GHI YR SKLTRILQ+SLGGNARTAIICT
Sbjct: 243 SAGSRLKEGCHINRSLLTLGTVIRKLSKGKNGHIPYRDSKLTRILQNSLGGNARTAIICT 302
Query: 326 VSPSLSHVEQTRNTLSFATNAKEVINTARVNMV-----------------EGELRNPEPE 368
+SP+ SH+EQ+RNTL FAT AKEV A+VN+V E EL+N P
Sbjct: 303 MSPARSHLEQSRNTLLFATCAKEVTTNAQVNLVVSEKALVKQLQRELARMENELKNLGPA 362
Query: 369 HAGLRS-----LLAEKELKIQQMEKDMEDLRRQRDLAQCQLDLERRANKVQKGSS 418
A S +L +KE I +ME+ + +L+ QRD+AQ +++ ++ ++ SS
Sbjct: 363 SASSTSDFYALMLKQKEELIAKMEEQIHELKWQRDVAQSRVENLLKSTAEERSSS 417
>At2g21300 putative kinesin heavy chain
Length = 581
Score = 377 bits (967), Expect = e-104
Identities = 206/414 (49%), Positives = 275/414 (65%), Gaps = 36/414 (8%)
Query: 28 REEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNPNQERPS--TSYTFDRVFPPA 85
REEKI V VR+RPLN KE + W+C++D T++++N +E + ++Y+FDRV+
Sbjct: 21 REEKILVLVRLRPLNEKEILANEAADWECINDTTVLYRNTLREGSTFPSAYSFDRVYRGE 80
Query: 86 CSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGITENAIRDIYECIKNTPD 145
C T++VY++G K+VALS + GIN++IFAYGQTSSGKT+TM GITE A+ DI++ I D
Sbjct: 81 CPTRQVYEDGPKEVALSVVKGINSSIFAYGQTSSGKTYTMSGITEFAVADIFDYIFKHED 140
Query: 146 RDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENLFEEVARDAQHLRHLIGI 205
R FV+K SA+EIYNE + DLL+ +S PLR+ DD EK VE EE RD HL+ LI +
Sbjct: 141 RAFVVKFSAIEIYNEAIRDLLSPDSTPLRLRDDPEKGAAVEKATEETLRDWNHLKELISV 200
Query: 206 CEAHRQVGETTLNDKSSRSHQIIRLTVESFHRESPDHVKSYI-----------------A 248
CEA R++GET+LN++SSRSHQII+L S S++ A
Sbjct: 201 CEAQRKIGETSLNERSSRSHQIIKLVKLFSITGSKAQRFSFLFLLSALSKLLTLCRQLKA 260
Query: 249 SLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTR 308
LNF+DLAGSERASQ + G RLKEG HIN+SLL L VIR+LS+G+ GHI+YR SKLTR
Sbjct: 261 LLNFIDLAGSERASQALSAGARLKEGCHINRSLLTLGTVIRKLSNGRQGHINYRDSKLTR 320
Query: 309 ILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMV---------- 358
ILQ LGGNARTAI+CT+SP+ SHVEQTRNTL FA AKEV A++N+V
Sbjct: 321 ILQPCLGGNARTAIVCTLSPARSHVEQTRNTLLFACCAKEVTTKAQINVVMSDKALVKQL 380
Query: 359 -------EGELRNPEPEHAGLRSLLAEKELKIQQMEKDMEDLRRQRDLAQCQLD 405
E ELRNP P + + ++ +Q +K + ++ +QRD+AQ +L+
Sbjct: 381 QRELARLESELRNPAPATSSCDCGVTLRKKDLQIQKKQLAEMTKQRDIAQSRLE 434
>At4g38950 kinesin like protein
Length = 834
Score = 370 bits (949), Expect = e-102
Identities = 208/420 (49%), Positives = 274/420 (64%), Gaps = 62/420 (14%)
Query: 28 REEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNPNQERPS--TSYTFDRVFPPA 85
REEKI V VR+RPLN+KE A + W+C++D TI+++N +E + ++Y+FD+V+
Sbjct: 10 REEKILVLVRLRPLNQKEIAANEAADWECINDTTILYRNTLREGSNFPSAYSFDKVYRGE 69
Query: 86 CSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGITENAIRDIYECIKNTPD 145
C T++VY++G K++ALS + GIN +IFAYGQTSSGKT+TM GITE A+ DI++ I +
Sbjct: 70 CPTRQVYEDGTKEIALSVVKGINCSIFAYGQTSSGKTYTMTGITEFAVADIFDYIFQHEE 129
Query: 146 RDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENLFEEVARDAQHLRHLIGI 205
R F +K SA+EIYNE + DLL+ + LR+ DD EK TVVE EE RD HL+ L+ I
Sbjct: 130 RAFSVKFSAIEIYNEAIRDLLSSDGTSLRLRDDPEKGTVVEKATEETLRDWNHLKELLSI 189
Query: 206 CE--------------------AHRQVGETTLNDKSSRSHQIIRLTVESFHRESPDHVKS 245
CE A R++GET+LN++SSRSHQ+IRL
Sbjct: 190 CEGKTQKTCFLKSKLTCFYIFAAQRKIGETSLNERSSRSHQMIRL--------------- 234
Query: 246 YIASLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSK 305
NF+DLAGSERASQ + GTRLKEG HIN+SLL L VIR+LS G+ GHI++R SK
