

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124966.5 - phase: 0
(1309 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
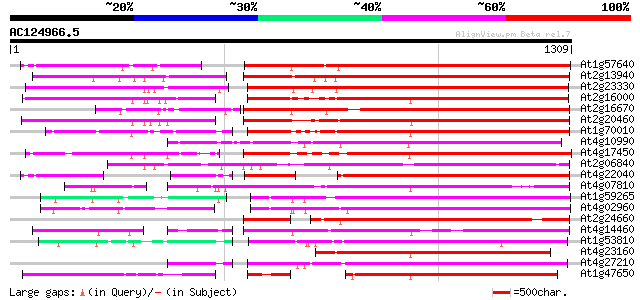
Score E
Sequences producing significant alignments: (bits) Value
At1g57640 731 0.0
At2g13940 putative retroelement pol polyprotein 726 0.0
At2g23330 putative retroelement pol polyprotein 691 0.0
At2g16000 putative retroelement pol polyprotein 672 0.0
At2g16670 putative retroelement pol polyprotein 671 0.0
At2g20460 putative retroelement pol polyprotein 668 0.0
At1g70010 hypothetical protein 655 0.0
At4g10990 putative retrotransposon polyprotein 641 0.0
At4g17450 retrotransposon like protein 634 0.0
At2g06840 putative retroelement pol polyprotein 629 e-180
At4g22040 LTR retrotransposon like protein 605 e-173
At4g07810 putative polyprotein 591 e-168
At1g59265 polyprotein, putative 579 e-165
At4g02960 putative polyprotein of LTR transposon 578 e-165
At2g24660 putative retroelement pol polyprotein 571 e-163
At4g14460 retrovirus-related like polyprotein 552 e-157
At1g53810 533 e-151
At4g23160 putative protein 531 e-151
At4g27210 putative protein 521 e-148
At1g47650 hypothetical protein 514 e-145
>At1g57640
Length = 1444
Score = 731 bits (1888), Expect = 0.0
Identities = 381/783 (48%), Positives = 525/783 (66%), Gaps = 27/783 (3%)
Query: 548 QTYL-FYFGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNMN 606
Q+YL F GEC+L+AAYLIN+ P+ +L+ KSP+++L + P YS LRVFG LC+A N N
Sbjct: 666 QSYLPIQFWGECILSAAYLINRTPSMLLQGKSPYEMLYKTAPKYSHLRVFGSLCYAHNQN 725
Query: 607 IQ-HKFDERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQ--------- 656
+ KF RS+ +FVGYP QKG+R++D++ ++ +VSRDV F ET FPY
Sbjct: 726 HKGDKFAARSRRCVFVGYPHGQKGWRLFDLEEQKFFVSRDVIFQETEFPYSKMSCNEEDE 785
Query: 657 ----DIQSPPF-NNAISINTQILDNEFDDLFTGSPTHPNIPPENSHNDNSNDTIVTISTP 711
D PPF AI T I N + + I PE + +S V++S+
Sbjct: 786 RVLVDCVGPPFIEEAIGPRTIIGRNIGEATVGPNVATGPIIPEINQESSSPSEFVSLSSL 845
Query: 712 EDDASSNPPSFSAESLSNNPPSTEMNPPNHRHSQRIRNPPIHLKDYICKINNV------- 764
+ +S+ + LS+ P+ P R S R P+ LK+++ +V
Sbjct: 846 DPFLASSTVQTADLPLSSTTPA----PIQLRRSSRQTQKPMKLKNFVTNTVSVESISPEA 901
Query: 765 TSKINFPLENYLSLSNLSNSHRAFLINIIENKEPKSYSQAMKSVEWRDAMAKEIQALESN 824
+S +P+E Y+ ++SH+AFL + EP +Y++AM WR+AM+ EI++L N
Sbjct: 902 SSSSLYPIEKYVDCHRFTSSHKAFLAAVTAGMEPTTYNEAMVDKAWREAMSAEIESLRVN 961
Query: 825 NTWILCPLPEGKSAIGCKWIYKIKYHSDGSIDRYKARLVAKGYSQVQGIDYHDTFAPVAK 884
T+ + LP GK A+G KW+YKIKY SDG+I+RYKARLV G Q +G+DY +TFAPVAK
Sbjct: 962 QTFSIVNLPPGKRALGNKWVYKIKYRSDGAIERYKARLVVLGNCQKEGVDYDETFAPVAK 1021
Query: 885 LVTVRLLLSIAAIKNWPLYQFDVNNAFLQGDLSEEVYMKLPPGFSHKKKPCVCKLNKSIY 944
+ TVRL L +AA ++W ++Q DV+NAFL GDL EEVYMKLP GF VC+L+KS+Y
Sbjct: 1022 MSTVRLFLGVAAARDWHVHQMDVHNAFLHGDLKEEVYMKLPQGFQCDDPSKVCRLHKSLY 1081
Query: 945 GLKQASRQWFSKFSTTLIQKGFRQSISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENAI 1004
GLKQA R WFSK S+ L Q GF QS+SDYSLF+YN D + VLVYVDD+II+G+ +A+
Sbjct: 1082 GLKQAPRCWFSKLSSALKQYGFTQSLSDYSLFSYNNDGIFVHVLVYVDDLIISGSCPDAV 1141
Query: 1005 SKIKKFLAQSFSIKDLGNLSYILGIEVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDFP 1064
++ K +L F +KDLG L Y LGIEVSR+ +G +L QRKY LDI+S+ G+ G RPS FP
Sbjct: 1142 AQFKSYLESCFHMKDLGLLKYFLGIEVSRNAQGFYLSQRKYVLDIISEMGLLGARPSAFP 1201
Query: 1065 MEQHLRLRPNDGTPLSDPTVYRRHVGRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDAA 1124
+EQ+ +L + LSD + YRR VGRL+YL VTRP++ Y+V+TL+QFMQ+P H++AA
Sbjct: 1202 LEQNHKLSLSTSPLLSDSSRYRRLVGRLIYLVVTRPELSYSVHTLAQFMQNPRQDHWNAA 1261
Query: 1125 TQVLRYLKGSVGKGLFLSASSSINLVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTK 1184
+V+RYLK + G+G+ LS++S++ + G+ DSD+A CP TRRS TGYF LG PISWKTK
Sbjct: 1262 IRVVRYLKSNPGQGILLSSTSTLQINGWCDSDYAACPLTRRSLTGYFVQLGDTPISWKTK 1321
Query: 1185 KQPTISRSSAEAEYRSLATLSSELQWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVF 1244
KQPT+SRSSAEAEYR++A L+ EL WLK +L DLG+ H Q + I+ DS++AI ++ NPV
Sbjct: 1322 KQPTVSRSSAEAEYRAMAFLTQELMWLKRVLYDLGVSHVQAMRIFSDSKSAIALSVNPVQ 1381
Query: 1245 HERTKHIEIDCHFVREKIKSGLIAPSYIRSSDQLADIFTKPLGGDAYKRILGKLGVIEIS 1304
HERTKH+E+DCHF+R+ I G+IA S++ S QLADI TK LG + L KLG++++
Sbjct: 1382 HERTKHVEVDCHFIRDAILDGIIATSFVPSHKQLADILTKALGEKEVRYFLRKLGILDVH 1441
Query: 1305 IPP 1307
PP
Sbjct: 1442 APP 1444
Score = 180 bits (457), Expect = 4e-45
Identities = 135/495 (27%), Positives = 223/495 (44%), Gaps = 93/495 (18%)
Query: 26 MSDIPPSENSPLSSISSPITVHSTTNPDPCVIHHSDNPSTVLVTPLLTGDNYGSWSRAVT 85
M+D PP S I V T +P +DN V+ P+L +NY W+
Sbjct: 1 MADAPPPP-------PSVIEVRRTISPYDLTA--ADNSGAVISHPILKTNNYEEWACGFK 51
Query: 86 MALRAKNKFGFVDGSLTIPKK-KEDILTWQRCNDLVASWILNSVSTEIRPSILYAETAEQ 144
ALR++ KFGF+DG++ P D+ W N L+ SW+ ++ +E+ +I + + A
Sbjct: 52 TALRSRKKFGFLDGTIPQPLDGSPDLEDWLTINALLVSWMKMTIDSELLTNISHRDVARD 111
Query: 145 IWSDLQDRFSQSNAPKIYQLKQSISALKQEGMYVSLYFTQLKSLWDELNSIALVNPCICG 204
+W ++ RFS SN PK ++K ++ KQEGM V Y+ +L +WD +NS + C CG
Sbjct: 112 LWEQIRKRFSVSNGPKNQKMKADLATCKQEGMTVEGYYGKLNKIWDNINSYRPLRICKCG 171
Query: 205 NAKSIL--DQQ---NQDRAMEFLQGVHD-RFSAVRSQILLMDPFPSIQRIYNIVRQEEKQ 258
L DQ+ D ++L G+++ +F +RS + P P ++ +YNIVRQEE
Sbjct: 172 RCICNLGTDQEKYREDDMVHQYLYGLNETKFHTIRSSLTSRVPLPGLEEVYNIVRQEEDM 231
Query: 259 QE---------------INFRPVPAVESAALQTSKAPYRTPGKRQRPFCEHCNRHGHTIT 303
+ RP V S S+ + + C HCNR GH+
Sbjct: 232 VNNRSSNEERTDVTAFAVQMRPRSEVISEKFANSEK------LQNKKLCTHCNRGGHSPE 285
Query: 304 TCYQIHGFPS-NPSKPQKKTETSPSTSANQ------------------------------ 332
C+ + G+P +P+ K+ ++ STS +
Sbjct: 286 NCFVLIGYPEWWGDRPRGKSNSNGSTSRGRGRFGPGFNGGQPRPTYVNVVMTGPFPSSEH 345
Query: 333 ---------------LSSAQYHKLLTLLAKEDNVGSSVNLAGT----AFTCIPF-SWIID 372
L+ Q+ ++ LL N G S N + + TC F SWI+D
Sbjct: 346 VNRVITDSDRDAVSGLTDEQWRGVVKLL----NAGRSDNKSNAHETQSGTCSLFTSWILD 401
Query: 373 SGASNHICTSLSLFSSYYPVNNQISVQQPDGSQALVKHIGTINCSPSLILTNVYHVPTFK 432
+GAS+H+ +L L S + + + + DG++ + GT+ LIL +V++V +
Sbjct: 402 TGASHHMTGNLELLSDMRSM-SPVLIILADGNKRVAVSEGTVRLGSHLILKSVFYVKELE 460
Query: 433 FNLMSVTQLTESLNC 447
+L+SV Q+ + +C
Sbjct: 461 SDLISVGQMMDENHC 475
>At2g13940 putative retroelement pol polyprotein
Length = 1501
Score = 726 bits (1875), Expect = 0.0
Identities = 376/804 (46%), Positives = 513/804 (63%), Gaps = 48/804 (5%)
Query: 546 FKQTYLFYFGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNM 605
F+ + F GE +LTAAYLIN+ P+ IL ++P++VL GS P YS LRVFG C+ +
Sbjct: 702 FQASLPIKFWGESILTAAYLINRTPSSILSGRTPYEVLHGSKPVYSQLRVFGSACYVHRV 761
Query: 606 NI-QHKFDERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDIQSPPFN 664
+ KF +RS+ IFVGYPF +KG+++YD++ + VSRDV F E VFPY + N
Sbjct: 762 TRDKDKFGQRSRSCIFVGYPFGKKGWKVYDIERNEFLVSRDVIFREEVFPYAGV-----N 816
Query: 665 NAISINTQILDNEFDDLFTGSPTHPNIPPENSHNDN---SNDTIVTIST----------- 710
++ +T + DD + P ++ + + D +V ++
Sbjct: 817 SSTLASTSLPTVSEDDDWAIPPLEVRGSIDSVETERVVCTTDEVVLDTSVSDSEIPNQEF 876
Query: 711 -PEDDASSNPPSFSAESLSNNPPST---------EMNPPNHRHSQRIRNPPIHLKDYIC- 759
P+D S+P S S N P + ++PP R S+R +PP L DY+
Sbjct: 877 VPDDTPPSSPLSVSPSGSPNTPTTPIVVPVASPIPVSPPKQRKSKRATHPPPKLNDYVLY 936
Query: 760 -----------------KINNVTSKINFPLENYLSLSNLSNSHRAFLINIIENKEPKSYS 802
+ + V K FPL +Y+S + S+SHRA+L I +N EPK +
Sbjct: 937 NAMYTPSSIHALPADPSQSSTVPGKSLFPLTDYVSDAAFSSSHRAYLAAITDNVEPKHFK 996
Query: 803 QAMKSVEWRDAMAKEIQALESNNTWILCPLPEGKSAIGCKWIYKIKYHSDGSIDRYKARL 862
+A++ W DAM E+ ALE N TW + LP GK AIG +W++K KY+SDG+++RYKARL
Sbjct: 997 EAVQIKVWNDAMFTEVDALEINKTWDIVDLPPGKVAIGSQWVFKTKYNSDGTVERYKARL 1056
Query: 863 VAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFLQGDLSEEVYM 922
V +G QV+G DY +TFAPV ++ TVR LL A W +YQ DV+NAFL GDL EEVYM
Sbjct: 1057 VVQGNKQVEGEDYKETFAPVVRMTTVRTLLRNVAANQWEVYQMDVHNAFLHGDLEEEVYM 1116
Query: 923 KLPPGFSHKKKPCVCKLNKSIYGLKQASRQWFSKFSTTLIQKGFRQSISDYSLFTYNCDQ 982
KLPPGF H VC+L KS+YGLKQA R WF K S +L++ GF QS DYSLF+Y +
Sbjct: 1117 KLPPGFRHSHPDKVCRLRKSLYGLKQAPRCWFKKLSDSLLRFGFVQSYEDYSLFSYTRNN 1176
Query: 983 TTIFVLVYVDDIIITGNNENAISKIKKFLAQSFSIKDLGNLSYILGIEVSRSKKGIFLCQ 1042
+ VL+YVDD++I GN+ + K K +L++ FS+KDLG L Y LGIEVSR +GIFL Q
Sbjct: 1177 IELRVLIYVDDLLICGNDGYMLQKFKDYLSRCFSMKDLGKLKYFLGIEVSRGPEGIFLSQ 1236
Query: 1043 RKYTLDILSDSGMTGCRPSDFPMEQHLRLRPNDGTPLSDPTVYRRHVGRLLYLTVTRPDI 1102
RKY LD+++DSG G RP+ P+EQ+ L +DG LSDP YRR VGRLLYL TRP++
Sbjct: 1237 RKYALDVIADSGNLGSRPAHTPLEQNHHLASDDGPLLSDPKPYRRLVGRLLYLLHTRPEL 1296
