

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124966.6 + phase: 0
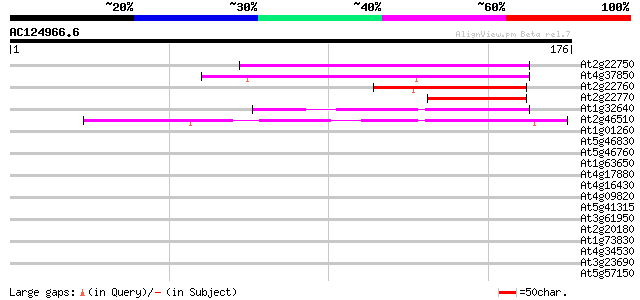
(176 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g22750 putative bHLH transcription factor (bHLH018) 60 5e-10
At4g37850 putative bHLH transcription factor (bHLH025) 56 1e-08
At2g22760 putative bHLH transcription factor (bHLH019) 49 1e-06
At2g22770 putative bHLH transcription factor (bHLH020) 48 3e-06
At1g32640 transcription factor like protein, BHLH6 41 4e-04
At2g46510 putative bHLH transcription factor 40 5e-04
At1g01260 transcription factor MYC7E, putative bHLH transcriptio... 39 0.001
At5g46830 putative transcription factor bHLH28 39 0.002
At5g46760 putative transcription factor BHLH5 38 0.003
At1g63650 putative transcription factor (BHLH2) 37 0.005
At4g17880 putative transcription factor BHLH4 37 0.007
At4g16430 putative transcription factor BHLH3 37 0.007
At4g09820 putative protein 37 0.007
At5g41315 bHLH transcription factor-like protein 34 0.035
At3g61950 bHLH transcription factor (bHLH067) like protein 33 0.060
At2g20180 putative bHLH transcription factor (bHLH015) 33 0.060
At1g73830 bHLH transcription factor (bHLH050) like protein 33 0.10
At4g34530 putative bHLH transcription factor (bHLH063) 32 0.13
At3g23690 bHLH transcription factor like protein (bHLH077) 32 0.18
At5g57150 unknown protein 32 0.23
>At2g22750 putative bHLH transcription factor (bHLH018)
Length = 304
Score = 60.5 bits (145), Expect = 5e-10
Identities = 36/91 (39%), Positives = 51/91 (55%)
Query: 73 PSQYILSFENSTMQPSPNSDIATCSSIMVQETTTLNNNVSSELPKIIKKRTKNLRSSCEM 132
PS ILSFE + + ++ S +E + + + + K+ RS
Sbjct: 65 PSSRILSFEKTGLHVMNHNSPNLIFSPKDEEIGLPEHKKAELIIRGTKRAQSLTRSQSNA 124
Query: 133 QDHIMAERKRRQVLTERFIALSATIPGLKKV 163
QDHI+AERKRR+ LT+RF+ALSA IPGLKK+
Sbjct: 125 QDHILAERKRREKLTQRFVALSALIPGLKKM 155
>At4g37850 putative bHLH transcription factor (bHLH025)
Length = 304
Score = 55.8 bits (133), Expect = 1e-08
Identities = 40/118 (33%), Positives = 56/118 (46%), Gaps = 15/118 (12%)
Query: 61 EYTWATNSQQLPP-----------SQYILSFENSTMQPSPNSDIATCSSIMVQETTTLNN 109
E T+ + S LPP S ILSFE+ + T + +
Sbjct: 40 ETTYISPSSHLPPNSKPHHIHRHSSSRILSFEDYGSNDMEHEYSPTYLNSIFSPKLEAQV 99
Query: 110 NVSSELPKIIKKRTKNL----RSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKKV 163
+ + +K TK R+ QDHI+AERKRR+ LT+RF+ALSA +PGLKK+
Sbjct: 100 QPHQKSDEFNRKGTKRAQPFSRNQSNAQDHIIAERKRREKLTQRFVALSALVPGLKKM 157
>At2g22760 putative bHLH transcription factor (bHLH019)
Length = 295
Score = 48.9 bits (115), Expect = 1e-06
Identities = 25/54 (46%), Positives = 37/54 (68%), Gaps = 6/54 (11%)
Query: 115 LPKIIKKRTKN------LRSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKK 162
+ K++ + TK RS ++H++AERKRR+ L+E+FIALSA +PGLKK
Sbjct: 94 MDKLVGRGTKRKTCSHGTRSPVLAKEHVLAERKRREKLSEKFIALSALLPGLKK 147