Sbjct: 235 -----NFIDLAGSERASQAMSAGTRLKEGCHINRSLLTLGTVIRKLSKGRQGHINFRDSK 289
Query: 306 LTRILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMV------- 358
LTRILQ LGGNARTAIICT+SP+ SHVE T+NTL FA AKEV AR+N+V
Sbjct: 290 LTRILQPCLGGNARTAIICTLSPARSHVELTKNTLLFACCAKEVTTKARINVVMSDKALL 349
Query: 359 ----------EGELRNPEPEHAG---LRSLLAEKELKIQQMEKDMEDLRRQRDLAQCQLD 405
E ELRNP A + +K+L+IQ+MEK++ +LR+QRDLAQ +L+
Sbjct: 350 KQLQRELARLETELRNPASSPASNCDCAMTVRKKDLQIQKMEKEIAELRKQRDLAQSRLE 409
>At5g42490 kinesin heavy chain-like protein
Length = 1087
Score = 280 bits (717), Expect = 1e-75
Identities = 153/266 (57%), Positives = 195/266 (72%), Gaps = 5/266 (1%)
Query: 30 EKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNPNQER---PSTSYTFDRVFPPAC 86
EKI V+VR+RP N KE+A D+ W+C+++ TIV N ER PST YTFD+VF
Sbjct: 8 EKILVSVRVRPQNEKEKARNDICDWECVNNTTIVCNNNLPERSLFPST-YTFDKVFGFDS 66
Query: 87 STQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGITENAIRDIYECIKNTPDR 146
T++VY++GAK+VAL L GIN++IFAYGQTSSGKT+TM GIT+ A+ DI+ I+ DR
Sbjct: 67 PTKQVYEDGAKEVALCVLGGINSSIFAYGQTSSGKTYTMCGITKFAMDDIFCYIQKHTDR 126
Query: 147 DFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENLFEEVARDAQHLRHLIGIC 206
F LK SA+EIYNE V DLL+ ++ R+LDD E+ TVVE L EE +D HL L+ +C
Sbjct: 127 KFTLKFSAIEIYNEAVRDLLSGDNNQRRLLDDPERGTVVEKLIEETIQDRTHLEELLTVC 186
Query: 207 EAHRQVGETTLNDKSSRSHQIIRLTVESFHRE-SPDHVKSYIASLNFVDLAGSERASQTN 265
E R++GET+LN+ SSRSHQI+RLT+ES RE SPD + AS+ F+DLAGSERASQT
Sbjct: 187 ETQRKIGETSLNEVSSRSHQILRLTIESTGREYSPDSSSTLAASVCFIDLAGSERASQTL 246
Query: 266 TCGTRLKEGSHINKSLLQLALVIRQL 291
+ GTRLKEG HIN+SLL L VIR+L
Sbjct: 247 SAGTRLKEGCHINRSLLTLGTVIRKL 272
>At1g21730 kinesin-related protein (MKRP1)
Length = 890
Score = 271 bits (693), Expect = 7e-73
Identities = 188/505 (37%), Positives = 278/505 (54%), Gaps = 37/505 (7%)
Query: 9 TPSSKIRRKLADTPGGSKIREEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNPN 68
+PS+ + +K++E I VT+R RPL+ +E D IAW D TI N
Sbjct: 54 SPSTSSAAASSTAVASTKLKEN-ITVTIRFRPLSPREVNNGDEIAWYADGDYTI----RN 108
Query: 69 QERPSTSYTFDRVFPPACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRG- 127
+ PS Y FDRVF P +T++VYD A+ V A+SGIN T+FAYG TSSGKT TM G
Sbjct: 109 EYNPSLCYGFDRVFGPPTTTRRVYDIAAQQVVSGAMSGINGTVFAYGVTSSGKTHTMHGE 168
Query: 128 -----ITENAIRDIYECIKNTPDRDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKV 182
I A++D++ I+ TP+R+F+L++S LEIYNE + DLL+ LRI +D++
Sbjct: 169 QRSPGIIPLAVKDVFSIIQETPEREFLLRVSYLEIYNEVINDLLDPTGQNLRIREDSQG- 227
Query: 183 TVVENLFEEVARDAQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESF-HRESPD 241
T VE + +EV H LI E HR VG +N SSRSH + LT+ES H + D
Sbjct: 228 TYVEGIKDEVVLSPAHALSLIASGEEHRHVGSNNVNLFSSRSHTMFTLTIESSPHGKGDD 287
Query: 242 HVKSYIASLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISY 301
++ L+ +DLAGSE +S+T G R KEGS INKSLL L VI +L+ K+ HI Y
Sbjct: 288 GEDVSLSQLHLIDLAGSE-SSKTEITGQRRKEGSSINKSLLTLGTVISKLTDTKAAHIPY 346
Query: 302 RTSKLTRILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMVEGE 361
R SKLTR+LQS+L G+ R ++ICT++P+ S E+T NTL FA K V A N + E
Sbjct: 347 RDSKLTRLLQSTLSGHGRVSLICTITPASSTSEETHNTLKFAQRCKHVEIKASRNKIMDE 406
Query: 362 LRNPEPEHAGLRSLLAEKELKIQQMEKDMEDLR--RQRDLAQCQLDLE---RRANKVQKG 416