Query: 1103 QYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVGKGLFLSASSSINLVGYADSDWAGCPT 1162
Y+V+ L+QFMQ+P +HFDAA +V+RYLKGS G+G+ L+A + L Y DSDW CP
Sbjct: 1297 SYSVHVLAQFMQNPREAHFDAALRVVRYLKGSPGQGILLNADPDLTLEVYCDSDWQSCPL 1356
Query: 1163 TRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEAEYRSLATLSSELQWLKYLLSDLGIDH 1222
TRRS + Y +LG +PISWKTKKQ T+S SSAEAEYR+++ E++WL+ LL +LGI+
Sbjct: 1357 TRRSISAYVVLLGGSPISWKTKKQDTVSHSSAEAEYRAMSYALKEIKWLRKLLKELGIEQ 1416
Query: 1223 PQPITIYCDSQAAIHIAENPVFHERTKHIEIDCHFVREKIKSGLIAPSYIRSSDQLADIF 1282
P +YCDS+AAIHIA NPVFHERTKHIE DCH VR+ ++ G+I ++R+++QLAD+F
Sbjct: 1417 STPARLYCDSKAAIHIAANPVFHERTKHIESDCHSVRDAVRDGIITTQHVRTTEQLADVF 1476
Query: 1283 TKPLGGDAYKRILGKLGVIEISIP 1306
TK LG + + ++ KLGV + P
Sbjct: 1477 TKALGRNQFLYLMSKLGVQNLHTP 1500
Score = 213 bits (542), Expect = 6e-55
Identities = 152/519 (29%), Positives = 252/519 (48%), Gaps = 72/519 (13%)
Query: 54 PCVIHHSDNPSTVLVTPLLTGDNYGSWSRAVTMALRAKNKFGFVDGSLTIPKKKE-DILT 112
P + SDNP V+ + L GDNY W+ + AL+AK K GF++G++ P + +
Sbjct: 31 PYTLASSDNPGAVISSVELNGDNYNQWATEMLNALQAKRKTGFINGTIPRPPPNDPNYEN 90
Query: 113 WQRCNDLVASWILNSVSTEIRPSILYAETAEQIWSDLQDRFSQSNAPKIYQLKQSISALK 172
W N ++ WI S+ +++ ++ + A +W DL+ RFS N +I+Q++ +S+ +
Sbjct: 91 WTAVNSMIVGWIRTSIEPKVKATVTFISDAHLLWKDLKQRFSVGNKVRIHQIRAQLSSCR 150
Query: 173 QEGMYVSLYFTQLKSLWDELN-----SIALVNPCICGNAKSILDQQNQDRAMEFLQGVHD 227
Q+G V Y+ +L +LW+E N ++ C CG ++ +++ +F+ G+ +
Sbjct: 151 QDGQAVIEYYGRLSNLWEEYNIYKPVTVCTCGLCRCGATSEPTKEREEEKIHQFVLGLDE 210
Query: 228 -RFSAVRSQILLMDPFPSIQRIYN-IVRQ----------EEKQQEINF---RPVPAVESA 272
RF + + ++ MDP PS+ IY+ ++R+ E+K++ + F R S
Sbjct: 211 SRFGGLCATLINMDPLPSLGEIYSRVIREEQRLASVHVREQKEEAVGFLARREQLDHHSR 270
Query: 273 ALQTSKAPYRTPGKRQ------RPFCEHCNRHGHTITTCYQIHGFP-------------- 312
+S T G R R C +C R GH C+QI GFP
Sbjct: 271 VDASSSRSEHTGGSRSNSIIKGRVTCSNCGRTGHEKKECWQIVGFPDWWSERNGGRGSNG 330
Query: 313 -------SNPSKPQKKTETSPSTSANQL----SSAQYHKLLTLLAKE-DNVGSSVN---- 356
SN + Q + + +TS+N + ++ ++L+ L KE N GS+ N
Sbjct: 331 RGRGGRGSNGGRGQGQVMAAHATSSNSSVFPEFTEEHMRVLSQLVKEKSNSGSTSNNNSD 390
Query: 357 -LAG-TAFTCIPFSWIIDSGASNHICTSLSLFSSYYPVNNQISVQQPDGSQALVKHIGTI 414
L+G T I I+DSGAS+H+ +LS ++ PV V DGS+A +G +
Sbjct: 391 RLSGKTKLGDI----ILDSGASHHMTGTLSSLTNVVPV-PPCPVGFADGSKAFALSVGVL 445
Query: 415 NCSPSLILTNVYHVPTFKFNLMSVTQLTESLNCDAIFSSSGCVFQDQATKKMIGRGSARN 474
S ++ LTNV VP+ L+SV++L + C A F+ + C QD+++K +IG G R
Sbjct: 446 TLSNTVSLTNVLFVPSLNCTLISVSKLLKQTQCLATFTDTLCFLQDRSSKTLIGSGEERG 505
Query: 475 ELYYLNQDLVSNKH----DKD----HYCLSHPLSSLFSS 505
+YYL + H D D H L HP S+ SS
Sbjct: 506 GVYYLTDVTPAKIHTANVDSDQALWHQRLGHPSFSVLSS 544
>At2g23330 putative retroelement pol polyprotein
Length = 1496
Score = 691 bits (1783), Expect = 0.0
Identities = 367/809 (45%), Positives = 500/809 (61%), Gaps = 60/809 (7%)
Query: 554 FGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAK-NMNIQHKFD 612
F GE +LTA +LIN+ P+ +LK K+P+++L G PSY LR FGCLC+A + KF
Sbjct: 683 FWGESILTATHLINRTPSAVLKGKTPYELLFGERPSYDMLRSFGCLCYAHIRPRNKDKFT 742
Query: 613 ERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDIQ---------SPPF 663
RS+ +F+GYP +K +R+YD++T +I+ SRDV+FHE ++PY +PP
Sbjct: 743 SRSRKCVFIGYPHGKKAWRVYDLETGKIFASRDVRFHEDIYPYATATQSNVPLPPPTPPM 802
Query: 664 NNA---ISINTQILDNEFDDLFTGSPTHPNI---PPENSHNDNSNDT---------IVTI 708
N + I+TQ+ D + SP PP + S T IV
Sbjct: 803 VNDDWFLPISTQVDSTNVDSSSSSSPAQSGSIDQPPRSIDQSPSTSTNPVPEEIGSIVPS 862
Query: 709 STPEDDASSNPPSFSA----ESLSNNPPSTEMNPPNHR---HSQRIRNPPIHLKDYICKI 761
S+P + SA E LS ST +P R + + LKD++
Sbjct: 863 SSPSRSIDRSTSDLSASDTTELLSTGESSTPSSPGLPELLGKGCREKKKSVLLKDFVTNT 922
Query: 762 ----------------------------NNVTSKINFPLENYLSLSNLSNSHRAFLINII 793
++V+ K +PL ++L+ S S +H AF+ I+
Sbjct: 923 TSKKKTASHNIHSPSQVLPSGLPTSLSADSVSGKTLYPLSDFLTNSGYSANHIAFMAAIL 982
Query: 794 ENKEPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLPEGKSAIGCKWIYKIKYHSDG 853
++ EPK + A+ EW +AM+KEI ALE+N+TW + LP GK AI KW+YK+KY+SDG
Sbjct: 983 DSNEPKHFKDAILIKEWCEAMSKEIDALEANHTWDITDLPHGKKAISSKWVYKLKYNSDG 1042
Query: 854 SIDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFLQ 913
+++R+KARLV G Q +G+D+ +TFAPVAKL TVR +L++AA K+W ++Q DV+NAFL
Sbjct: 1043 TLERHKARLVVMGNHQKEGVDFKETFAPVAKLTTVRTILAVAAAKDWEVHQMDVHNAFLH 1102
Query: 914 GDLSEEVYMKLPPGFSHKKKPCVCKLNKSIYGLKQASRQWFSKFSTTLIQKGFRQSISDY 973
GDL EEVYM+LPPGF VC+L KS+YGLKQA R WFSK ST L GF QS DY
Sbjct: 1103 GDLEEEVYMRLPPGFKCSDPSKVCRLRKSLYGLKQAPRCWFSKLSTALRNIGFTQSYEDY 1162
Query: 974 SLFTYNCDQTTIFVLVYVDDIIITGNNENAISKIKKFLAQSFSIKDLGNLSYILGIEVSR 1033
SLF+ T I VLVYVDD+I+ GNN +AI + K L + F +KDLG L Y LG+EVSR
Sbjct: 1163 SLFSLKNGDTIIHVLVYVDDLIVAGNNLDAIDRFKSQLHKCFHMKDLGKLKYFLGLEVSR 1222
Query: 1034 SKKGIFLCQRKYTLDILSDSGMTGCRPSDFPMEQHLRLRPNDGTPLSDPTVYRRHVGRLL 1093
G L QRKY LDI+ ++G+ GC+PS P+ + +L G ++P YRR VGR +
Sbjct: 1223 GPDGFCLSQRKYALDIVKETGLLGCKPSAVPIALNHKLASITGPVFTNPEQYRRLVGRFI 1282
Query: 1094 YLTVTRPDIQYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVGKGLFLSASSSINLVGYA 1153
YLT+TRPD+ YAV+ LSQFMQ+P +H++AA +++RYLKGS +G+FL + SS+ + Y
Sbjct: 1283 YLTITRPDLSYAVHILSQFMQAPLVAHWEAALRLVRYLKGSPAQGIFLRSDSSLIINAYC 1342
Query: 1154 DSDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEAEYRSLATLSSELQWLKY 1213
DSD+ CP TRRS + Y LG +PISWKTKKQ T+S SSAEAEYR++A EL+WLK
Sbjct: 1343 DSDYNACPLTRRSLSAYVVYLGDSPISWKTKKQDTVSYSSAEAEYRAMAYTLKELKWLKA 1402
Query: 1214 LLSDLGIDHPQPITIYCDSQAAIHIAENPVFHERTKHIEIDCHFVREKIKSGLIAPSYIR 1273
LL DLG+ H P+ ++CDS+AAIHIA NPVFHERTKHIE DCH VR+ + LI +I
Sbjct: 1403 LLKDLGVHHSSPMKLHCDSEAAIHIAANPVFHERTKHIESDCHKVRDAVLDKLITTEHIY 1462
Query: 1274 SSDQLADIFTKPLGGDAYKRILGKLGVIE 1302
+ DQ+AD+ TK L ++R+L LGV +
Sbjct: 1463 TEDQVADLLTKSLPRPTFERLLSTLGVTD 1491
Score = 239 bits (609), Expect = 1e-62
Identities = 162/527 (30%), Positives = 253/527 (47%), Gaps = 58/527 (11%)
Query: 37 LSSISSPITVHSTTNPDPC---VIHHSDNPSTVLVTPLLTGDNYGSWSRAVTMALRAKNK 93
+SS S+ +T S++N +I+ SDNP ++ + +L +NY WS + LRAK K
Sbjct: 1 MSSASALVTSTSSSNTSSTTAYLINASDNPGALISSVVLKENNYAEWSEELQNFLRAKQK 60
Query: 94 FGFVDGSLTIPKKKEDILTWQRCNDLVASWILNSVSTEIRPSILYAETAEQIWSDLQDRF 153
GF+DGS+ P ++ W N ++ WI S+ IR ++ + A Q+W +L+ RF
Sbjct: 61 LGFIDGSIPKPAADPELSLWIAINSMIVGWIRTSIDPTIRSTVGFVSEASQLWENLRRRF 120
Query: 154 SQSNAPKIYQLKQSISALKQEGMYVSLYFTQLKSLWDELNSIALVNPCICGNAKSILDQQ 213
S N + LK I+A Q+G V Y+ +L LW+EL + C C A I ++
Sbjct: 121 SVGNGVRKTLLKDEIAACTQDGQPVLAYYGRLIKLWEELQNYKSGRECKCEAASDIEKER 180
Query: 214 NQDRAMEFLQGVHDRFSAVRSQILLMDPFPSIQRIYNIVRQEEKQQEINFRPVPAVESAA 273
DR +FL G+ RFS++RS I ++P P + ++Y+ V +EE+ + R V++ A
Sbjct: 181 EDDRVHKFLLGLDSRFSSIRSSITDIEPLPDLYQVYSRVVREEQNLNAS-RTKDVVKTEA 239
Query: 274 LQTSKAPYRTPGKRQRP--FCEHCNRHGHTITTCYQIHGFP-----SNPSKPQKKT---- 322
+ S TP R + FC HCNR GH +T C+ +HG+P NP + Q T
Sbjct: 240 IGFSVQSSTTPRFRDKSTLFCTHCNRKGHEVTQCFLVHGYPDWWLEQNPQENQPSTRGRG 299
Query: 323 --------------ETSPST----SANQLSSA----------QYHKLLTLLAKEDNVGSS 354
++P+T AN +A Q +L++LL + SS
Sbjct: 300 SNGRGSSSGRGGNRSSAPTTRGRGRANNAQAAAPTVSGDGNDQIAQLISLLQAQRPSSSS 359
Query: 355 VNLAGTAFTCIPFSWIIDSGASNHICTSLSLFSSYYPVNNQISVQQPDGSQALVKHIGTI 414
L+G TC+ +ID+GAS+H+ S+ + + V +PDG + GT+
Sbjct: 360 ERLSGN--TCLT-DGVIDTGASHHMTGDCSILVDVFDITPS-PVTKPDGKASQATKCGTL 415
Query: 415 NCSPSLILTNVYHVPTFKFNLMSVTQLTESLNCDAIFSSSGCVFQDQATKKMIGRGSARN 474
S L +V VP F L+SV++L + + AIF+ + C QD+ + +IG G R
Sbjct: 416 LLHDSYKLHDVLFVPDFDCTLISVSKLLKQTSSIAIFTDTFCFLQDRFLRTLIGAGEERE 475
Query: 475 ELYYLNQDLVSNKH----------DKDHYCLSHPLSS-LFSSKHCNK 510
+YY L H D H L HP +S L S CN+
Sbjct: 476 GVYYFTGVLAPRVHKASSDFAISGDLWHRRLGHPSTSVLLSLPECNR 522
>At2g16000 putative retroelement pol polyprotein
Length = 1454
Score = 672 bits (1735), Expect = 0.0
Identities = 338/754 (44%), Positives = 495/754 (64%), Gaps = 51/754 (6%)
Query: 556 GECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNMNIQ-HKFDER 614
G+CVLTA +LIN+ P+ +L K+P+++L G+ P Y LR FGCLC++ Q HKF R
Sbjct: 741 GDCVLTAVFLINRTPSQLLMNKTPYEILTGTAPVYEQLRTFGCLCYSSTSPKQRHKFQPR 800
Query: 615 SKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDIQSPPFNNAISINTQIL 674
S+ +F+GYP KGY++ D+++ +++SR+VQFHE VFP ++P +++ + T ++
Sbjct: 801 SRACLFLGYPSGYKGYKLMDLESNTVFISRNVQFHEEVFPL--AKNPGSESSLKLFTPMV 858
Query: 675 DNEFDDLFTGSPTHPNIPPENSHNDNSNDTIVTISTPEDDASSNPPSFSAESLSNNPPST 734
P I + +H+ PS +S+ PP
Sbjct: 859 -----------PVSSGIISDTTHS---------------------PSSLPSQISDLPPQI 886