>At2g22770 putative bHLH transcription factor (bHLH020)
Length = 320
Score = 47.8 bits (112), Expect = 3e-06
Identities = 21/31 (67%), Positives = 27/31 (86%)
Query: 132 MQDHIMAERKRRQVLTERFIALSATIPGLKK 162
+++H++AERKRRQ L ER IALSA +PGLKK
Sbjct: 130 LKEHVLAERKRRQKLNERLIALSALLPGLKK 160
>At1g32640 transcription factor like protein, BHLH6
Length = 623
Score = 40.8 bits (94), Expect = 4e-04
Identities = 27/87 (31%), Positives = 44/87 (50%), Gaps = 11/87 (12%)
Query: 77 ILSFENSTMQPSPNSDIATCSSIMVQETTTLNNNVSSELPKIIKKRTKNLRSSCEMQDHI 136
+LSF + T S +SD+ E + + + PK ++ N R E +H+
Sbjct: 406 VLSFGDKTAGESDHSDL---------EASVVKEVAVEKRPKKRGRKPANGRE--EPLNHV 454
Query: 137 MAERKRRQVLTERFIALSATIPGLKKV 163
AER+RR+ L +RF AL A +P + K+
Sbjct: 455 EAERQRREKLNQRFYALRAVVPNVSKM 481
>At2g46510 putative bHLH transcription factor
Length = 662
Score = 40.4 bits (93), Expect = 5e-04
Identities = 39/157 (24%), Positives = 67/157 (41%), Gaps = 24/157 (15%)
Query: 24 PSFSFESGNSQSSTIQYMNNIVDNVCVTNTVH---ENIAQEYTWATNSQQLPPSQYILSF 80
P+F + ++ +N +VDN + ++A +TN+QQ
Sbjct: 301 PTFGYTPQRDDVKVLENVNMVVDNNNYKTQIEFAGSSVAASSNPSTNTQQ--------EK 352
Query: 81 ENSTMQPSPNSDIATCSSIMVQETTTLNNNVSSELPKIIKKRTKNLRSSCEMQDHIMAER 140
S + P S +A + V V + P+ ++ N R E +H+ AER
Sbjct: 353 SESCTEKRPVSLLAGAGIVSV---------VDEKRPRKRGRKPANGRE--EPLNHVEAER 401
Query: 141 KRRQVLTERFIALSATIPGLKKV--SFLYKPTISLIK 175
+RR+ L +RF AL + +P + K+ + L IS IK
Sbjct: 402 QRREKLNQRFYALRSVVPNISKMDKASLLGDAISYIK 438
>At1g01260 transcription factor MYC7E, putative bHLH transcription
factor (AtbHLH013)
Length = 590
Score = 38.9 bits (89), Expect = 0.001
Identities = 25/98 (25%), Positives = 48/98 (48%), Gaps = 4/98 (4%)
Query: 66 TNSQQLPPSQYILSFENSTMQPSPNSDIATCSSIMVQETTTLNNNVSSELPKIIKKRTKN 125
++ + LPP+Q + F ++ + S N+ + + P+ +R N
Sbjct: 369 SSQRLLPPAQMQIDFSAASSRASENNSDGEGGGEWADAVGA--DESGNNRPRKRGRRPAN 426
Query: 126 LRSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKKV 163
R+ E +H+ AER+RR+ L +RF AL + +P + K+
Sbjct: 427 GRA--EALNHVEAERQRREKLNQRFYALRSVVPNISKM 462
>At5g46830 putative transcription factor bHLH28
Length = 511
Score = 38.5 bits (88), Expect = 0.002
Identities = 30/106 (28%), Positives = 49/106 (45%), Gaps = 16/106 (15%)
Query: 74 SQYILSFE--NSTMQPSPNSDIATCSSIMVQETTTLN-NNVSSELPKII---------KK 121
++Y L+F +ST+ +P D+ + + Q N N S ++ ++ KK
Sbjct: 267 NRYNLNFSTSSSTLARAPCGDVLSFGENVKQSFENRNPNTYSDQIQNVVPHATVMLEKKK 326
Query: 122 RTKNLRSSCEMQD----HIMAERKRRQVLTERFIALSATIPGLKKV 163
K R +D H+ AER RR+ L RF AL A +P + K+
Sbjct: 327 GKKRGRKPAHGRDKPLNHVEAERMRREKLNHRFYALRAVVPNVSKM 372
>At5g46760 putative transcription factor BHLH5
Length = 592
Score = 37.7 bits (86), Expect = 0.003
Identities = 25/87 (28%), Positives = 42/87 (47%), Gaps = 4/87 (4%)
Query: 77 ILSFENSTMQPSPNSDIATCSSIMVQETTTLNNNVSSELPKIIKKRTKNLRSSCEMQDHI 136
+LSF + +SD + + +V+E V K K+ K E +H+