+SL+ + + +I +++++ LR Q DLA +L ++ R + +
Sbjct: 407 -----------KSLIKKYQKEISCLQEELTQLRHGNQDDLADRKLQVKLQSRLEDDEEAK 455
Query: 417 SSDYGPSSQVVRCLSFAEENELAIGKHTPERTETVSRQAMLKNLLA-SPD---PSILVDE 472
++ G ++ + + + ++ L P+ + RQA ++ LA PD ++ D
Sbjct: 456 AALMGRIQRLTKLILVSTKSSLQAASVKPDH---IWRQAFGEDELAYLPDRRRENMADDG 512
Query: 473 IQKLEHRQLQLCEDANRALEVLHKD 497
L+ D N +L+ + KD
Sbjct: 513 AVSTVSEHLKEPRDGNSSLDEMTKD 537
>At2g21380 putative kinesin heavy chain
Length = 1058
Score = 265 bits (677), Expect = 5e-71
Identities = 160/374 (42%), Positives = 224/374 (59%), Gaps = 28/374 (7%)
Query: 30 EKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNPNQERPSTSYTFDRVFPPACSTQ 89
+ I VTVR RP++ +E D I W DK + N+ P T+Y FD+VF P +T
Sbjct: 103 DSISVTVRFRPMSEREYQRGDEIVWYPDADKMV----RNEYNPLTAYAFDKVFGPQSTTP 158
Query: 90 KVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRG------ITENAIRDIYECIKNT 143
+VYD AK V +A+ G+N T+FAYG TSSGKT TM G I AI+D++ I+ T
Sbjct: 159 EVYDVAAKPVVKAAMEGVNGTVFAYGVTSSGKTHTMHGDQDFPGIIPLAIKDVFSIIQET 218
Query: 144 PDRDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENLFEEVARDAQHLRHLI 203
R+F+L++S LEIYNE + DLL+ LRI +D++ T VE + EEV H I
Sbjct: 219 TGREFLLRVSYLEIYNEVINDLLDPTGQNLRIREDSQG-TYVEGIKEEVVLSPGHALSFI 277
Query: 204 GICEAHRQVGETTLNDKSSRSHQIIRLTVESF-HRESPDHVKSYIASLNFVDLAGSERAS 262
E HR VG N SSRSH I L +ES H + D V + LN +DLAGSE +S
Sbjct: 278 AAGEEHRHVGSNNFNLMSSRSHTIFTLMIESSAHGDQYDGV--IFSQLNLIDLAGSE-SS 334
Query: 263 QTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSSLGGNARTAI 322
+T T G R KEG++INKSLL L VI +L+ GK+ H+ +R SKLTR+LQSSL G+ ++
Sbjct: 335 KTETTGLRRKEGAYINKSLLTLGTVIGKLTEGKTTHVPFRDSKLTRLLQSSLSGHGHVSL 394
Query: 323 ICTVSPSLSHVEQTRNTLSFATNAKEV-INTARVNMVEGELRNPEPEHAGLRSLLAEKEL 381
ICTV+P+ S E+T NTL FA+ AK + IN +R +++ +SL+ + +
Sbjct: 395 ICTVTPASSSTEETHNTLKFASRAKRIEINASRNKIIDE------------KSLIKKYQK 442
Query: 382 KIQQMEKDMEDLRR 395
+I ++ +++ LRR
Sbjct: 443 EISTLKVELDQLRR 456
>At5g06670 kinesin heavy chain-like protein
Length = 997
Score = 262 bits (669), Expect = 4e-70
Identities = 180/513 (35%), Positives = 276/513 (53%), Gaps = 48/513 (9%)
Query: 8 ATPSSKIRRKLADTPGGSKIREEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNP 67
++P+S R P +E + VTVR RPL+ +E + IAW D +TIV
Sbjct: 43 SSPTSSSVRSKPQLPPKPLQSKENVTVTVRFRPLSPREIRKGEEIAWYA-DGETIVRNEN 101
Query: 68 NQERPSTSYTFDRVFPPACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRG 127
NQ S +Y +DRVF P +T+ VYD A+ V A++G+N TIFAYG TSSGKT TM G
Sbjct: 102 NQ---SIAYAYDRVFGPTTTTRNVYDVAAQHVVNGAMAGVNGTIFAYGVTSSGKTHTMHG 158
Query: 128 ------ITENAIRDIYECIKN----------TPDRDFVLKISALEIYNETVIDLLNRESG 171
I A++D + I+ TP R+F+L++S EIYNE V DLLN
Sbjct: 159 NQRSPGIIPLAVKDAFSIIQEVLLNFSFFSQTPRREFLLRVSYFEIYNEVVNDLLNPAGQ 218
Query: 172 PLRILDDTEKVTVVENLFEEVARDAQHLRHLIGICEA---HRQVGETTLNDKSSRSHQII 228
LRI +D E+ T +E + EEV H+ LI E HR +G T+ N SSRSH +
Sbjct: 219 NLRIRED-EQGTYIEGIKEEVVLSPAHVLSLIAAGEVMTEHRHIGSTSFNLLSSRSHTMF 277
Query: 229 RLTVESFHR-ESPDHVKSYIASLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALV 287
LT+ES ++ + +++ LN +DLAGSE +S+ T G R KEGS+INKSLL L V
Sbjct: 278 TLTIESSPLGDNNEGGAVHLSQLNLIDLAGSE-SSKAETSGLRRKEGSYINKSLLTLGTV 336
Query: 288 IRQLSSGKSGHISYRTSKLTRILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNAK 347