Query: 735 EMNPPNHRHSQRIRNPPIHLKDYICKINNVTSKINFPLENYLSLSNLSNSHRAFLINIIE 794
SQR+R PP HL DY C N + S +P+ + +S S +S SH ++ NI +
Sbjct: 887 S--------SQRVRKPPAHLNDYHC--NTMQSDHKYPISSTISYSKISPSHMCYINNITK 936
Query: 795 NKEPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLPEGKSAIGCKWIYKIKYHSDGS 854
P +Y++A + EW +A+ EI A+E NTW + LP+GK A+GCKW++ +K+ +DG+
Sbjct: 937 IPIPTNYAEAQDTKEWCEAVDAEIGAMEKTNTWEITTLPKGKKAVGCKWVFTLKFLADGN 996
Query: 855 IDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFLQG 914
++RYKARLVAKGY+Q +G+DY DTF+PVAK+ T++LLL ++A K W L Q DV+NAFL G
Sbjct: 997 LERYKARLVAKGYTQKEGLDYTDTFSPVAKMTTIKLLLKVSASKKWFLKQLDVSNAFLNG 1056
Query: 915 DLSEEVYMKLPPGFSHKK-----KPCVCKLNKSIYGLKQASRQWFSKFSTTLIQKGFRQS 969
+L EE++MK+P G++ +K V +L +SIYGLKQASRQWF KFS++L+ GF+++
Sbjct: 1057 ELEEEIFMKIPEGYAERKGIVLPSNVVLRLKRSIYGLKQASRQWFKKFSSSLLSLGFKKT 1116
Query: 970 ISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENAISKIKKFLAQSFSIKDLGNLSYILGI 1029
D++LF D + VLVYVDDI+I +E A +++ + L Q F ++DLG+L Y LG+
Sbjct: 1117 HGDHTLFLKMYDGEFVIVLVYVDDIVIASTSEAAAAQLTEELDQRFKLRDLGDLKYFLGL 1176
Query: 1030 EVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDFPMEQHLRLRPNDGTPLSDPTVYRRHV 1089
EV+R+ GI +CQRKY L++L +GM C+P PM +L++R +DG + D YRR V
Sbjct: 1177 EVARTTAGISICQRKYALELLQSTGMLACKPVSVPMIPNLKMRKDDGDLIEDIEQYRRIV 1236
Query: 1090 GRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVGKGLFLSASSSINL 1149
G+L+YLT+TRPDI +AVN L QF +P ++H AA +VL+Y+KG+VG+GLF SASS + L
Sbjct: 1237 GKLMYLTITRPDITFAVNKLCQFSSAPRTTHLTAAYRVLQYIKGTVGQGLFYSASSDLTL 1296
Query: 1150 VGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEAEYRSLATLSSELQ 1209
G+ADSDWA C +RRSTT + +G + ISW++KKQ T+SRSSAEAEYR+LA + E+
Sbjct: 1297 KGFADSDWASCQDSRRSTTSFTMFVGDSLISWRSKKQHTVSRSSAEAEYRALALATCEMV 1356
Query: 1210 WLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVFHERTKHIEIDCHFVREKIKSGLIAP 1269
WL LL L P PI +Y DS AAI+IA NPVFHERTKHI++DCH VRE++ +G +
Sbjct: 1357 WLFTLLVSLQASPPVPI-LYSDSTAAIYIATNPVFHERTKHIKLDCHTVRERLDNGELKL 1415
Query: 1270 SYIRSSDQLADIFTKPLGGDAYKRILGKLGVIEI 1303
++R+ DQ+ADI TKPL ++ + K+ ++ I
Sbjct: 1416 LHVRTEDQVADILTKPLFPYQFEHLKSKMSILNI 1449
Score = 255 bits (652), Expect = 1e-67
Identities = 159/511 (31%), Positives = 251/511 (49%), Gaps = 63/511 (12%)
Query: 30 PPSENSP---LSSISSPITVHSTTNP--DPCVIHHSDNPSTVLVTPLLTGDNYGSWSRAV 84
PP NSP S S +T + +P P +H +D+P +++ L NYG WS A+
Sbjct: 34 PP--NSPPVNRSGASRALTSSESGDPTQSPFFLHSADHPGLNIISHRLDETNYGDWSVAM 91
Query: 85 TMALRAKNKFGFVDGSLTIPKKKE-DILTWQRCNDLVASWILNSVSTEIRPSILYAETAE 143
++L AKNK GF+DG+L+ P + + + W RCN +V SW+LNSVS +I SIL A
Sbjct: 92 LISLDAKNKTGFIDGTLSRPLESDLNFRLWSRCNSMVKSWLLNSVSPQIYRSILRMNDAS 151
Query: 144 QIWSDLQDRFSQSNAPKIYQLKQSISALKQEGMYVSLYFTQLKSLWDELNSI-ALVNPCI 202
IW DL RF+ +N P+ Y L Q I +Q + +S Y+T+LK+LWD+L+S AL PC
Sbjct: 152 DIWRDLNSRFNVTNLPRTYNLTQEIQDFRQGTLSLSEYYTRLKTLWDQLDSTEALDEPCT 211
Query: 203 CGNAKSILDQQNQDRAMEFLQGVHDRFSAVRSQILLMDPFPSIQRIYNIVRQEEKQQEIN 262
CG A + + Q + ++FL G+++ ++ VR QI+ PS+ +Y+I+ Q+ QQ +
Sbjct: 212 CGKAMRLQQKAEQAKIVKFLAGLNESYAIVRRQIIAKKALPSLGEVYHILDQDNSQQSFS 271
Query: 263 FRPVPAVESAALQTSKAPYRTPG--------KRQRPFCEHCNRHGHTITTCYQIHGFP-- 312
P + +++P P + RP C NR GH CY+ HGFP
Sbjct: 272 NVVAPPAAFQVSEITQSPSMDPTVCYVQNGPNKGRPICSFYNRVGHIAERCYKKHGFPPG 331
Query: 313 --------SNPSKPQ---------KKTETSPSTSANQLSSAQYHKLLTLLAKE------- 348
KP+ + TS + LS Q + + + + +
Sbjct: 332 FTPKGKAGEKLQKPKPLAANVAESSEVNTSLESMVGNLSKEQLQQFIAMFSSQLQNTPPS 391
Query: 349 ----------DNVGSSVNLAGTAF---------TCIPFSWIIDSGASNHICTSLSLFSSY 389
DN+G + + +F T +W+IDSGA++H+ SLFSS
Sbjct: 392 TYATASTSQSDNLGICFSPSTYSFIGILTVARHTLSSATWVIDSGATHHVSHDRSLFSSL 451
Query: 390 YPVNNQISVQQPDGSQALVKHIGTINCSPSLILTNVYHVPTFKFNLMSVTQLTESLNCDA 449
+ +V P G + +GT+ + ++L NV +P F+ NL+S++ LT+ +
Sbjct: 452 -DTSVLSAVNLPTGPTVKISGVGTLKLNDDILLKNVLFIPEFRLNLISISSLTDDIGSRV 510
Query: 450 IFSSSGCVFQDQATKKMIGRGSARNELYYLN 480
IF + C QD +M+G+G LY L+
Sbjct: 511 IFDKNSCEIQDLIKGRMLGQGRRVANLYLLD 541
>At2g16670 putative retroelement pol polyprotein
Length = 1333
Score = 671 bits (1730), Expect = 0.0
Identities = 361/795 (45%), Positives = 489/795 (61%), Gaps = 61/795 (7%)
Query: 546 FKQTYLFYFGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNM 605
F+ F GECVLTAAYLIN+ P+ +L +P++ L P + LRVFG LC+A N
Sbjct: 565 FQANLPIQFWGECVLTAAYLINRTPSSVLNDSTPYERLHKKQPRFDHLRVFGSLCYAHNR 624
Query: 606 NI-QHKFDERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPY--------- 655
N KF ERS+ +FVGYP QKG+R++D++ + +VSRDV F E FP+
Sbjct: 625 NRGGDKFAERSRRCVFVGYPHGQKGWRLFDLEQNEFFVSRDVVFSELEFPFRISHEQNVI 684
Query: 656 ----QDIQSPPFNNAISINTQILDN--EFDDLFTGSPTHPNIPPENSHNDNSN--DTIV- 706
+ + +P + I + N + SP P+ S + S+ DT V
Sbjct: 685 EEEEEALWAPIVDGLIEEEVHLGQNAGPTPPICVSSPISPSATSSRSEHSTSSPLDTEVV 744
Query: 707 ---TISTPEDDASSNPPSFSAESLSNNPPSTEMN------PPNHRHSQRIRNPPIHLKDY 757
ST + S+P + LS P+T PP R S R + PP+ LKD+
Sbjct: 745 PTPATSTTSASSPSSPTNLQFLPLSRAKPTTAQAVAPPAVPPPRRQSTRNKAPPVTLKDF 804
Query: 758 ICK---INNVTSKIN---FPLENYLSLSNLSNSHRAFLINIIENKEPKSYSQAMKSVEWR 811
+ SK+N + L+ S SH ++
Sbjct: 805 VVNTTVCQESPSKLNSILYQLQKRDDTRRFSASHTTYV---------------------- 842
Query: 812 DAMAKEIQALESNNTWILCPLPEGKSAIGCKWIYKIKYHSDGSIDRYKARLVAKGYSQVQ 871
I A E N+TW + LP GK AIG +W+YK+K++SDGS++RYKARLVA G Q +
Sbjct: 843 -----AIDAQEENHTWTIEDLPPGKRAIGSQWVYKVKHNSDGSVERYKARLVALGNKQKE 897
Query: 872 GIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFLQGDLSEEVYMKLPPGFSHK 931
G DY +TFAPVAK+ TVRL L +A +NW ++Q DV+NAFL GDL EEVYMKLPPGF
Sbjct: 898 GEDYGETFAPVAKMATVRLFLDVAVKRNWEIHQMDVHNAFLHGDLREEVYMKLPPGFEAS 957
Query: 932 KKPCVCKLNKSIYGLKQASRQWFSKFSTTLIQKGFRQSISDYSLFTYNCDQTTIFVLVYV 991
VC+L K++YGLKQA R WF K +T L + GF+QS++DYSLFT I +L+YV
Sbjct: 958 HPNKVCRLRKALYGLKQAPRCWFEKLTTALKRYGFQQSLADYSLFTLVKGSVRIKILIYV 1017
Query: 992 DDIIITGNNENAISKIKKFLAQSFSIKDLGNLSYILGIEVSRSKKGIFLCQRKYTLDILS 1051
DD+IITGN++ A + K++LA F +KDLG L Y LGIEV+RS GI++CQRKY LDI+S
Sbjct: 1018 DDLIITGNSQRATQQFKEYLASCFHMKDLGPLKYFLGIEVARSTTGIYICQRKYALDIIS 1077
Query: 1052 DSGMTGCRPSDFPMEQHLRLRPNDGTPLSDPTVYRRHVGRLLYLTVTRPDIQYAVNTLSQ 1111
++G+ G +P++FP+EQ+ +L + L+DP YRR VGRL+YL VTR D+ ++V+ L++
Sbjct: 1078 ETGLLGVKPANFPLEQNHKLGLSTSPLLTDPQRYRRLVGRLIYLAVTRLDLAFSVHILAR 1137
Query: 1112 FMQSPYSSHFDAATQVLRYLKGSVGKGLFLSASSSINLVGYADSDWAGCPTTRRSTTGYF 1171
FMQ P H+ AA +V+RYLK G+G+FL S + G+ DSDWAG P +RRS TGYF
Sbjct: 1138 FMQEPREDHWAAALRVVRYLKADPGQGVFLRRSGDFQITGWCDSDWAGDPMSRRSVTGYF 1197
Query: 1172 TMLGSNPISWKTKKQPTISRSSAEAEYRSLATLSSELQWLKYLLSDLGIDHPQPITIYCD 1231
G +PISWKTKKQ T+S+SSAEAEYR+++ L+SEL WLK LL LG+ H QP+ + CD
Sbjct: 1198 VQFGDSPISWKTKKQDTVSKSSAEAEYRAMSFLASELLWLKQLLFSLGVSHVQPMIMCCD 1257
Query: 1232 SQAAIHIAENPVFHERTKHIEIDCHFVREKIKSGLIAPSYIRSSDQLADIFTKPLGGDAY 1291
S++AI+IA NPVFHERTKHIEID HFVR++ G+I P ++ ++ QLADIFTKPLG D +
Sbjct: 1258 SKSAIYIATNPVFHERTKHIEIDYHFVRDEFVKGVITPRHVGTTSQLADIFTKPLGRDCF 1317
Query: 1292 KRILGKLGVIEISIP 1306
KLG+ + P
Sbjct: 1318 SAFRIKLGIRNLYAP 1332
Score = 84.0 bits (206), Expect = 5e-16
Identities = 98/392 (25%), Positives = 163/392 (41%), Gaps = 79/392 (20%)
Query: 201 CICGNAKSILDQQNQDRAMEFLQGVHDR-FSAVRSQILLMDPFPSIQRIYNIVRQEEK-- 257
C+C + +D+ EFL G+ D F VRS ++ P ++ +YNIVRQEE
Sbjct: 80 CVCDLGALQEKDREEDKVHEFLSGLDDALFRTVRSSLVSRIPVQPLEEVYNIVRQEEDLL 139
Query: 258 ---------QQEINFRPVPAVESAALQTSKAPYRTPG--KRQRPFCEHCNRHGHTITTCY 306
Q+E+N + A Q Y+ G K + C+HCNR GH +CY
Sbjct: 140 RNGANVLDDQREVN--------AFAAQMRPKLYQGRGDEKDKSMVCKHCNRSGHASESCY 191
Query: 307 QIHGFP----SNPSKPQKKTETSPSTSAN---QLSSAQYHKLLTL------------LAK 347
+ G+P P +T T+++ +A Y +T+ L
Sbjct: 192 AVIGYPEWWGDRPRSRSLQTRGRGGTNSSGGRGRGAAAYANRVTVPNHDTYEQANYALTD 251
Query: 348 EDNVGSSVNLAGTAFTCIPFSWIIDSG--ASNHICTSLSLFSSYYPVNNQISVQQPDGSQ 405
ED G + L + + I I++SG A+ T + + I + DG +
Sbjct: 252 EDRDGVN-GLTDSQWRTI--KSILNSGKDAATEKLTDM----------DPILIVLADGRE 298
Query: 406 ALVKHIGTINCSPSLILTNVYHVPTFKFNLMSVTQLTESLNCDAIFSSSGCVFQDQATKK 465
+ GT+ +L++ +V++V F+ +L+S+ QL + C S V QD+ ++
Sbjct: 299 RISVKEGTVRLGSNLVMISVFYVEEFQSDLISIGQLMDENRCVLQMSDRFLVVQDRTSRM 358
Query: 466 MIGRGSARNELYYLNQD----LVSNKHDKD----HYCLSHPLSSL----------FSSKH 507
++G G ++ V+ K +K+ H + HP + + SS H
Sbjct: 359 VMGAGRRVGGTFHFRSTEIAASVTVKEEKNYELWHSRMGHPAARVVSLIPESSVSVSSTH 418
Query: 508 CNK-FDLWHLRLVALQLRNKMEL-WNENIDIF 537
NK D+ H A Q RN L N+ + IF