Sbjct: 362 MLSFSTVVRSAANDSDHSDLEASVVKEAIV----VEPPEKKPRKRGRKPANGREEPLNHV 417
Query: 137 MAERKRRQVLTERFIALSATIPGLKKV 163
AER+RR+ L +RF +L A +P + K+
Sbjct: 418 EAERQRREKLNQRFYSLRAVVPNVSKM 444
>At1g63650 putative transcription factor (BHLH2)
Length = 596
Score = 37.0 bits (84), Expect = 0.005
Identities = 18/50 (36%), Positives = 30/50 (60%)
Query: 114 ELPKIIKKRTKNLRSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKKV 163
E+P + KK + E +H ++E+KRR+ L ERF+ L + IP + K+
Sbjct: 385 EVPLMNKKEELLPDTPEETGNHALSEKKRREKLNERFMTLRSIIPSISKI 434
>At4g17880 putative transcription factor BHLH4
Length = 589
Score = 36.6 bits (83), Expect = 0.007
Identities = 29/102 (28%), Positives = 49/102 (47%), Gaps = 7/102 (6%)
Query: 65 ATNSQQLPPS---QYILSFENSTMQPSPNSDIATCSSIMVQETTTLNNNVSSELPKIIKK 121
++N ++ P S + +LSF + S +SD+ V + N V K K+
Sbjct: 348 SSNKKRSPVSNNEEGMLSFTSVLPCDSNHSDLEAS----VAKEAESNRVVVEPEKKPRKR 403
Query: 122 RTKNLRSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKKV 163
K E +H+ AER+RR+ L +RF +L A +P + K+
Sbjct: 404 GRKPANGREEPLNHVEAERQRREKLNQRFYSLRAVVPNVSKM 445
>At4g16430 putative transcription factor BHLH3
Length = 467
Score = 36.6 bits (83), Expect = 0.007
Identities = 17/44 (38%), Positives = 26/44 (58%)
Query: 120 KKRTKNLRSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKKV 163
K+ K E +H+ AER+RR+ L +RF AL A +P + K+
Sbjct: 306 KRGRKPANGREEALNHVEAERQRREKLNQRFYALRAVVPNISKM 349
>At4g09820 putative protein
Length = 379
Score = 36.6 bits (83), Expect = 0.007
Identities = 17/41 (41%), Positives = 27/41 (65%)
Query: 123 TKNLRSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKKV 163
TK+ R E H++AER+RR+ L E+FI L + +P + K+
Sbjct: 213 TKDKRLPREDLSHVVAERRRREKLNEKFITLRSMVPFVTKM 253
>At5g41315 bHLH transcription factor-like protein
Length = 637
Score = 34.3 bits (77), Expect = 0.035
Identities = 19/54 (35%), Positives = 32/54 (59%), Gaps = 5/54 (9%)
Query: 114 ELPKIIKKRTKNLRSSCEMQD----HIMAERKRRQVLTERFIALSATIPGLKKV 163
++P++ +K K + S E +D H + E+KRR+ L ERF+ L IP + K+
Sbjct: 418 DVPRVHQKE-KLMLDSPEARDETGNHAVLEKKRREKLNERFMTLRKIIPSINKI 470
>At3g61950 bHLH transcription factor (bHLH067) like protein
Length = 358
Score = 33.5 bits (75), Expect = 0.060
Identities = 27/100 (27%), Positives = 50/100 (50%), Gaps = 15/100 (15%)
Query: 65 ATNSQQLPPSQYILSFENSTMQPSPNSDIATCSSIMVQETTTLNNNVSSELPKIIKKR-- 122
A +SQ++P SQ ++ +ST P S + +N+ + E+ + +KR
Sbjct: 113 AVSSQEIPFSQANMTLPSSTSSPL---------SAHSRRKRKINHLLPQEMTREKRKRRK 163
Query: 123 TKNLRSSCEMQD----HIMAERKRRQVLTERFIALSATIP 158
TK +++ E+++ HI ER RR+ + E +L A +P
Sbjct: 164 TKPSKNNEEIENQRINHIAVERNRRRQMNEHINSLRALLP 203
>At2g20180 putative bHLH transcription factor (bHLH015)
Length = 478
Score = 33.5 bits (75), Expect = 0.060
Identities = 21/61 (34%), Positives = 29/61 (47%)
Query: 102 QETTTLNNNVSSELPKIIKKRTKNLRSSCEMQDHIMAERKRRQVLTERFIALSATIPGLK 161