I +L+ ++ H+ YR SKLTR+L+SSL G+ R ++ICTV+P+ S+ E+T NTL FA AK
Sbjct: 337 ISKLTDRRASHVPYRDSKLTRLLESSLSGHGRVSLICTVTPASSNSEETHNTLKFAHRAK 396
Query: 348 EVINTARVNMVEGELRNPEPEHAGLRSLLAEKELKIQQMEKDMEDLR-------RQRDLA 400
+ A N + E +SL+ + + +I+Q+++++E L+ + +D++
Sbjct: 397 HIEIQAAQNKIIDE-----------KSLIKKYQYEIRQLKEELEQLKQGIKPVSQLKDIS 445
Query: 401 QCQLDLERRANKVQKGSSDYGPSSQVVRCLSFAEENELAIGKHTPERTETVSRQAMLKNL 460
+D+ K+++ ++ L+ + + TP+ + R +
Sbjct: 446 GDDIDIVLLKQKLEEEEDAKAALLSRIQRLT----KLILVSNKTPQTSRFSYRADPRRRH 501
Query: 461 LASPDPSILVDEIQKLEHRQLQLCEDANRALEV 493
+ I+ ++ L H++ L +D N L V
Sbjct: 502 SFGEEELIMHGQLAYLPHKRRDLTDDENLELYV 534
>At1g59540 zcf125 kinesin-like protein
Length = 823
Score = 258 bits (659), Expect = 6e-69
Identities = 159/402 (39%), Positives = 230/402 (56%), Gaps = 20/402 (4%)
Query: 30 EKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNPNQERPSTSYTFDRVFPPACSTQ 89
EKI V VR+RP + A W D++ + K+ + + S+ FD VF + +
Sbjct: 2 EKICVAVRVRPPAPENGASL----WKVEDNRISLHKSLDTPITTASHAFDHVFDESSTNA 57
Query: 90 KVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRG------ITENAIRDIYECIKNT 143
VY+ KD+ +A+ G N T FAYGQTSSGKTFTM G I ++RD++E I
Sbjct: 58 SVYELLTKDIIHAAVEGFNGTAFAYGQTSSGKTFTMTGSETDPGIIRRSVRDVFERIHMI 117
Query: 144 PDRDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENLFEEVARDAQHLRHLI 203
DR+F++++S +EIYNE + DLL E+ L+I + E+ V L EE+ DA+ + LI
Sbjct: 118 SDREFLIRVSYMEIYNEEINDLLAVENQRLQIHEHLERGVFVAGLKEEIVSDAEQILKLI 177
Query: 204 GICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRESPDHVKSYIASLNFVDLAGSERASQ 263
E +R GET +N SSRSH I R+ +ES +++ ++ LN VDLAGSER ++
Sbjct: 178 DSGEVNRHFGETNMNVHSSRSHTIFRMVIESRGKDNSSSDAIRVSVLNLVDLAGSERIAK 237
Query: 264 TNTCGTRLKEGSHINKSLLQLALVIRQLSSGKS--GHISYRTSKLTRILQSSLGGNARTA 321
T G RL+EG +INKSL+ L VI +LS HI YR SKLTRILQ +LGGNA+T
Sbjct: 238 TGAGGVRLQEGKYINKSLMILGNVINKLSDSTKLRAHIPYRDSKLTRILQPALGGNAKTC 297
Query: 322 IICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMVEGE---LRNPEPEHAGLRSLLAE 378
IICT++P H+E+++ TL FA+ AK + N A+VN + + L+ + E LR L
Sbjct: 298 IICTIAPEEHHIEESKGTLQFASRAKRITNCAQVNEILTDAALLKRQKLEIEELRMKLQG 357
Query: 379 K-----ELKIQQMEKDMEDLRRQRDLAQCQLDLERRANKVQK 415
E +I + M + + + QL+ E+R K Q+
Sbjct: 358 SHAEVLEQEILNLSNQMLKYELECERLKTQLEEEKRKQKEQE 399
>At3g12020 unknown protein
Length = 956
Score = 255 bits (651), Expect = 5e-68
Identities = 166/386 (43%), Positives = 226/386 (58%), Gaps = 41/386 (10%)
Query: 22 PGGSKIREEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNPNQERPSTSYTFDRV 81
P ++ +E + VTVR RPL+ +E + +AW D +TIV N+ P+ +Y +DRV
Sbjct: 60 PQTAQRSKENVTVTVRFRPLSPREIRQGEEVAWYA-DGETIV---RNEHNPTIAYAYDRV 115
Query: 82 FPPACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRG------ITENAIRD 135
F P +T+ VYD A V A+ GIN TIFAYG TSSGKT TM G I A++D
Sbjct: 116 FGPTTTTRNVYDIAAHHVVNGAMEGINGTIFAYGVTSSGKTHTMHGDQRSPGIIPLAVKD 175
Query: 136 IYECIKNTPDRDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENLFEEVARD 195
++ F+L+IS +EIYNE V DLLN LRI +D + T VE + EEV
Sbjct: 176 AFK---------FLLRISYMEIYNEVVNDLLNPAGHNLRIREDKQG-TFVEGIKEEVVLS 225
Query: 196 AQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRESPDHVKS-----YIASL 250
H LI E R VG T N SSRSH I LT+ES SP KS +++ L