Sbjct: 419 LNKACDVCHR---AKQTRNSFPLSINKTLRIF 447
Score = 35.0 bits (79), Expect = 0.28
Identities = 20/63 (31%), Positives = 32/63 (50%), Gaps = 14/63 (22%)
Query: 40 ISSPITVHSTTN----PDPCV----------IHHSDNPSTVLVTPLLTGDNYGSWSRAVT 85
+ +P V++ TN P+P V + ++NP V+ PLLTG NY W+ ++
Sbjct: 2 VGAPANVNTNTNTNPIPNPTVEVRRTILPYDLTSANNPCAVISHPLLTGSNYDEWACSMK 61
Query: 86 MAL 88
AL
Sbjct: 62 TAL 64
>At2g20460 putative retroelement pol polyprotein
Length = 1461
Score = 668 bits (1723), Expect = 0.0
Identities = 343/766 (44%), Positives = 483/766 (62%), Gaps = 55/766 (7%)
Query: 546 FKQTYLFYFGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNM 605
F+ + G+CVLTA +LIN+ P+ +L K+P +VL G P YS L+ FGCLC++
Sbjct: 742 FQSNMSLPYWGDCVLTAVFLINRTPSALLSNKTPFEVLTGKLPDYSQLKTFGCLCYSSTS 801
Query: 606 NIQ-HKFDERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDIQSPPFN 664
+ Q HKF RS+ +F+GYPF KGY++ D+++ +++SR+V+FHE +FP Q
Sbjct: 802 SKQRHKFLPRSRACVFLGYPFGFKGYKLLDLESNVVHISRNVEFHEELFPLASSQQ---- 857
Query: 665 NAISINTQILDNEFDDLFTGSPTHPNIPPENSHNDNSNDTIVTISTPEDDASSNPPSFSA 724
S T + TP D P S
Sbjct: 858 ------------------------------------SATTASDVFTPMD------PLSSG 875
Query: 725 ESLSNNPPSTEMNPPNHRHSQRIRNPPIHLKDYICKINNVTSKINFPLENYLSLSNLSNS 784
S++++ PS +++P +RI P HL+DY C N + P+ + LS S +S S
Sbjct: 876 NSITSHLPSPQISPSTQISKRRITKFPAHLQDYHCYFVNKDD--SHPISSSLSYSQISPS 933
Query: 785 HRAFLINIIENKEPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLPEGKSAIGCKWI 844
H ++ NI + P+SY +A S EW A+ +EI A+E +TW + LP GK A+GCKW+
Sbjct: 934 HMLYINNISKIPIPQSYHEAKDSKEWCGAIDQEIGAMERTDTWEITSLPPGKKAVGCKWV 993
Query: 845 YKIKYHSDGSIDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQ 904
+ +K+H+DGS++R+KAR+VAKGY+Q +G+DY +TF+PVAK+ TV+LLL ++A K W L Q
Sbjct: 994 FTVKFHADGSLERFKARIVAKGYTQKEGLDYTETFSPVAKMATVKLLLKVSASKKWYLNQ 1053
Query: 905 FDVNNAFLQGDLSEEVYMKLPPGFSHKKKPC-----VCKLNKSIYGLKQASRQWFSKFST 959
D++NAFL GDL E +YMKLP G++ K VC+L KSIYGLKQASRQWF KFS
Sbjct: 1054 LDISNAFLNGDLEETIYMKLPDGYADIKGTSLPPNVVCRLKKSIYGLKQASRQWFLKFSN 1113
Query: 960 TLIQKGFRQSISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENAISKIKKFLAQSFSIKD 1019
+L+ GF + D++LF I +LVYVDDI+I E A + + L SF +++
Sbjct: 1114 SLLALGFEKQHGDHTLFVRCIGSEFIVLLVYVDDIVIASTTEQAAQSLTEALKASFKLRE 1173
Query: 1020 LGNLSYILGIEVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDFPMEQHLRLRPNDGTPL 1079
LG L Y LG+EV+R+ +GI L QRKY L++L+ + M C+PS PM ++RL NDG L
Sbjct: 1174 LGPLKYFLGLEVARTSEGISLSQRKYALELLTSADMLDCKPSSIPMTPNIRLSKNDGLLL 1233
Query: 1080 SDPTVYRRHVGRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVGKGL 1139
D +YRR VG+L+YLT+TRPDI +AVN L QF +P ++H A +VL+Y+KG+VG+GL
Sbjct: 1234 EDKEMYRRLVGKLMYLTITRPDITFAVNKLCQFSSAPRTAHLAAVYKVLQYIKGTVGQGL 1293
Query: 1140 FLSASSSINLVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEAEYR 1199
F SA + L GY D+DW CP +RRSTTG+ +GS+ ISW++KKQPT+SRSSAEAEYR
Sbjct: 1294 FYSAEDDLTLKGYTDADWGTCPDSRRSTTGFTMFVGSSLISWRSKKQPTVSRSSAEAEYR 1353
Query: 1200 SLATLSSELQWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVFHERTKHIEIDCHFVR 1259
+LA S E+ WL LL L + PI +Y DS AA++IA NPVFHERTKHIEIDCH VR
Sbjct: 1354 ALALASCEMAWLSTLLLALRVHSGVPI-LYSDSTAAVYIATNPVFHERTKHIEIDCHTVR 1412
Query: 1260 EKIKSGLIAPSYIRSSDQLADIFTKPLGGDAYKRILGKLGVIEISI 1305
EK+ +G + ++++ DQ+ADI TKPL + +L K+ + I +
Sbjct: 1413 EKLDNGQLKLLHVKTKDQVADILTKPLFPYQFAHLLSKMSIQNIFV 1458
Score = 254 bits (648), Expect = 3e-67
Identities = 166/518 (32%), Positives = 242/518 (46%), Gaps = 65/518 (12%)
Query: 27 SDIPPSENSPLSSISSPITVHSTTNP--DPCVIHHSDNPSTVLVTPLLTGDNYGSWSRAV 84
SD PP S + S + +P P +H +D+P +++ L YG WS A+
Sbjct: 36 SDSPPMARSQTAGASRGVFSSGFDDPTQSPFFLHSADHPGLSIISHRLDETTYGDWSVAM 95
Query: 85 TMALRAKNKFGFVDGSLTIPKKKE-DILTWQRCNDLVASWILNSVSTEIRPSILYAETAE 143
++L AKNK GFVDGSL P + + + W RCN +V SW+LNSVS +I SIL A
Sbjct: 96 RISLDAKNKLGFVDGSLPRPLESDPNFRLWSRCNSMVKSWLLNSVSPQIYRSILRLNDAT 155
Query: 144 QIWSDLQDRFSQSNAPKIYQLKQSISALKQEGMYVSLYFTQLKSLWDELNSI-ALVNPCI 202
IW DL DRF+ +N P+ Y L Q I L+Q M +S Y+T LK+LWD+L+S AL +PC
Sbjct: 156 DIWRDLFDRFNLTNLPRTYNLTQEIQDLRQGTMSLSEYYTLLKTLWDQLDSTEALDDPCT 215
Query: 203 CGNAKSILDQQNQDRAMEFLQGVHDRFSAVRSQILLMDPFPSIQRIYNIVRQEEKQQEIN 262
CG A + + + + M+FL G+++ ++ VR QI+ PS+ +Y+I+ Q+ Q+
Sbjct: 216 CGKAVRLYQKAEKAKIMKFLAGLNESYAIVRRQIIAKKALPSLAEVYHILDQDNSQKGFF 275
Query: 263 FRPVPAVESAALQTSKAPYRTP--------GKRQRPFCEHCNRHGHTITTCYQIHGFP-- 312
P + S +P +P + RP C CNR GH CY+ HGFP
Sbjct: 276 NVVAPPAAFQVSEVSHSPITSPEIMYVQSGPNKGRPTCSFCNRVGHIAERCYKKHGFPPG 335
Query: 313 --------SNPSKPQ---KKTETSP-------STSANQLSSAQYHKLLTLLAK------- 347
P KPQ + SP T A S Q L+ L +
Sbjct: 336 FTPKGKSSDKPPKPQAVAAQVTLSPDKMTGQLETLAGNFSPDQIQNLIALFSSQLQPQIV 395
Query: 348 ---------EDNVGSSVNLAGTAFTCIPF----------------SWIIDSGASNHICTS 382
E + SV +G F+ + +W+IDSGA++H+
Sbjct: 396 SPQTASSQHEASSSQSVAPSGILFSPSTYCFIGILAVSHNSLSSDTWVIDSGATHHVSHD 455
Query: 383 LSLFSSYYPVNNQISVQQPDGSQALVKHIGTINCSPSLILTNVYHVPTFKFNLMSVTQLT 442
LF + + V P G + +GT+ + +IL NV +P F+ NL+S++ LT
Sbjct: 456 RKLFQT-LDTSIVSFVNLPTGPNVRISGVGTVLINKDIILQNVLFIPEFRLNLISISSLT 514
Query: 443 ESLNCDAIFSSSGCVFQDQATKKMIGRGSARNELYYLN 480
L IF S C QD +G G LY L+
Sbjct: 515 TDLGTRVIFDPSCCQIQDLTKGLTLGEGKRIGNLYVLD 552
>At1g70010 hypothetical protein
Length = 1315
Score = 655 bits (1690), Expect = 0.0
Identities = 338/758 (44%), Positives = 481/758 (62%), Gaps = 38/758 (5%)
Query: 554 FGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNM-NIQHKFD 612
+ G+C+LTA YLIN+LP PIL+ K P +VL + P+Y ++VFGCLC+A +HKF
Sbjct: 588 YWGDCILTAVYLINRLPAPILEDKCPFEVLTKTVPTYDHIKVFGCLCYASTSPKDRHKFS 647
Query: 613 ERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDIQSPPFNNAISINTQ 672
R+K F+GYP KGY++ D++T I VSR V FHE +FP+ S +Q
Sbjct: 648 PRAKACAFIGYPSGFKGYKLLDLETHSIIVSRHVVFHEELFPFLG----------SDLSQ 697
Query: 673 ILDNEFDDLFTGSPTHPNIPPENSHNDNSNDTIVTISTPEDDASSNPPSFSAESLSNNPP 732
N F DL +P P + +D+ N P D +SS E L + P
Sbjct: 698 EEQNFFPDL------NPTPPMQRQSSDHVN--------PSDSSSS------VEILPSANP 737
Query: 733 STEMNPPNHRHSQRIRNPPIHLKDYICKINNVTSKINFPLENYLSLSNLSNSHRAFLINI 792
+ + P+ + S R P +L+DY C ++V S + +LS +++ + FL +
Sbjct: 738 TNNVPEPSVQTSHRKAKKPAYLQDYYC--HSVVSSTPHEIRKFLSYDRINDPYLTFLACL 795
Query: 793 IENKEPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLPEGKSAIGCKWIYKIKYHSD 852
+ KEP +Y++A K WRDAM E LE +TW +C LP K IGC+WI+KIKY+SD
Sbjct: 796 DKTKEPSNYTEAEKLQVWRDAMGAEFDFLEGTHTWEVCSLPADKRCIGCRWIFKIKYNSD 855
Query: 853 GSIDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFL 912
GS++RYKARLVA+GY+Q +GIDY++TF+PVAKL +V+LLL +AA L Q D++NAFL
Sbjct: 856 GSVERYKARLVAQGYTQKEGIDYNETFSPVAKLNSVKLLLGVAARFKLSLTQLDISNAFL 915
Query: 913 QGDLSEEVYMKLPPGFSHKKKP-----CVCKLNKSIYGLKQASRQWFSKFSTTLIQKGFR 967
GDL EE+YM+LP G++ ++ VC+L KS+YGLKQASRQW+ KFS+TL+ GF
Sbjct: 916 NGDLDEEIYMRLPQGYASRQGDSLPPNAVCRLKKSLYGLKQASRQWYLKFSSTLLGLGFI 975
Query: 968 QSISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENAISKIKKFLAQSFSIKDLGNLSYIL 1027
QS D++ F D + VLVY+DDIII NN+ A+ +K + F ++DLG L Y L
Sbjct: 976 QSYCDHTCFLKISDGIFLCVLVYIDDIIIASNNDAAVDILKSQMKSFFKLRDLGELKYFL 1035
Query: 1028 GIEVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDFPMEQHLRLRPNDGTPLSDPTVYRR 1087
G+E+ RS KGI + QRKY LD+L ++G GC+PS PM+ + + G + YRR
Sbjct: 1036 GLEIVRSDKGIHISQRKYALDLLDETGQLGCKPSSIPMDPSMVFAHDSGGDFVEVGPYRR 1095
Query: 1088 HVGRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVGKGLFLSASSSI 1147
+GRL+YL +TRPDI +AVN L+QF +P +H A ++L+Y+KG++G+GLF SA+S +
Sbjct: 1096 LIGRLMYLNITRPDITFAVNKLAQFSMAPRKAHLQAVYKILQYIKGTIGQGLFYSATSEL 1155
Query: 1148 NLVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEAEYRSLATLSSE 1207
L YA++D+ C +RRST+GY LG + I WK++KQ +S+SSAEAEYRSL+ + E
Sbjct: 1156 QLKVYANADYNSCRDSRRSTSGYCMFLGDSLICWKSRKQDVVSKSSAEAEYRSLSVATDE 1215
Query: 1208 LQWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVFHERTKHIEIDCHFVREKIKSGLI 1267
L WL L +L + +P ++CD++AAIHIA N VFHERTKHIE DCH VRE++ GL
Sbjct: 1216 LVWLTNFLKELQVPLSKPTLLFCDNEAAIHIANNHVFHERTKHIESDCHSVRERLLKGLF 1275
Query: 1268 APSYIRSSDQLADIFTKPLGGDAYKRILGKLGVIEISI 1305
+I + Q+AD FTKPL + R++ K+G++ I +
Sbjct: 1276 ELYHINTELQIADPFTKPLYPSHFHRLISKMGLLNIFV 1313
Score = 239 bits (610), Expect = 8e-63
Identities = 150/446 (33%), Positives = 231/446 (51%), Gaps = 46/446 (10%)
Query: 84 VTMALRAKNKFGFVDGSLTIPKKKED---ILTWQRCNDLVASWILNSVSTEIRPSILYAE 140