+ TTT SE K + T + + S + H ++ERKRR + ER AL IP
Sbjct: 256 EATTTDETESRSEETKQARVSTTSTKRSRAAEVHNLSERKRRDRINERMKALQELIPRCN 315
Query: 162 K 162
K
Sbjct: 316 K 316
>At1g73830 bHLH transcription factor (bHLH050) like protein
Length = 261
Score = 32.7 bits (73), Expect = 0.10
Identities = 37/153 (24%), Positives = 65/153 (42%), Gaps = 8/153 (5%)
Query: 10 LDEDKFQREIVQHQPSFSFESGNSQSSTIQYMNNIVDNVCVTNTVHENIAQEYTWATNSQ 69
LD F + H P S E+ Q I + +NI +N ++ + A ++
Sbjct: 41 LDSLFFHNQFPDHFPGKSLENNFHQG--IFFPSNIQNNEESSSQFDTKKRKSLMEAVSTS 98
Query: 70 QLPPSQYILSFENSTMQPSPNSDIATCSSIMVQETTTLNNNVSSELPKIIKKRTKNLRSS 129
+ S LS S+ Q S N +I+T ++ + + N E +++ R + +++
Sbjct: 99 ENSVSDQTLS--TSSAQVSINGNISTKNNSSRRGKRSKNREEEKER-EVVHVRARRGQAT 155
Query: 130 CEMQDHIMAERKRRQVLTERFIALSATIPGLKK 162
H +AER RR + ER L +PG K
Sbjct: 156 ---DSHSIAERVRRGKINERLKCLQDIVPGCYK 185
>At4g34530 putative bHLH transcription factor (bHLH063)
Length = 335
Score = 32.3 bits (72), Expect = 0.13
Identities = 19/62 (30%), Positives = 30/62 (47%), Gaps = 6/62 (9%)
Query: 109 NNVSSELPKIIKKRTKN------LRSSCEMQDHIMAERKRRQVLTERFIALSATIPGLKK 162
NN S++ K+ K+ K R H +AER RR+ ++ER L +PG K
Sbjct: 151 NNFSNDSSKVTKELEKTDYIHVRARRGQATDSHSIAERVRREKISERMKFLQDLVPGCDK 210
Query: 163 VS 164
++
Sbjct: 211 IT 212
>At3g23690 bHLH transcription factor like protein (bHLH077)
Length = 371
Score = 32.0 bits (71), Expect = 0.18
Identities = 26/101 (25%), Positives = 44/101 (42%), Gaps = 4/101 (3%)
Query: 68 SQQLPPSQYILSFENSTMQPSPNS--DIATCSSIMVQETTTLNNNVSSELPKIIKKRTKN 125
S L S +S EN + S D+A S V++ + +N P K +
Sbjct: 131 SSSLTASNSKVSGENGGSKGGKRSKQDVAGSSKNGVEKCDSKGDNKDDAKPPEAPKDYIH 190
Query: 126 LRS--SCEMQDHIMAERKRRQVLTERFIALSATIPGLKKVS 164
+R+ H +AER RR+ ++ER L +PG +++
Sbjct: 191 VRARRGQATDSHSLAERARREKISERMTLLQDLVPGCNRIT 231
>At5g57150 unknown protein
Length = 226
Score = 31.6 bits (70), Expect = 0.23
Identities = 17/50 (34%), Positives = 29/50 (58%), Gaps = 2/50 (4%)
Query: 128 SSCEMQDHIMAERKRRQVLTERFIALSATIPGLKKV--SFLYKPTISLIK 175
+S +I++ER RRQ L +R AL + +P + K+ + + K IS I+
Sbjct: 49 ASSPASKNIVSERNRRQKLNQRLFALRSVVPNITKMDKASIIKDAISYIE 98
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.127 0.354
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,728,430
Number of Sequences: 26719
Number of extensions: 138672
Number of successful extensions: 616
Number of sequences better than 10.0: 76
Number of HSP's better than 10.0 without gapping: 51
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 564
Number of HSP's gapped (non-prelim): 76
length of query: 176
length of database: 11,318,596
effective HSP length: 93
effective length of query: 83
effective length of database: 8,833,729
effective search space: 733199507
effective search space used: 733199507
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Medicago: description of AC124966.6