Sbjct: 226 PAHALSLIAAGEEQRHVGSTNFNLLSSRSHTIFTLTIES----SPLGDKSKGEAVHLSQL 281
Query: 251 NFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRIL 310
N VDLAGSE +S+ T G R KEGS+INKSLL L VI +L+ ++ H+ YR SKLTRIL
Sbjct: 282 NLVDLAGSE-SSKVETSGVRRKEGSYINKSLLTLGTVISKLTDVRASHVPYRDSKLTRIL 340
Query: 311 QSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMVEGELRNPEPEHA 370
QSSL G+ R ++ICTV+P+ S E+T NTL FA AK + A N + E
Sbjct: 341 QSSLSGHDRVSLICTVTPASSSSEETHNTLKFAHRAKHIEIQAEQNKIIDE--------- 391
Query: 371 GLRSLLAEKELKIQQMEKDMEDLRRQ 396
+SL+ + + +I+Q+++++E L+++
Sbjct: 392 --KSLIKKYQREIRQLKEELEQLKQE 415
>At3g10180 putative kinesin-like centromere protein
Length = 459
Score = 249 bits (637), Expect = 2e-66
Identities = 148/376 (39%), Positives = 223/376 (58%), Gaps = 38/376 (10%)
Query: 79 DRVFPPACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRG------ITENA 132
DR+F C T +VY+ K++ +A+ G N T+FAYGQT+SGKT TMRG + A
Sbjct: 45 DRIFREDCKTVQVYEARTKEIVSAAVRGFNGTVFAYGQTNSGKTHTMRGSPIEPGVIPLA 104
Query: 133 IRDIYECIKNTPDRDFVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENLFEEV 192
+ D+++ I R+F+L++S LEIYNE + DLL E L+I ++ EK V L EE+
Sbjct: 105 VHDLFDTIYQDASREFLLRMSYLEIYNEDINDLLAPEHRKLQIHENLEKGIFVAGLREEI 164
Query: 193 ARDAQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRESPDHVKS-----YI 247
Q + ++ E+HR +GET +N SSRSH I R+ +ES + + V + +
Sbjct: 165 VASPQQVLEMMEFGESHRHIGETNMNLYSSRSHTIFRMIIESRQKMQDEGVGNSCDAVRV 224
Query: 248 ASLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSG---KSGHISYRTS 304
+ LN VDLAGSERA++T G RLKEGSHINKSL+ L VI++LS G + GH+ YR S
Sbjct: 225 SVLNLVDLAGSERAAKTGAEGVRLKEGSHINKSLMTLGTVIKKLSEGVETQGGHVPYRDS 284
Query: 305 KLTRILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMV------ 358
KLTRILQ +LGGNA TAIIC ++ + H ++T+++L FA+ A V N A VN +
Sbjct: 285 KLTRILQPALGGNANTAIICNITLAPIHADETKSSLQFASRALRVTNCAHVNEILTDAAL 344
Query: 359 -----------EGELRNPEPEHA-----GLRSLLAEKELKIQQMEKDMEDLRRQRDLAQC 402
+L+ +H+ LR+ L + EL+ +++ ++E+ ++ + AQ
Sbjct: 345 LKRQKKEIEELRSKLKTSHSDHSEEEILNLRNTLLKSELERERIALELEEEKKAQ--AQR 402
Query: 403 QLDLERRANKVQKGSS 418
+ L+ +A K++ SS
Sbjct: 403 ERVLQEQAKKIKNLSS 418
>At2g28620 putative kinesin-like spindle protein
Length = 1076
Score = 204 bits (518), Expect = 1e-52
Identities = 133/362 (36%), Positives = 190/362 (51%), Gaps = 33/362 (9%)
Query: 25 SKIREEK---IRVTVRMRPLNRKEQAMYDLIAWDCLDDK--TIVFKNPNQERPSTSYTFD 79
SK +EK I+V VR RP N +E + C D K V +N ++ ++ FD
Sbjct: 41 SKNEKEKGVNIQVIVRCRPFNSEETRLQTPAVLTCNDRKKEVAVAQNIAGKQIDKTFLFD 100
Query: 80 RVFPPACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRG------------ 127
+VF P + +Y + + L G N TIFAYGQT +GKT+TM G
Sbjct: 101 KVFGPTSQQKDLYHQAVSPIVFEVLDGYNCTIFAYGQTGTGKTYTMEGGARKKNGEIPSD 160
Query: 128 --ITENAIRDIYECIKNTPDRDFVLKISALEIYNETVIDLL---------NRESGPLRIL 176
+ A++ I++ ++ ++ LK+S LE+YNE + DLL ++ PL ++
Sbjct: 161 AGVIPRAVKQIFDILEAQSAAEYSLKVSFLELYNEELTDLLAPEETKFADDKSKKPLALM 220
Query: 177 DDTEKVTVVENLFEEVARDAQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTV--ES 234
+D + V L EE+ A + ++ A R+ ET LN +SSRSH I +T+ +
Sbjct: 221 EDGKGGVFVRGLEEEIVSTADEIYKVLEKGSAKRRTAETLLNKQSSRSHSIFSVTIHIKE 280
Query: 235 FHRESPDHVKSYIASLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSG 294
E + VKS LN VDLAGSE S++ R +E INKSLL L VI L