+T ++ AKNK GFVDGS IPK +D W+RCN +V SW+LNSVS EI SILY
Sbjct: 1 MTTSIEAKNKLGFVDGS--IPKPDDDDPYCKIWRRCNSMVKSWLLNSVSKEIYTSILYFP 58
Query: 141 TAEQIWSDLQDRFSQSNAPKIYQLKQSISALKQEGMYVSLYFTQLKSLWDELNSIALVNP 200
TA IW DL RF +S+ P++Y+L+Q I +L+Q + +S Y T+ ++LW+EL S+ V
Sbjct: 59 TAAAIWKDLYTRFHKSSLPRLYKLRQQIHSLRQGNLDLSSYHTRTQTLWEELTSLQAVPR 118
Query: 201 CICGNAKSILDQQNQDRAMEFLQGVHDRFSAVRSQILLMDPFPSIQRIYNIVRQEEKQQE 260
+ + +L ++ +R ++FL G++D + VRSQIL+ PS+ ++N++ Q+E Q+
Sbjct: 119 TV----EDLLIERETNRVIDFLMGLNDCYDTVRSQILMKKTLPSLSEVFNMIDQDETQRS 174
Query: 261 INFRPVPAVESAALQTSKAPYR------TPGKRQRPFCEHCNRHGHTITTCYQIHGFPSN 314
P + S+ S + T K++RP C +C+R GH TCY+ HG+P++
Sbjct: 175 ARISTTPGMTSSVFPVSNQSSQSALNGDTYQKKERPVCSYCSRPGHVEDTCYKKHGYPTS 234
Query: 315 PSKPQKKTETSPSTSANQLSSAQYHKLLTLLAKEDNVGSSVNLAGTAFTCIPFSWIIDSG 374
QK + PS SAN + + V ++ +++ T ++
Sbjct: 235 FKSKQKFVK--PSISANAAIGS------------EEVVNNTSVSTGDLTTSQIQQLVSFL 280
Query: 375 ASNHICTSLSLFSSYYPVNNQISV-QQPDGSQALVKHIGTINCSPSLILTNVYHVPTFKF 433
+S S + P + ISV P S + G+++ LIL +V +P FKF
Sbjct: 281 SSKLQPPS----TPVQPEVHSISVSSDPSSSSTVCPISGSVHLGRHLILNDVLFIPQFKF 336
Query: 434 NLMSVTQLTESLNCDAIFSSSGCVFQDQATKKMIGRGSARNELYYLNQDLVSNKHDKDHY 493
NL+SV+ LT+S+ C F + CV QD + M+G G LY ++ D
Sbjct: 337 NLLSVSSLTKSMGCRIWFDETSCVLQDATRELMVGMGKQVANLYIVDLD----------- 385
Query: 494 CLSHP-LSSLFSSKHCNKFDLWHLRL 518
LSHP S + DLWH RL
Sbjct: 386 SLSHPGTDSSITVASVTSHDLWHKRL 411
>At4g10990 putative retrotransposon polyprotein
Length = 1203
Score = 641 bits (1654), Expect = 0.0
Identities = 371/959 (38%), Positives = 549/959 (56%), Gaps = 78/959 (8%)
Query: 368 SWIIDSGASNHICTSLSLFSSYYPVNNQISVQQPDGSQALVKHIGTINCSPSLILTNVYH 427
+WIIDSGAS+H+C+ L++F V+ ++V P+G++ + H GTI + +LIL NV
Sbjct: 98 AWIIDSGASSHVCSDLTMFRELIHVSG-VTVTLPNGTRVAITHTGTICITSTLILHNVLL 156
Query: 428 VPTFKFNLMSVTQLTESLNCDAIFSSSGCVFQDQATKKMIGRGSARNELYYLNQDLVSNK 487
VP FKFNL+SV L ++L+ A F + C Q+ MIGRG N LY L S
Sbjct: 157 VPDFKFNLISVCCLVKTLSYSAHFFADCCYIQELTRGLMIGRGKTYNNLYILETQRTS-- 214
Query: 488 HDKDHYCLSHPLSSLFSSKHCNKFDLWHLRLVALQLRNKMELWNENIDIFLMSQELYAFK 547
+ S P +S F+ + LWH RL +R+ + N ++ ++ +
Sbjct: 215 -----FSPSLPAASSFTGTVQDDCLLWHQRL---GIRHYLHYRN----LYFLTLVDDCTR 262
Query: 548 QTYLFYFGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNMNI 607
T+++ + ++ N P + + + + + S +++ F K +
Sbjct: 263 TTWVYMMKNKSEVS-----NIFPVFVKLIFTQYNAKIKAIRS-DNVKELAFTKFVKEQGM 316
Query: 608 QHKFDERSKP---GIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDIQSPPFN 664
H+F P + YP KGY++ D+++ I ++R+V FHET FP++
Sbjct: 317 IHQFSCAYTPQQNSVVERYPSGYKGYKVLDLESHSISITRNVVFHETKFPFK-------- 368
Query: 665 NAISINTQILDNEFDDLFTGS------PTH--PNIPPENSHNDNSNDTIVTISTPEDDAS 716
T E D+F S P H ++P ++ + N+ + S AS
Sbjct: 369 ------TSKFLKESVDMFPNSILPLPAPLHFVESMPLDDDLRADDNNA--STSNSASSAS 420
Query: 717 SNPPSFSAESLSNNPP-STEMNPPNHRHSQRIRNPPIHLKDYICKINNVTSKIN------ 769
S PP S + N + N +R P +L +Y C S ++
Sbjct: 421 SIPPLPSTVNTQNTDALDIDTNSVPIARPKRNAKAPAYLSEYHCNSVPFLSSLSPTTSTS 480
Query: 770 ----------------FPLENYLSLSNLSNSHRAFLINIIENKEPKSYSQAMKSVEWRDA 813
+P+ +S L+ +++ EPK+++QAMKS +W A
Sbjct: 481 IETPSSSIPPKKITTPYPMSTAISYDKLTPLFHSYICAYNVETEPKAFTQAMKSEKWTRA 540
Query: 814 MAKEIQALESNNTWILCPLPEGKSAIGCKWIYKIKYHSDGSIDRYKARLVAKGYSQVQGI 873
+E+ ALE N TWI+ L EGK+ +GCKW++ IKY+ DGSI+RYKARLVA+G++Q +GI
Sbjct: 541 ANEELHALEQNKTWIVESLTEGKNVVGCKWVFTIKYNPDGSIERYKARLVAQGFTQQEGI 600
Query: 874 DYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFLQGDLSEEVYMKLPPGF----- 928
DY +TF+PVAK +V+LLL +AA W L Q DV+NAFL G+L EE+YM LP G+
Sbjct: 601 DYMETFSPVAKFGSVKLLLGLAAATGWSLTQMDVSNAFLHGELDEEIYMSLPQGYTPPTG 660
Query: 929 -SHKKKPCVCKLNKSIYGLKQASRQWFSKFSTTLIQKGFRQSISDYSLFTYNCDQTTIFV 987
S KP VC+L KS+YGLKQASRQW+ + S+ + F QS +D ++F + I V
Sbjct: 661 ISLPSKP-VCRLLKSLYGLKQASRQWYKRLSSVFLGANFIQSPADNTMFVKVSCTSIIVV 719
Query: 988 LVYVDDIIITGNNENAISKIKKFLAQSFSIKDLGNLSYILGIEVSRSKKGIFLCQRKYTL 1047
LVYVDD++I N+ +A+ +K+ L F IKDLG + LG+E++RS +GI +CQRKY
Sbjct: 720 LVYVDDLMIASNDSSAVENLKELLRSEFKIKDLGPARFFLGLEIARSSEGISVCQRKYAQ 779
Query: 1048 DILSDSGMTGCRPSDFPMEQHLRLRPNDGTPLSDPTVYRRHVGRLLYLTVTRPDIQYAVN 1107
++L D G++GC+PS PM+ +L L GT L + T YR VGRLLYL +TRPDI +AV+
Sbjct: 780 NLLEDVGLSGCKPSSIPMDPNLHLTKEMGTLLPNATSYRELVGRLLYLCITRPDITFAVH 839
Query: 1108 TLSQFMQSPYSSHFDAATQVLRYLKGSVGKGLFLSASSSINLVGYADSDWAGCPTTRRST 1167
TLSQF+ +P H AA +VLRYLKG+ G+GL SASS + L G++D+DW C +RRS
Sbjct: 840 TLSQFLSAPTDIHMQAAHKVLRYLKGNPGQGLMYSASSELCLNGFSDADWGTCKDSRRSV 899
Query: 1168 TGYFTMLGSNPISWKTKKQPTISRSSAEAEYRSLATLSSELQWLKYLLSDLGIDHPQPIT 1227
TG+ LG++ I+WK+KKQ +SRSS E+EYRSLA + E+ WL+ LL DL + P
Sbjct: 900 TGFCIYLGTSLITWKSKKQSVVSRSSTESEYRSLAQATCEIIWLQQLLKDLHVTMTCPAK 959
Query: 1228 IYCDSQAAIHIAENPVFHERTKHIEIDCHFVREKIKSGLIAPSYIRSSDQLADIFTKPL 1286
++CD+++A+H+A NPVFHERTKHIEIDCH VR++IK+G + ++ + +QLADI TKPL
Sbjct: 960 LFCDNKSALHLATNPVFHERTKHIEIDCHTVRDQIKAGKLKTLHVPTGNQLADILTKPL 1018
>At4g17450 retrotransposon like protein
Length = 1433
Score = 634 bits (1636), Expect = 0.0
Identities = 328/772 (42%), Positives = 473/772 (60%), Gaps = 63/772 (8%)
Query: 546 FKQTYLFYFGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNM 605
F+ F G+CVLTA +LIN+LPTP+L KSP++ L PP+Y SL+ FGCLC++
Sbjct: 717 FQSNIPLEFWGDCVLTAVFLINRLPTPVLNNKSPYEKLKNIPPAYESLKTFGCLCYSSTS 776
Query: 606 NIQ-HKFDERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDIQSPPFN 664
Q HKF+ R++ +F+GYP KGY++ D++T + +SR V FHE +FP+
Sbjct: 777 PKQRHKFEPRARACVFLGYPLGYKGYKLLDIETHAVSISRHVIFHEDIFPF--------- 827
Query: 665 NAISINTQILDN--EFDDLFTGSPTHPNIPPENSHNDNSNDTIVTISTPEDDASSNPPSF 722
I++ I D+ +F L ++P E T + + P D SS+
Sbjct: 828 ----ISSTIKDDIKDFFPLLQFPARTDDLPLEQ--------TSIIDTHPHQDVSSSKALV 875
Query: 723 SAESLSNNPPSTEMNPPNHRHSQRIRNPPIHLKDYICKINNVTSKINFPLENYLSLSNLS 782
+ LS +R + PP HL+D+ C NN T +
Sbjct: 876 PFDPLS----------------KRQKKPPKHLQDFHC-YNNTTEPFH------------- 905
Query: 783 NSHRAFLINIIENKEPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLPEGKSAIGCK 842
AF+ NI P+ YS+A W DAM +EI A+ NTW + LP K AIGCK
Sbjct: 906 ----AFINNITNAVIPQRYSEAKDFKAWCDAMKEEIGAMVRTNTWSVVSLPPNKKAIGCK 961
Query: 843 WIYKIKYHSDGSIDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPL 902
W++ IK+++DGSI+RYKARLVAKGY+Q +G+DY +TF+PVAKL +VR++L +AA W +
Sbjct: 962 WVFTIKHNADGSIERYKARLVAKGYTQEEGLDYEETFSPVAKLTSVRMMLLLAAKMKWSV 1021
Query: 903 YQFDVNNAFLQGDLSEEVYMKLPPGFSHK-----KKPCVCKLNKSIYGLKQASRQWFSKF 957
+Q D++NAFL GDL EE+YMK+PPG++ +C+L+KSIYGLKQASRQW+ K
Sbjct: 1022 HQLDISNAFLNGDLDEEIYMKIPPGYADLVGEALPPHAICRLHKSIYGLKQASRQWYLKL 1081
Query: 958 STTLIQKGFRQSISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENAISKIKKFLAQSFSI 1017
S TL GF++S +D++LF + + VLVYVDDI+I N+++A+++ L F +
Sbjct: 1082 SNTLKGMGFQKSNADHTLFIKYANGVLMGVLVYVDDIMIVSNSDDAVAQFTAELKSYFKL 1141
Query: 1018 KDLGNLSYILGIEVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDFPMEQHLRLRPNDGT 1077
+DLG Y LGIE++RS+KGI +CQRKY L++LS +G G +PS P++ ++L DG
Sbjct: 1142 RDLGAAKYFLGIEIARSEKGISICQRKYILELLSTTGFLGSKPSSIPLDPSVKLNKEDGV 1201
Query: 1078 PLSDPTVYRRHVGRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVGK 1137
PL+D T YR+ VG+L+YL +TRPDI YAVNTL QF +P S H A +VLRYLKG+VG+
Sbjct: 1202 PLTDSTSYRKLVGKLMYLQITRPDIAYAVNTLCQFSHAPTSVHLSAVHKVLRYLKGTVGQ 1261
Query: 1138 GLFLSASSSINLVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEAE 1197
GLF SA +L GY DSD+ C +RR Y +G +SWK+KKQ T+S S+AEAE
Sbjct: 1262 GLFYSADDKFDLRGYTDSDFGSCTDSRRCVAAYCMFIGDYLVSWKSKKQDTVSMSTAEAE 1321
Query: 1198 YRSLATLSSELQWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVFHERTKHIEIDCHF 1257
+R+++ + E+ WL L D + P +YCD+ AA+HI N VFHERTK +E+DC+
Sbjct: 1322 FRAMSQGTKEMIWLSRLFDDFKVPFIPPAYLYCDNTAALHIVNNSVFHERTKFVELDCYK 1381
Query: 1258 VREKIKSGLIAPSYIRSSDQLADIFTKPLGGDAYKRILGKLGVIEISIPPPT 1309
RE ++SG + ++ + +Q+AD TK + + +++GK+GV I P P+
Sbjct: 1382 TREAVESGFLKTMFVETGEQVADPLTKAIHPAQFHKLIGKMGVCNIFAPLPS 1433
Score = 232 bits (592), Expect = 9e-61
Identities = 156/516 (30%), Positives = 245/516 (47%), Gaps = 110/516 (21%)
Query: 38 SSISSPITVHSTTNPDPCVIHHSDNPSTVLVTPLLTGDNYGSWSRAVTMALRAKNKFGFV 97
+SI S HS P +H SD+P +V+ +L G NY +WS A+ M+L AKNK FV
Sbjct: 56 ASIESYDNAHS-----PYFLHSSDHPGLNIVSHILDGTNYNNWSIAMRMSLDAKNKLSFV 110
Query: 98 DGSLTIPKKKEDILT-WQRCNDLVASWILNSVSTEIRPSILYAETAEQIWSDLQDRFSQS 156
DGSL P + + W RCN +V +W+LN V+ ++W+DL RF S
Sbjct: 111 DGSLPRPDVSDRMFKIWSRCNSMVKTWLLNVVT--------------EMWNDLFSRFRVS 156