Sbjct: 281 CTPEGEEIVKS--GKLNLVDLAGSENISRSGAREGRAREAGEINKSLLTLGRVINALVE- 337
Query: 295 KSGHISYRTSKLTRILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEVINTAR 354
SGHI YR SKLTR+L+ SLGG +T +I TVSPS+ +E+T +TL +A AK + N
Sbjct: 338 HSGHIPYRESKLTRLLRDSLGGKTKTCVIATVSPSVHCLEETLSTLDYAHRAKHIKNKPE 397
Query: 355 VN 356
VN
Sbjct: 398 VN 399
>At3g17360 kinesin-like protein
Length = 2008
Score = 201 bits (512), Expect = 7e-52
Identities = 133/352 (37%), Positives = 180/352 (50%), Gaps = 42/352 (11%)
Query: 28 REEKIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNPNQERPSTSYTFDRVFPPACS 87
++ ++V +R+RPL E+A N P +TFD V S
Sbjct: 165 KDHNVQVLIRLRPLGTMERA------------------NQGYGHPEARFTFDHVASETIS 206
Query: 88 TQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRG--------------ITENAI 133
+K++ + + LSG N+ +FAYGQT SGKT+TM G +T
Sbjct: 207 QEKLFRVAGLPMVENCLSGYNSCVFAYGQTGSGKTYTMMGEISEAEGSLGEDCGVTARIF 266
Query: 134 RDIYECIKNTPD--RD----FVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVEN 187
++ IK + RD F K S LEIYNE + DLL S L++ +D K VEN
Sbjct: 267 EYLFSRIKMEEEERRDENLKFSCKCSFLEIYNEQITDLLEPSSTNLQLREDLGKGVYVEN 326
Query: 188 LFEEVARDAQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRESPDHVKSYI 247
L E R + L+ +R++ T +N +SSRSH + T+ES E +S
Sbjct: 327 LVEHNVRTVSDVLKLLLQGATNRKIAATRMNSESSRSHSVFTCTIESLW-EKDSLTRSRF 385
Query: 248 ASLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQL---SSGKSGHISYRTS 304
A LN VDLAGSER + G RLKE ++INKSL L LVI L + GK H+ YR S
Sbjct: 386 ARLNLVDLAGSERQKSSGAEGDRLKEAANINKSLSTLGLVIMSLVDLAHGKHRHVPYRDS 445
Query: 305 KLTRILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVN 356
+LT +LQ SLGGN++T II VSPSL +T +TL FA AK + N A+VN
Sbjct: 446 RLTFLLQDSLGGNSKTMIIANVSPSLCSTNETLSTLKFAQRAKLIQNNAKVN 497
>At3g49650 kinesin-like protein
Length = 813
Score = 197 bits (500), Expect = 2e-50
Identities = 154/496 (31%), Positives = 252/496 (50%), Gaps = 44/496 (8%)
Query: 32 IRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNPNQERP----------STSYTFDRV 81
+ V V+ RPL KE+ D++ + + K +V +P+ + Y FD
Sbjct: 15 LTVAVKCRPLMEKERGR-DIVRVN--NSKEVVVLDPDLSKDYLDRIQNRTKEKKYCFDHA 71
Query: 82 FPPACSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRGITEN------AIRD 135
F P + + VY V S + G+NAT+FAYG T SGKT+TM G + ++
Sbjct: 72 FGPESTNKNVY-RSMSSVISSVVHGLNATVFAYGSTGSGKTYTMVGTRSDPGLMVLSLNT 130
Query: 136 IYECIKNTPDRD-FVLKISALEIYNETVIDLLNRESGPLRILDDTEKVTVVENLFEEVAR 194
I++ IK+ D F + S LE+YNE + DLL + SG L + +D E+ VV L
Sbjct: 131 IFDMIKSDKSSDEFEVTCSYLEVYNEVIYDLLEKSSGHLELREDPEQGIVVAGLRSIKVH 190
Query: 195 DAQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRESPDHVKSYIASLNFVD 254
A + L+ + + R+ T +N SSRSH ++ + V+ + ++ +A VD
Sbjct: 191 SADRILELLNLGNSRRKTESTEMNGTSSRSHAVLEIAVKRRQKNQNQVMRGKLA---LVD 247
Query: 255 LAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQL-SSGKSG--HISYRTSKLTRILQ 311
LAGSERA++TN G +L++G++IN+SLL LA I L K G ++ YR SKLTRIL+
Sbjct: 248 LAGSERAAETNNGGQKLRDGANINRSLLALANCINALGKQHKKGLAYVPYRNSKLTRILK 307
Query: 312 SSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEV-------INTARVNMVEGE--L 362
L GN++T ++ T+SP+ S T NTL +A AKE+ I T +M + + +
Sbjct: 308 DGLSGNSQTVMVATISPADSQYHHTVNTLKYADRAKEIKTHIQKNIGTIDTHMSDYQRMI 367