Query: 157 NAPKIYQLKQSISALKQEGMYVSLYFTQLKSLWDELNS--IALVNPCICGNAKSILDQQN 214
N P+ YQL+QSI LKQ + +S Y+T+ K+LW++L + + V C C + K +L++
Sbjct: 157 NLPRKYQLEQSIHTLKQGNLDLSTYYTKKKTLWEQLANTRVLTVRKCNCEHVKELLEEAE 216
Query: 215 QDRAMEFLQGVHDRFSAVRSQILLMDPFPSIQRIYNIVRQEEKQQEINFRPVPAVESAAL 274
R ++FL G++D F+ +R QIL M P P + IYN++ Q+E Q+ + P + +AA
Sbjct: 217 TSRIIQFLMGLNDNFAHIRGQILNMKPRPGLTEIYNMLDQDESQRLVG-NPTLSNPTAAF 275
Query: 275 QTSKAPY------RTPGKRQRPFCEHCNRHGHTITTCYQIHGFP---------------- 312
Q +P G ++P C +CN+ GH + CY+ HG+P
Sbjct: 276 QVQASPIIDSQVNMAQGSYKKPKCSYCNKLGHLVDKCYKKHGYPPGSKWTKGQTIGSTNL 335
Query: 313 -SNPSKPQKKTETSPSTSANQLSSAQYHKLLTLLAKEDNVGSSVNLAGTAFTCI------ 365
S +P +T + S + S+ Q +++ L+ + ++ S+ + T+ I
Sbjct: 336 ASTQLQPVNETPNEKTDSYEEFSTDQIQTMISYLSTKLHIASASPMPTTSSASISASPSV 395
Query: 366 ---------------------------------PFSWIIDSGASNHICTSLSLFSSYYPV 392
P W+IDSGA++H+ + L+ ++ +
Sbjct: 396 PMISQISGTFLSLFSNAYYDMLISSVSQEPAVSPRGWVIDSGATHHVTHNRDLYLNFRSL 455
Query: 393 NNQISVQQPDGSQALVKHIGTINCSPSLILTNVYHVPTFKFNLMSVTQLTESLNCDAIFS 452
N V+ P+ + IG I S ++ L NV ++P FKFNL+S +LT+ L
Sbjct: 456 ENTF-VRLPNDCTVKIAGIGFIQLSDAISLHNVLYIPEFKFNLIS--ELTKEL------- 505
Query: 453 SSGCVFQDQATKKMIGRGSARNELYYLNQDLVSNKH 488
MIGRGS LY L D N H
Sbjct: 506 -------------MIGRGSQVGNLYVL--DFNENNH 526
>At2g06840 putative retroelement pol polyprotein
Length = 1102
Score = 629 bits (1622), Expect = e-180
Identities = 414/1190 (34%), Positives = 595/1190 (49%), Gaps = 213/1190 (17%)
Query: 228 RFSAVRSQILLMDPFPSIQRIYNIVRQEEKQQEINFRPVPAVESAAL----QTSKAPY-- 281
RF+ +RS+I DP PS R+Y+ V + ++ ++ R S A+ +T AP
Sbjct: 14 RFAPIRSKITDEDPLPSHNRVYSRVIRGQQNLDVA-RSKETTNSEAINFSVKTPSAPQVA 72
Query: 282 --RTPGKRQRPFCEHCNRHGHTITTCYQIHGFP------------------------SNP 315
P R R C HC+R GH +T C+ +HGFP + P
Sbjct: 73 AVYAPKPRDRS-CTHCHRQGHDVTDCFLVHGFPEWYYEQKGGSRVSSDNREVVSRLENKP 131
Query: 316 SKPQKKTETSPSTSANQLSSA-----------QYHKLLTLLAKEDNVGSSVNLAGTAFTC 364
+K + ++ +++SA Q +L++LL + +S L+G TC
Sbjct: 132 AKREGRSSKGNGRGRGRVNSARAPLSSSNGSDQITQLISLLQAQRPKSTSERLSGN--TC 189
Query: 365 IPFSWIIDSGASNHICTSLSLFSSYYPVNNQISVQQPDGSQALVKHIGTINCSPSLILTN 424
+ IIDSGAS+H+ S+ + + +V +PDG + T+ S S L +
Sbjct: 190 LT-DVIIDSGASHHMTGDCSILVDVFDIIPS-AVTKPDGKASCATKCVTLLLSSSYKLQD 247
Query: 425 VYHVPTFKFNLMSVTQLTESLNCDAIFSSSGCVFQDQATKKMIGRGSARNELYYLNQDLV 484
V VP F L+SV++L + ++ +IG G R +YY LV
Sbjct: 248 VLFVPDFDCTLISVSKLLKQTG--------------PFSRTLIGAGEVRERVYYFTGVLV 293
Query: 485 SNKHDKD----------HYCLSHPLSSLFSSK---HCNKFDLWHLRLVALQLRNKMELWN 531
++ H H L HP +S S H + DL + R K L +
Sbjct: 294 ASAHKTSSDSTSSGALWHRRLGHPSTSFLLSLPECHQSSKDLGKIDSCDTCSRAKQTLPS 353
Query: 532 ENIDIFLMSQELY-------------------AFKQTYLFYFGGECVLTA---------- 562
+ M+ + ++ Q + CV T
Sbjct: 354 LIRNFCAMADRQFRKPVRSIRSDNGTEFMCHTSYFQEHGILHETSCVDTPQQNARVERKH 413
Query: 563 AYLINKLPTPILK--FKSPHQV---------LLGSPPSYSSLRVFGCLCFAK-NMNIQHK 610
+++N T + + F +P V L G PSY LR FGCLC+A + K
Sbjct: 414 RHILNVARTCLFQGNFPTPSPVLKGKTPYEVLFGKQPSYDMLRTFGCLCYAHIRPRDKDK 473
Query: 611 FDERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDIQSPPFNNAISIN 670
F RS+ IF+GYP HET P
Sbjct: 474 FASRSRKCIFIGYP------------------------HETATP---------------- 493
Query: 671 TQILDNEFDDLFTGSPTHPNIPPENSHNDNSNDTIVTISTPEDDASSNPPSFSAESLSNN 730
TH +I P ++ +D +N + TP+ + +P S S+ + +N
Sbjct: 494 ---------------NTHDSIDPTSTSSDENNTPPEPV-TPQAEQPHSPSSISSPHIVHN 537
Query: 731 PPSTEMNPPNHRH-------------SQRIRNPPIHLKDYIC-KINNVTSKINFPLENYL 776
S N H R ++PP++LKDY+ K+++ + L +
Sbjct: 538 KGSVHSRHLNEDHDSSSPGLPELLGKGHRPKHPPVYLKDYVAHKVHSSPHTSSPGLSDSN 597
Query: 777 SLSNLSNSHRAFLINIIENKEPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLPEGK 836
+S +H AF+ I+++ E + + EW DAM KEI+ALE+N+TW + LP GK
Sbjct: 598 VSPTVSANHIAFMAAILDSNEQNHFKDDVLIKEWCDAMQKEIEALEANHTWDVTDLPHGK 657
Query: 837 SAIGCKWIYKIKYHSDGSIDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAA 896
AI KW+YK+K++SDG+++R+KARLV G Q +GID+ +TFAPVAK+ TVRLLL++AA
Sbjct: 658 KAISSKWVYKLKFNSDGTLERHKARLVVMGNHQKEGIDFKETFAPVAKMTTVRLLLAVAA 717
Query: 897 IKNWPLYQFDVNNAFLQGDLSEEVYMKLPPGFSHKKKPCVCKLNKSIYGLKQASRQWFSK 956
K+W ++Q DV+NAFL GDL +E S+YGLKQA R WF+K
Sbjct: 718 AKDWDVFQMDVHNAFLHGDLEQE----------------------SLYGLKQAPRCWFAK 755
Query: 957 FSTTLIQKGFRQSISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENAISKIKKFLAQSFS 1016
ST L + GF QS DYSLF+ N D T I LVYVDD II GNN AI K+ L + F
Sbjct: 756 LSTALRKLGFTQSYEDYSLFSLNRDGTVIHFLVYVDDFIIVGNNLKAIDHFKEHLHKCFH 815
Query: 1017 IKDLGNLSYILGIEVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDFPMEQHLRLRPNDG 1076
+KDLG L Y LG+EVSR G L Q+KY LDI++++G+ G +PS PME H +L
Sbjct: 816 MKDLGKLKYFLGLEVSRGADGFCLSQQKYALDIINEAGLLGYKPSAVPMELHHKLGSISS 875
Query: 1077 TPLSDPTVYRRHVGRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVG 1136
+P YRR V R +YLT+TRPD+ YAV+ LSQFMQ+P +H+ A +++RYLKGS
Sbjct: 876 PVFDNPAQYRRLVDRFIYLTITRPDLSYAVHILSQFMQTPLEAHWHATLRLVRYLKGSPD 935
Query: 1137 KGLFLSASSSINLVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEA 1196
+G+ L + +++L Y DSD+ CP TRRS + Y LG PISWKTKKQ T+S SSAEA
Sbjct: 936 QGILLRSDRALSLTAYCDSDYNPCPRTRRSLSAYVLYLGDTPISWKTKKQDTVSSSSAEA 995
Query: 1197 EYRSLATLSSELQWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVFHERTKHIEIDCH 1256
EYR++A E++WLK L++ LG+DH QPI ++CDSQAAIHIA NPVFHERTKHIE DCH
Sbjct: 996 EYRAMAYTLKEIKWLKALMTTLGVDHTQPILLFCDSQAAIHIAANPVFHERTKHIEKDCH 1055
Query: 1257 FVREKIKSGLIAPSYIRSSDQLADIFTKPLGGDAYKRILGKLGVIEISIP 1306
VR+ + +I+ +I ++ D+ TK L ++R+L LG +P
Sbjct: 1056 QVRDAVTDKVISTPHISTT----DLLTKALPRPTFERLLSTLGTCNYDLP 1101
>At4g22040 LTR retrotransposon like protein
Length = 1109
Score = 605 bits (1561), Expect = e-173
Identities = 295/543 (54%), Positives = 397/543 (72%), Gaps = 1/543 (0%)
Query: 764 VTSKINFPLENYLSLSNLSNSHRAFLINIIENKEPKSYSQAMKSVEWRDAMAKEIQALES 823
+ + FP +S + ++SH+AFL + EP +Y++AM WR+AM+ EI++L
Sbjct: 567 IFQETEFPYSK-MSCNRFTSSHKAFLAAVTAGMEPTTYNEAMVDKAWREAMSAEIESLRV 625
Query: 824 NNTWILCPLPEGKSAIGCKWIYKIKYHSDGSIDRYKARLVAKGYSQVQGIDYHDTFAPVA 883
N T+ + LP GK A+G KW+YKIKY SDG+I+RYKARLV G Q +G+DY +TFAPVA
Sbjct: 626 NQTFSIVNLPPGKRALGNKWVYKIKYRSDGAIERYKARLVVLGNCQKEGVDYDETFAPVA 685
Query: 884 KLVTVRLLLSIAAIKNWPLYQFDVNNAFLQGDLSEEVYMKLPPGFSHKKKPCVCKLNKSI 943
K+ TVRL L +AA ++W ++Q DV+NAFL GDL EEVYMKLP GF VC+L+KS+
Sbjct: 686 KMSTVRLFLGVAAARDWHVHQMDVHNAFLHGDLKEEVYMKLPQGFQCDDPSKVCRLHKSL 745
Query: 944 YGLKQASRQWFSKFSTTLIQKGFRQSISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENA 1003
YGLKQA R WFSK S+ L Q GF QS+SDYSLF+YN D + VLVYVDD+II+G+ +A
Sbjct: 746 YGLKQAPRCWFSKLSSALKQYGFTQSLSDYSLFSYNNDGVFVHVLVYVDDLIISGSCPDA 805
Query: 1004 ISKIKKFLAQSFSIKDLGNLSYILGIEVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDF 1063
+++ K +L F +KDLG L Y LGIEVSR+ +G +L QRKY LDI+S+ G+ G RPS F
Sbjct: 806 VAQFKSYLESCFHMKDLGLLKYFLGIEVSRNAQGFYLSQRKYVLDIISEMGLLGARPSAF 865
Query: 1064 PMEQHLRLRPNDGTPLSDPTVYRRHVGRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDA 1123
P+EQ+ +L + LSD + YRR VGRL+YL VTRP++ Y+V+TL+QFMQ+P H++A
Sbjct: 866 PLEQNHKLSLSTSPLLSDSSRYRRLVGRLIYLAVTRPELSYSVHTLAQFMQNPRQDHWNA 925
Query: 1124 ATQVLRYLKGSVGKGLFLSASSSINLVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKT 1183
A +V+RYLK + G+G+ LS++S++ + G+ DSD+A CP TRRS TGYF LG PISWKT
Sbjct: 926 AIRVVRYLKSNPGQGILLSSTSTLQINGWCDSDYAACPLTRRSLTGYFVQLGDTPISWKT 985
Query: 1184 KKQPTISRSSAEAEYRSLATLSSELQWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPV 1243
KKQPTISRSSAEAEYR++A L+ EL WLK +L DLG+ H Q + I+ DS++AI ++ NPV
Sbjct: 986 KKQPTISRSSAEAEYRAMAFLTQELMWLKRVLYDLGVSHVQAMRIFSDSKSAIALSVNPV 1045
Query: 1244 FHERTKHIEIDCHFVREKIKSGLIAPSYIRSSDQLADIFTKPLGGDAYKRILGKLGVIEI 1303
HERTKH+E+DCHF+R+ I G+IA S++ S QLADI TK LG + L KLG++++
Sbjct: 1046 QHERTKHVEVDCHFIRDAILDGIIATSFVPSHKQLADILTKALGEKEVRYFLRKLGILDV 1105
Query: 1304 SIP 1306
P
Sbjct: 1106 HAP 1108
Score = 119 bits (299), Expect = 9e-27
Identities = 59/121 (48%), Positives = 85/121 (69%), Gaps = 2/121 (1%)
Query: 548 QTYL-FYFGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNMN 606
Q+YL F GEC+L+AAYLIN+ P+ +L+ KSP+++L + P YS LRVFG LC+A N N
Sbjct: 466 QSYLPIQFWGECILSAAYLINRTPSMLLQGKSPYEMLYKTAPKYSHLRVFGSLCYAHNQN 525
Query: 607 IQ-HKFDERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDIQSPPFNN 665
+ KF RS+ +FVGYP QKG+R++D++ ++ +VSRDV F ET FPY + F +
Sbjct: 526 HKGDKFAARSRRCVFVGYPHGQKGWRLFDLEEQKFFVSRDVIFQETEFPYSKMSCNRFTS 585
Query: 666 A 666
+
Sbjct: 586 S 586
Score = 105 bits (262), Expect = 2e-22
Identities = 61/195 (31%), Positives = 98/195 (49%), Gaps = 10/195 (5%)
Query: 26 MSDIPPSENSPLSSISSPITVHSTTNPDPCVIHHSDNPSTVLVTPLLTGDNYGSWSRAVT 85
M+D PP S I V T P +DN V+ P+L +NY W+
Sbjct: 1 MADAPPPP-------PSVIEVRRTILPYDLTA--ADNSGAVISHPILKTNNYEEWACGFK 51
Query: 86 MALRAKNKFGFVDGSLTIPKK-KEDILTWQRCNDLVASWILNSVSTEIRPSILYAETAEQ 144
ALR++ KFGF+DG++ P D+ W N L+ SW+ ++ +E+ +I + + A
Sbjct: 52 TALRSRKKFGFLDGTIPQPLDGSPDLEDWLTINALLVSWMKMTIDSELLTNISHRDVARD 111
Query: 145 IWSDLQDRFSQSNAPKIYQLKQSISALKQEGMYVSLYFTQLKSLWDELNSIALVNPCICG 204
+W ++ RFS SN PK ++K ++ KQEGM V Y+ +L +WD +NS + C
Sbjct: 112 LWEQIRKRFSVSNGPKNQKMKADLATCKQEGMTVEGYYGKLNKIWDNINSYRPLRICASH 171
Query: 205 NAKSILDQQNQDRAM 219
+ L+ + R+M
Sbjct: 172 HMTGNLELLSGMRSM 186
Score = 45.8 bits (107), Expect = 2e-04
Identities = 36/144 (25%), Positives = 62/144 (43%), Gaps = 16/144 (11%)
Query: 375 ASNHICTSLSLFSSYYPVNNQISVQQPDGSQALVKHIGTINCSPSLILTNVYHVPTFKFN 434
AS+H+ +L L S ++ + + DG++ + GT+ LIL +V++V + +
Sbjct: 169 ASHHMTGNLELLSGMRSMS-PVLIILADGNKRVAVSEGTVRLGSHLILKSVFYVKELESD 227
Query: 435 LMSVTQLTESLNCDAIFSSSGCVFQDQATKKMIGRGSARNELYYLNQDLVSNKHDKDHYC 494
L+SV Q+ + +C + V QD+ T+ + G G N + H
Sbjct: 228 LISVGQMMDENHCVVQLADHFLVIQDRTTRMVTGIGKRENGSFCFRG---MENAAAVHTS 284
Query: 495 LSHPLSSLFSSKHCNKFDLWHLRL 518
+ P FDLWH RL
Sbjct: 285 VKAP------------FDLWHRRL 296
>At4g07810 putative polyprotein
Length = 1366
Score = 591 bits (1523), Expect = e-168
Identities = 361/986 (36%), Positives = 545/986 (54%), Gaps = 104/986 (10%)
Query: 368 SWIIDSGASNHICTSLSLFSSYYPVNNQISVQQPDGSQALVKHIGTINCSPSLILTNVYH 427
+W +D+ AS HIC LSLF + Y +++ ++ P+ + + T+ + LIL V++
Sbjct: 437 TWSLDTRASCHICCDLSLFCNVYHIDHT-NITLPNNIKISINIAETVKLNDRLILHLVFY 495
Query: 428 VPTFKFNLMS---------VTQLTESLNCDAIFSSSGCVFQDQATKKMIGRGSARNELY- 477
VP+F FNL+S V L+ +L+ S C + +K + S N++Y
Sbjct: 496 VPSFHFNLISRLGHPSMSRVQALSSNLHIPQKLSEFHCKICHLSKQKCLSFVS-NNKIYE 554
Query: 478 ------YLNQDLVS------NKHDKDHYCLSHPLSS-----LFSSKHCNKFDLWHLRLVA 520
+++ D+ + C + S L F ++H A
Sbjct: 555 EPFPLIHIDSDVTTIFPEFLKLVQTQFGCTVKSIRSDNAPELQFKDLLATFGIFHYHSCA 614
Query: 521 LQLRNKMELWNENIDIFLMSQELYAFKQTYLFYFGGECVLTAAYLINKLPTPILKFKSPH 580
+ + + + +++ LY F+ + ECV TAA+LIN+ PTP L+ KSP+
Sbjct: 615 YTPQQNYVVERNHQHLLNVARSLY-FQSNIPLAYWPECVSTAAFLINRTPTPNLEHKSPY 673
Query: 581 QVLLGSPPSYSSLRVFGCLCFAK-NMNIQHKFDERSKPGIFVGYPFNQKGYRIYDMKTRQ 639
+VL P Y+SLRVF CLC+A + + +HKF ER+ +F+GY KGY+I D+++
Sbjct: 674 EVLYKKLPDYNSLRVFCCLCYASTHQHERHKFTERATSCVFIGYESGFKGYKILDLESNT 733
Query: 640 IYVSRDVQFHETVFPYQD---IQSPPF--NNAISINTQILDNEFD--DLFT--GSPTHPN 690
+ V+R+V FHET+FP+ D Q+ F ++ + I+ + +N F D F P
Sbjct: 734 VSVTRNVVFHETIFPFIDKHSTQNVSFFDDSVLPISEKQKENRFQIYDYFNVLNLEVCPV 793
Query: 691 IPPENSHNDNSNDTIVTISTPEDDASSNPPSFSAESLSNNPPSTEMNPPNHRHSQRIRNP 750
I P ++ +ST +++N+ +M+ N ++
Sbjct: 794 IEPTTVPAHTHTRSLAPLST---------------TVTNDQFGNDMD--NTLMPRKETRA 836
Query: 751 PIHLKDYICK--INNVTSKIN---FPLENYLSLSNLSNSHRAFLINIIENKEPKSYSQAM 805
P +L Y C + +S ++ L ++LS LSN +R F II KEP ++ +A
Sbjct: 837 PSYLSQYHCSNVLKEPSSSLHGTAHSLSSHLSYDKLSNEYRLFCFAIIAEKEPTTFKEAA 896
Query: 806 KSVEWRDAMAKEIQALESNNTWILCPLPEGKSAIGCKWIYKIKYHSDGSIDRYKARLVAK 865
+W DAM E+ AL S +T +C L +GK AIGCKW++KIKY SDG+I+RYKARLVA
Sbjct: 897 LLQKWLDAMNVELDALVSTSTREICSLHDGKRAIGCKWVFKIKYKSDGTIERYKARLVAN 956
Query: 866 GYSQVQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFLQGDLSEEVYMKLP 925
GY+Q +G+DY DTF+P+AKL +VRL+L++AAI NW + Q DV NAFL GD EE+YM+LP
Sbjct: 957 GYTQQEGVDYIDTFSPIAKLTSVRLILALAAIHNWSISQMDVTNAFLHGDFEEEIYMQLP 1016
Query: 926 PGFSHKK-----KPCVCKLNKSIYGLKQASRQWFSKFSTTLIQKGFRQSISDYSLFTYNC 980
G++ +K K VC+L KS+YGLKQASRQWF KFS LIQ GF QS+ D +LF
Sbjct: 1017 QGYTPRKGELLPKRPVCRLVKSLYGLKQASRQWFHKFSGVLIQNGFMQSLFDPTLFVRVR 1076
Query: 981 DQTTIFVLVYVDDIIITGNNENAISKIKKFLAQSFSIKDLGNLSYILGIEVSRSKKGIFL 1040
+ T + +LVYVDDI++ N ++A+ ++K+ LA+ F +KDLG Y LG+E++RSK+GI +
Sbjct: 1077 EDTFLALLVYVDDIMLVSNKDSAVIEVKQILAKEFKLKDLGQKRYFLGLEIARSKEGISI 1136
Query: 1041 CQRKYTLDILSDSGMTGCRPSDFPMEQHLRLRPNDGTPLSDPTVYRRHVGRLLYLTVTRP 1100
QRKY L++L + G GC+P PME +L+L DG L D + YR+ +GRL+YLTVTRP
Sbjct: 1137 SQRKYALELLEEFGFLGCKPVPTPMELNLKLSQEDGALLLDASHYRKLIGRLVYLTVTRP 1196
Query: 1101 DIQYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVGKGLFLSASSSINLVGYADSDWAGC 1160
DI +AVN L+Q+M +P H AA ++LRYLK G+G+F ASS++ +AD+DW+ C
Sbjct: 1197 DICFAVNKLNQYMSAPREPHLMAARRILRYLKNDPGQGVFYPASSTLTFRAFADADWSNC 1256
Query: 1161 PTTRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEAEYRSLATLSSELQWLKYLLSDLGI 1220
P + S + F + + S EA WL L
Sbjct: 1257 PESSISISIVFWL-----------------KLSTEA-------------WLVLSL----- 1281
Query: 1221 DHPQPITIYCDSQAAIHIAENPVFHERTKHIEIDCHFVREKIKSGLIAPSYIRSSDQLAD 1280
P I +Y D ++A+HIA+N VFHE TK+ D H VREK+ G I ++ + + D
Sbjct: 1282 --PDTIFVYYDDESALHIAKNSVFHESTKNFLHDIHVVREKVAVGFIKTLHVDTEHNIVD 1339
Query: 1281 IFTKPLGGDAYKRILGKLGVIEISIP 1306
+ TKPL + +L K+G+ + P
Sbjct: 1340 LLTKPLTALRFNYLLSKMGLHHLYSP 1365
Score = 105 bits (261), Expect = 2e-22
Identities = 58/206 (28%), Positives = 95/206 (45%), Gaps = 17/206 (8%)
Query: 129 STEIRPSILYAETAEQIWSDLQDRFSQSNAPKIYQLKQSISALKQEGMYVSLYFTQLKSL 188
S ++ ++Y A +W L+ R QSN KIY ++ + L Q + +S Y+T+L
Sbjct: 64 SGDLGSGMVYFYDAHLLWLKLEGRSRQSNLSKIYSVQNQLDRLHQGSLDLSAYYTRLTVT 123
Query: 189 --------WDELNSI-----ALVNPCICGNAKSILDQQNQDRAMEFLQGVHDRFSAVRSQ 235
W+EL + C CG+ + + + FL +++ F R Q
Sbjct: 124 FIRSQCLSWEELKNFEELPSCTCGKCTCGSNDRWIQLYEKHNIVRFLMRLNESFIQARRQ 183
Query: 236 ILLMDPFPSIQRIYNIVRQEEKQQEINFRPV---PAVESAALQTSKAPYRTPGKRQRPFC 292
IL+MDP P +YN + Q+++Q+ N P P +++ Q + GK RP C
Sbjct: 184 ILMMDPLPEFTNLYNFISQDDQQRSFNSMPTTEKPVFQASITQQKPKFFNQQGK-SRPLC 242
Query: 293 EHCNRHGHTITTCYQIHGFPSNPSKP 318
+C GHT CY++HG+P P
Sbjct: 243 TYCGLLGHTNARCYKLHGYPPGYKVP 268
>At1g59265 polyprotein, putative
Length = 1466
Score = 579 bits (1492), Expect = e-165
Identities = 327/826 (39%), Positives = 456/826 (54%), Gaps = 97/826 (11%)
Query: 562 AAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFA--KNMNIQHKFDERSKPGI 619
A YLIN+LPTP+L+ +SP Q L G+ P+Y LRVFGC C+ + N QHK D++S+ +
Sbjct: 657 AVYLINRLPTPLLQLESPFQKLFGTSPNYDKLRVFGCACYPWLRPYN-QHKLDDKSRQCV 715
Query: 620 FVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDI--------------------- 658
F+GY Q Y ++T ++Y+SR V+F E FP+ +
Sbjct: 716 FLGYSLTQSAYLCLHLQTSRLYISRHVRFDENCFPFSNYLATLSPVQEQRRESSCVWSPH 775
Query: 659 ---------------------------QSPPFNNAISINTQILDNEFDDLFTGSP----- 686
S PF N+ +++ LD+ F F SP
Sbjct: 776 TTLPTRTPVLPAPSCSDPHHAATPPSSPSAPFRNS-QVSSSNLDSSFSSSFPSSPEPTAP 834
Query: 687 -----------------THP--NIPPENSHNDNSNDTIVTISTPEDDASSNPP---SFSA 724
TH N N N++ + ++STP +SS+P S S+
Sbjct: 835 RQNGPQPTTQPTQTQTQTHSSQNTSQNNPTNESPSQLAQSLSTPAQSSSSSPSPTTSASS 894
Query: 725 ESLSNNPPSTEMNPPNHRHSQRIRNPPIHLKDYICKINNVTSKINFPLENYLSLSNLS-N 783
S S PPS ++PP PP+ I NN + + N
Sbjct: 895 SSTSPTPPSILIHPP----------PPLA---QIVNNNNQAPLNTHSMGTRAKAGIIKPN 941
Query: 784 SHRAFLINIIENKEPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLPEGK-SAIGCK 842
+ +++ EP++ QA+K WR+AM EI A N+TW L P P + +GC+
Sbjct: 942 PKYSLAVSLAAESEPRTAIQALKDERWRNAMGSEINAQIGNHTWDLVPPPPSHVTIVGCR 1001
Query: 843 WIYKIKYHSDGSIDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPL 902
WI+ KY+SDGS++RYKARLVAKGY+Q G+DY +TF+PV K ++R++L +A ++WP+
Sbjct: 1002 WIFTKKYNSDGSLNRYKARLVAKGYNQRPGLDYAETFSPVIKSTSIRIVLGVAVDRSWPI 1061
Query: 903 YQFDVNNAFLQGDLSEEVYMKLPPGFSHKKKP-CVCKLNKSIYGLKQASRQWFSKFSTTL 961
Q DVNNAFLQG L+++VYM PPGF K +P VCKL K++YGLKQA R W+ + L
Sbjct: 1062 RQLDVNNAFLQGTLTDDVYMSQPPGFIDKDRPNYVCKLRKALYGLKQAPRAWYVELRNYL 1121
Query: 962 IQKGFRQSISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENAISKIKKFLAQSFSIKDLG 1021
+ GF S+SD SLF ++ +++LVYVDDI+ITGN+ + L+Q FS+KD
Sbjct: 1122 LTIGFVNSVSDTSLFVLQRGKSIVYMLVYVDDILITGNDPTLLHNTLDNLSQRFSVKDHE 1181
Query: 1022 NLSYILGIEVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDFPMEQHLRLRPNDGTPLSD 1081
L Y LGIE R G+ L QR+Y LD+L+ + M +P PM +L GT L+D
Sbjct: 1182 ELHYFLGIEAKRVPTGLHLSQRRYILDLLARTNMITAKPVTTPMAPSPKLSLYSGTKLTD 1241
Query: 1082 PTVYRRHVGRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVGKGLFL 1141
PT YR VG L YL TRPDI YAVN LSQFM P H A ++LRYL G+ G+FL