Query: 363 RNPEPEHAGLRSLLAEKE--LKIQQMEKDMEDLRRQRDLAQCQLDLERRANKVQKGSSDY 420
N + E + L++ LAEKE L I+ E+ +E D Q+ E +++ + +
Sbjct: 368 DNLQSEVSQLKTQLAEKESQLSIKPFERGVERELSWLDGLSHQIS-ENVQDRINLQKALF 426
Query: 421 GPSSQVVRCLSFAEENELAIGKHTPER--TETVS--RQAMLKNLLASPDPSILVD-EIQK 475
+R + + + AI K E+ E +S RQ +L N+ + + + +I++
Sbjct: 427 ELEETNLRNRTELQHLDDAIAKQATEKDVVEALSSRRQVILDNIRDNDEAGVNYQRDIEE 486
Query: 476 LEHRQLQLCEDANRAL 491
E + +L + N A+
Sbjct: 487 NEKHRCELQDMLNEAI 502
>At3g45850 kinesin-related protein - like
Length = 1058
Score = 196 bits (497), Expect = 4e-50
Identities = 163/587 (27%), Positives = 259/587 (43%), Gaps = 95/587 (16%)
Query: 1 MVKTPMAATP--SSKIRRKLADTPGGSKIREEK-----IRVTVRMRPLNRKEQAMYDLIA 53
+V A TP S K R+ + S R +K ++V +R RPL+ E ++ +
Sbjct: 11 IVSLSPAQTPRSSDKSARESRSSESNSTNRNDKEKGVNVQVILRCRPLSEDEARIHTPVV 70
Query: 54 WDCLDDKTIVFKNPN--QERPSTSYTFDRVFPPACSTQKVYDEGAKDVALSALSGINATI 111
C +++ V + + + FD+VF PA + +YD+ + L G N TI
Sbjct: 71 ISCNENRREVAATQSIAGKHIDRHFAFDKVFGPASQQKDLYDQAICPIVFEVLEGYNCTI 130
Query: 112 FAYGQTSSGKTFTMRG--------------ITENAIRDIYECIKNTPDRDFVLKISALEI 157
FAYGQT +GKT+TM G + A++ I++ ++ ++ +K++ LE+
Sbjct: 131 FAYGQTGTGKTYTMEGGARKKNGEFPSDAGVIPRAVKQIFDILE-AQGAEYSMKVTFLEL 189
Query: 158 YNETVIDLLNRESGPLRILDDTEKVTV-----------VENLFEEVARDAQHLRHLIGIC 206
YNE + DLL E ++ +D+ K ++ V L EE+ A + ++
Sbjct: 190 YNEEISDLLAPEE-TIKFVDEKSKKSIALMEDGKGSVFVRGLEEEIVSTANEIYKILEKG 248
Query: 207 EAHRQVGETTLNDKSSRSHQIIRLTVESFHRESPDHVKSYIASLNFVDLAGSERASQTNT 266
A R+ ET LN +SSRSH I +T+ LN VDLAGSE S++
Sbjct: 249 SAKRRTAETLLNKQSSRSHSIFSITIHIKENTPEGEEMIKCGKLNLVDLAGSENISRSGA 308
Query: 267 CGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSSLGGNARTAIICTV 326
R +E INKSLL L VI L SGHI YR SKLTR+L+ SLGG +T +I T+
Sbjct: 309 REGRAREAGEINKSLLTLGRVINALVE-HSGHIPYRDSKLTRLLRESLGGKTKTCVIATI 367
Query: 327 SPSLSHVEQTRNTLSFATNAKEVINTARVN------------------------------ 356
SPS+ +E+T +TL +A AK + N +N
Sbjct: 368 SPSIHCLEETLSTLDYAHRAKNIKNKPEINQKMMKSAVMKDLYSEIDRLKQEVYAAREKN 427
Query: 357 ---MVEGELRNPEPEHAGLRSLLAEKELKIQQMEK---DMEDLRRQRDLAQCQLD--LER 408
+ + E E + + EL+ + +K D+++L + + +L LE+
Sbjct: 428 GIYIPKDRYIQEEAEKKAMAEKIERLELQSESKDKRVVDLQELYNSQQILTAELSEKLEK 487
Query: 409 RANKVQKGS-SDYGPSSQVVRCLSFAEENELAIGKHTPERTETVSRQAMLKNLL--ASPD 465
K+++ S + + + + +E E I V R L+ L AS D
Sbjct: 488 TEKKLEETEHSLFDLEEKYRQANATIKEKEFVISNLLKSEKSLVERAFQLRTELESASSD 547
Query: 466 PSILVDEIQKLEHRQLQLCEDANR------------ALEVLHKDFAT 500
S L +I++ + ED NR LE+LHK A+
Sbjct: 548 VSNLFSKIERKDK-----IEDGNRFLIQKFQSQLTQQLELLHKTVAS 589
>At5g65930 kinesin-like calmodulin-binding protein
Length = 1260
Score = 194 bits (494), Expect = 8e-50
Identities = 129/379 (34%), Positives = 208/379 (54%), Gaps = 26/379 (6%)
Query: 31 KIRVTVRMRPLNRKEQAMYDLIAWDCLDDKTIVFKNPNQERPSTSYTFDRVFPPACSTQK 90
KIRV R+RPLN KE + + +D+ T+ ++P ++ + +DRVF S
Sbjct: 888 KIRVYCRIRPLNEKESSEREKQMLTTVDEFTV--EHPWKDDKRKQHIYDRVFDMRASQDD 945
Query: 91 VYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRG------ITENAIRDIYECIKNTP 144
++++ K + SA+ G N IFAYGQT SGKTFT+ G +T A ++++ +K
Sbjct: 946 IFED-TKYLVQSAVDGYNVCIFAYGQTGSGKTFTIYGHESNPGLTPRATKELFNILKRDS 1004
Query: 145 DR-DFVLKISALEIYNETVIDLLNRESG---PLRILDDTEKVTVVENLFEEVARDAQHLR 200
R F LK +E+Y +T++DLL +S L I D++ + VEN+ + LR
Sbjct: 1005 KRFSFSLKAYMVELYQDTLVDLLLPKSARRLKLEIKKDSKGMVFVENVTTIPISTLEELR 1064
Query: 201 HLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRESPDHVKSYIASLNFVDLAGSER 260
++ R V T +N++SSRSH I+ + +ES ++ + L+FVDLAGSER
Sbjct: 1065 MILERGSERRHVSGTNMNEESSRSHLILSVVIESIDLQTQSAARG---KLSFVDLAGSER 1121
Query: 261 ASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISYRTSKLTRILQSSLGGNART 320
++ + G +LKE INKSL L VI LSSG HI YR KLT ++ SLGGNA+T
Sbjct: 1122 VKKSGSAGCQLKEAQSINKSLSALGDVIGALSSGNQ-HIPYRNHKLTMLMSDSLGGNAKT 1180
Query: 321 AIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMVEGELRNPEPEHAGLRSLLA--E 378
+ VSP+ S++++T N+L +A+ + ++N ++ E+ L+ L+A +
Sbjct: 1181 LMFVNVSPAESNLDETYNSLLYASRVRTIVNDPSKHISSKEM-------VRLKKLVAYWK 1233
Query: 379 KELKIQQMEKDMEDLRRQR 397
++ + E+D+ D+ R
Sbjct: 1234 EQAGKKGEEEDLVDIEEDR 1252
>At2g37420 putative kinesin heavy chain
Length = 1022
Score = 194 bits (494), Expect = 8e-50
Identities = 132/435 (30%), Positives = 215/435 (49%), Gaps = 42/435 (9%)
Query: 28 REEKIRVTVRMRPLNRKEQ--AMYDLIAWDCLDDKTIVFKNPNQERPSTSYTFDRVFPPA 85
+E ++V +R +PL+ +EQ ++ +I+ + + + V ++ + FD+VF P
Sbjct: 45 KEVNVQVILRCKPLSEEEQKSSVPRVISCNEMRREVNVLHTIANKQVDRLFNFDKVFGPK 104
Query: 86 CSTQKVYDEGAKDVALSALSGINATIFAYGQTSSGKTFTMRG--------------ITEN 131
+ +YD+ + L G + T+FAYGQT +GKT+TM G +
Sbjct: 105 SQQRSIYDQAIAPIVHEVLEGFSCTVFAYGQTGTGKTYTMEGGMRKKGGDLPAEAGVIPR 164
Query: 132 AIRDIYECIKNTPDRDFVLKISALEIYNETVIDLL----------NRESGPLRILDDTEK 181
A+R I++ ++ + D+ +K++ LE+YNE V DLL +++ P+ +++D +
Sbjct: 165 AVRHIFDTLE-AQNADYSMKVTFLELYNEEVTDLLAQDDSSRSSEDKQRKPISLMEDGKG 223
Query: 182 VTVVENLFEEVARDAQHLRHLIGICEAHRQVGETTLNDKSSRSHQIIRLTVESFHRESPD 241
V+ L EEV A + L+ + R+ +T LN +SSRSH + +TV D
Sbjct: 224 SVVLRGLEEEVVYSANDIYALLERGSSKRRTADTLLNKRSSRSHSVFTITVHIKEESMGD 283
Query: 242 HVKSYIASLNFVDLAGSERASQTNTCGTRLKEGSHINKSLLQLALVIRQLSSGKSGHISY 301
LN VDLAGSE ++ R +E INKSLL L VI L S H+ Y
Sbjct: 284 EELIKCGKLNLVDLAGSENILRSGARDGRAREAGEINKSLLTLGRVINALVE-HSSHVPY 342
Query: 302 RTSKLTRILQSSLGGNARTAIICTVSPSLSHVEQTRNTLSFATNAKEVINTARVNMVEGE 361
R SKLTR+L+ SLGG +T II T+SPS +E+T +TL +A AK + N N
Sbjct: 343 RDSKLTRLLRDSLGGKTKTCIIATISPSAHSLEETLSTLDYAYRAKNIKNKPEAN----- 397
Query: 362 LRNPEPEHAGLRSLLAEKELKIQQMEKDMEDLRRQRDLAQCQLDLERRANKVQKGSSDYG 421
+ LL + L++++M+ ED+R RD + ER + + +
Sbjct: 398 ------QKLSKAVLLKDLYLELERMK---EDVRAARDKNGVYIAHERYTQEEVEKKARIE 448
Query: 422 PSSQVVRCLSFAEEN 436
Q+ L+ +E N
Sbjct: 449 RIEQLENELNLSESN 463
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.130 0.357
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,337,201
Number of Sequences: 26719
Number of extensions: 452343
Number of successful extensions: 1971
Number of sequences better than 10.0: 111
Number of HSP's better than 10.0 without gapping: 68
Number of HSP's successfully gapped in prelim test: 43
Number of HSP's that attempted gapping in prelim test: 1630
Number of HSP's gapped (non-prelim): 141
length of query: 552
length of database: 11,318,596
effective HSP length: 104
effective length of query: 448
effective length of database: 8,539,820
effective search space: 3825839360
effective search space used: 3825839360
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC124967.12