Sbjct: 1242 PTEYRGIVGSLQYLAFTRPDISYAVNRLSQFMHMPTEEHLQALKRILRYLAGTPNHGIFL 1301
Query: 1142 SASSSINLVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEAEYRSL 1201
++++L Y+D+DWAG ST GY LG +PISW +KKQ + RSS EAEYRS+
Sbjct: 1302 KKGNTLSLHAYSDADWAGDKDDYVSTNGYIVYLGHHPISWSSKKQKGVVRSSTEAEYRSV 1361
Query: 1202 ATLSSELQWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVFHERTKHIEIDCHFVREK 1261
A SSE+QW+ LL++LGI +P IYCD+ A ++ NPVFH R KHI ID HF+R +
Sbjct: 1362 ANTSSEMQWICSLLTELGIRLTRPPVIYCDNVGATYLCANPVFHSRMKHIAIDYHFIRNQ 1421
Query: 1262 IKSGLIAPSYIRSSDQLADIFTKPLGGDAYKRILGKLGVIEISIPP 1307
++SG + ++ + DQLAD TKPL A++ K+GV +PP
Sbjct: 1422 VQSGALRVVHVSTHDQLADTLTKPLSRTAFQNFASKIGVTR--VPP 1465
Score = 106 bits (264), Expect = 1e-22
Identities = 113/490 (23%), Positives = 185/490 (37%), Gaps = 90/490 (18%)
Query: 72 LTGDNYGSWSRAVTMALRAKNKFGFVDGSLTIP----------KKKEDILTWQRCNDLVA 121
LT NY WSR V GF+DGS T+P + D W+R + L+
Sbjct: 26 LTSTNYLMWSRQVHALFDGYELAGFLDGSTTMPPATIGTDAAPRVNPDYTRWKRQDKLIY 85
Query: 122 SWILNSVSTEIRPSILYAETAEQIWSDLQDRFSQSNAPKIYQLKQSISALKQEGMYVSLY 181
S +L ++S ++P++ A TA QIW L+ ++ + + QL+ + + + Y
Sbjct: 86 SAVLGAISMSVQPAVSRATTAAQIWETLRKIYANPSYGHVTQLRTQLKQWTKGTKTIDDY 145
Query: 182 FTQLKSLWDEL----------NSIALVNPCICGNAKSILDQQNQDRAMEFLQGVHDRFSA 231
L + +D+L + V + K ++DQ L +H+R
Sbjct: 146 MQGLVTRFDQLALLGKPMDHDEQVERVLENLPEEYKPVIDQIAAKDTPPTLTEIHERLLN 205
Query: 232 VRSQILLMDPFPSIQRIYNIV------------------RQEEKQQEINFRPVPAVESAA 273
S+IL + I N V R + + N +P
Sbjct: 206 HESKILAVSSATVIPITANAVSHRNTTTTNNNNNGNRNNRYDNRNNNNNSKP-------- 257
Query: 274 LQTSKAPYRTPGKRQRPF---CEHCNRHGHTITTCYQIHGFPSNPSKPQKKTETSP-STS 329
Q S + + +P+ C+ C GH+ C Q+ F S+ + Q + +P
Sbjct: 258 WQQSSTNFHPNNNQSKPYLGKCQICGVQGHSAKRCSQLQHFLSSVNSQQPPSPFTPWQPR 317
Query: 330 ANQLSSAQYHKLLTLLAKEDNVGSSVNLAGTAFTCIPFSWIIDSGASNHICTSLSLFSSY 389
AN + Y SS N W++DSGA++HI + + S +
Sbjct: 318 ANLALGSPY--------------SSNN------------WLLDSGATHHITSDFNNLSLH 351
Query: 390 YPVNNQISVQQPDGSQALVKHIGTINCSPS---LILTNVYHVPTFKFNLMSVTQLTESLN 446
P V DGS + H G+ + S L L N+ +VP NL+SV +L +
Sbjct: 352 QPYTGGDDVMVADGSTIPISHTGSTSLSTKSRPLNLHNILYVPNIHKNLISVYRLCNANG 411
Query: 447 CDAIFSSSGCVFQDQATKKMIGRGSARNELY---YLNQDLVS--------NKHDKDHYCL 495
F + +D T + +G ++ELY + VS H H L
Sbjct: 412 VSVEFFPASFQVKDLNTGVPLLQGKTKDELYEWPIASSQPVSLFASPSSKATHSSWHARL 471
Query: 496 SHPLSSLFSS 505
HP S+ +S
Sbjct: 472 GHPAPSILNS 481
>At4g02960 putative polyprotein of LTR transposon
Length = 1456
Score = 578 bits (1491), Expect = e-165
Identities = 328/818 (40%), Positives = 461/818 (56%), Gaps = 77/818 (9%)
Query: 562 AAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFA--KNMNIQHKFDERSKPGI 619
A YLIN+LPTP+L+ +SP Q L G PP+Y L+VFGC C+ + N +HK +++SK
Sbjct: 636 AVYLINRLPTPLLQLQSPFQKLFGQPPNYEKLKVFGCACYPWLRPYN-RHKLEDKSKQCA 694
Query: 620 FVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPY----------QDIQS--------- 660
F+GY Q Y + T ++Y SR VQF E FP+ Q+ +S
Sbjct: 695 FMGYSLTQSAYLCLHIPTGRLYTSRHVQFDERCFPFSTTNFGVSTSQEQRSDSAPNWPSH 754
Query: 661 -------------------------PPFNNAISINTQILDNEFDDLFTGSPT-------- 687
PP + + TQ+ + SP+
Sbjct: 755 TTLPTTPLVLPAPPCLGPHLDTSPRPPSSPSPLCTTQVSSSNLPSSSISSPSSSEPTAPS 814
Query: 688 ----HPNIPPENSHNDNSNDTIVTISTPEDDASSNPPSFSA--ESLSNNP----PSTEMN 737
P P + N NSN I+ P + ++P S +S ++P PST ++
Sbjct: 815 HNGPQPTAQPHQTQNSNSNSPILNNPNPNSPSPNSPNQNSPLPQSPISSPHIPTPSTSIS 874
Query: 738 PPNHRHSQRIRNPPIHLKDYICKINNVTSKINFPLENYLSLSNLSNSHR------AFLIN 791
PN S PP+ I V ++ P+ + + + R ++ +
Sbjct: 875 EPNSPSSSSTSTPPLPPVLPAPPIIQVNAQA--PVNTHSMATRAKDGIRKPNQKYSYATS 932
Query: 792 IIENKEPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLPEGK-SAIGCKWIYKIKYH 850
+ N EP++ QAMK WR AM EI A N+TW L P P + +GC+WI+ K++
Sbjct: 933 LAANSEPRTAIQAMKDDRWRQAMGSEINAQIGNHTWDLVPPPPPSVTIVGCRWIFTKKFN 992
Query: 851 SDGSIDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNA 910
SDGS++RYKARLVAKGY+Q G+DY +TF+PV K ++R++L +A ++WP+ Q DVNNA
Sbjct: 993 SDGSLNRYKARLVAKGYNQRPGLDYAETFSPVIKSTSIRIVLGVAVDRSWPIRQLDVNNA 1052
Query: 911 FLQGDLSEEVYMKLPPGFSHKKKP-CVCKLNKSIYGLKQASRQWFSKFSTTLIQKGFRQS 969
FLQG L++EVYM PPGF K +P VC+L K+IYGLKQA R W+ + T L+ GF S
Sbjct: 1053 FLQGTLTDEVYMSQPPGFVDKDRPDYVCRLRKAIYGLKQAPRAWYVELRTYLLTVGFVNS 1112
Query: 970 ISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENAISKIKKFLAQSFSIKDLGNLSYILGI 1029
ISD SLF ++ I++LVYVDDI+ITGN+ + L+Q FS+K+ +L Y LGI
Sbjct: 1113 ISDTSLFVLQRGRSIIYMLVYVDDILITGNDTVLLKHTLDALSQRFSVKEHEDLHYFLGI 1172
Query: 1030 EVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDFPMEQHLRLRPNDGTPLSDPTVYRRHV 1089
E R +G+ L QR+YTLD+L+ + M +P PM +L + GT L DPT YR V
Sbjct: 1173 EAKRVPQGLHLSQRRYTLDLLARTNMLTAKPVATPMATSPKLTLHSGTKLPDPTEYRGIV 1232
Query: 1090 GRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVGKGLFLSASSSINL 1149
G L YL TRPD+ YAVN LSQ+M P H++A +VLRYL G+ G+FL ++++L
Sbjct: 1233 GSLQYLAFTRPDLSYAVNRLSQYMHMPTDDHWNALKRVLRYLAGTPDHGIFLKKGNTLSL 1292
Query: 1150 VGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEAEYRSLATLSSELQ 1209
Y+D+DWAG ST GY LG +PISW +KKQ + RSS EAEYRS+A SSELQ
Sbjct: 1293 HAYSDADWAGDTDDYVSTNGYIVYLGHHPISWSSKKQKGVVRSSTEAEYRSVANTSSELQ 1352
Query: 1210 WLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVFHERTKHIEIDCHFVREKIKSGLIAP 1269
W+ LL++LGI P IYCD+ A ++ NPVFH R KHI +D HF+R +++SG +
Sbjct: 1353 WICSLLTELGIQLSHPPVIYCDNVGATYLCANPVFHSRMKHIALDYHFIRNQVQSGALRV 1412
Query: 1270 SYIRSSDQLADIFTKPLGGDAYKRILGKLGVIEISIPP 1307
++ + DQLAD TKPL A++ K+GVI+ +PP
Sbjct: 1413 VHVSTHDQLADTLTKPLSRVAFQNFSRKIGVIK--VPP 1448
Score = 107 bits (268), Expect = 3e-23
Identities = 102/432 (23%), Positives = 176/432 (40%), Gaps = 62/432 (14%)
Query: 72 LTGDNYGSWSRAVTMALRAKNKFGFVDGSL----------TIPKKKEDILTWQRCNDLVA 121
LT NY WSR V GF+DGS +P+ D W+R + L+
Sbjct: 26 LTSTNYLMWSRQVHALFDGYELAGFLDGSTPMPPATIGTDAVPRVNPDYTRWRRQDKLIY 85
Query: 122 SWILNSVSTEIRPSILYAETAEQIWSDLQDRFSQSNAPKIYQLKQSISALKQEGMYVSLY 181
S IL ++S ++P++ A TA QIW L+ ++ + + QL+ I+ Q +
Sbjct: 86 SAILGAISMSVQPAVSRATTAAQIWETLRKIYANPSYGHVTQLR-FITRFDQLAL----- 139
Query: 182 FTQLKSLWDELNSIALVNPCICGNAKSILDQQNQDRAMEFLQGVHDRFSAVRSQILLMDP 241
L D + V + + K ++DQ L +H+R S++L ++
Sbjct: 140 ---LGKPMDHDEQVERVLENLPDDYKPVIDQIAAKDTPPSLTEIHERLINRESKLLALNS 196
Query: 242 FPSIQRIYNIV---------RQEEKQQEINFRPVPAVESAALQTSKAPYRTPGKRQRPF- 291
+ N+V Q + N+ S + Q S + R+ ++ +P+
Sbjct: 197 AEVVPITANVVTHRNTNTNRNQNNRGDNRNYNN-NNNRSNSWQPSSSGSRSDNRQPKPYL 255
Query: 292 --CEHCNRHGHTITTCYQIHGFPSNPSKPQKKTETSP-STSANQLSSAQYHKLLTLLAKE 348
C+ C+ GH+ C Q+H F S ++ Q + +P AN ++ Y+
Sbjct: 256 GRCQICSVQGHSAKRCPQLHQFQSTTNQQQSTSPFTPWQPRANLAVNSPYN--------- 306
Query: 349 DNVGSSVNLAGTAFTCIPFSWIIDSGASNHICTSLSLFSSYYPVNNQISVQQPDGSQALV 408
+W++DSGA++HI + + S + P V DGS +
Sbjct: 307 -----------------ANNWLLDSGATHHITSDFNNLSFHQPYTGGDDVMIADGSTIPI 349
Query: 409 KHIGTIN---CSPSLILTNVYHVPTFKFNLMSVTQLTESLNCDAIFSSSGCVFQDQATKK 465
H G+ + S SL L V +VP NL+SV +L + F + +D T
Sbjct: 350 THTGSASLPTSSRSLDLNKVLYVPNIHKNLISVYRLCNTNRVSVEFFPASFQVKDLNTGV 409
Query: 466 MIGRGSARNELY 477
+ +G ++ELY
Sbjct: 410 PLLQGKTKDELY 421
>At2g24660 putative retroelement pol polyprotein
Length = 1156
Score = 571 bits (1472), Expect = e-163
Identities = 303/631 (48%), Positives = 396/631 (62%), Gaps = 54/631 (8%)
Query: 701 SNDTIVTISTPEDDASS----NPPSFSAESLSNNPPSTEMNPPNHRHSQRIRNPPIHLKD 756
S DT+ I D S+ N P F +S P E P R Q IR + L+D
Sbjct: 525 SGDTLAQIDDSPDIVSTPNRNNQPLFVVDS-----PFVEATSPRQRKRQ-IRQS-VRLQD 577
Query: 757 YICKINNVTSKIN---------------------FPLENYLSLSNLSNSHRAFLINIIEN 795
Y+ N S IN +PL +Y+S S H+AFL I N
Sbjct: 578 YVL-YNATVSPINPHALPDSSSQSSSMVQGTSSLYPLSDYVSDDCFSAGHKAFLAAITAN 636
Query: 796 KEPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLPEGKSAIGCKWIYKIKYHSDGSI 855
EPK + +A++ W DAM KE+ ALE N TW + LP GK AIG +W+YK KY++DGSI
Sbjct: 637 DEPKHFKEAVRIKVWNDAMFKEVDALEINKTWDIVDLPPGKVAIGSQWVYKTKYNADGSI 696
Query: 856 DRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFLQGD 915
+RYKARLV +G QV+G DY++TFAPV K+ TVR LL + A W +YQ DVNNAFL GD
Sbjct: 697 ERYKARLVVQGNKQVEGEDYNETFAPVVKMTTVRTLLRLVAANQWEVYQMDVNNAFLHGD 756
Query: 916 LSEEVYMKLPPGFSHKKKPCVCKLNKSIYGLKQASRQWFSKFSTTLIQKGFRQSISDYSL 975
L EEVYMKLPPGF H VC+L KS+YGLKQA R WF K S L++ GF Q DYS
Sbjct: 757 LDEEVYMKLPPGFRHSHPDKVCRLRKSLYGLKQAPRCWFKKLSDALLRFGFVQGHEDYSF 816
Query: 976 FTYNCDQTTIFVLVYVDDIIITGNNENAISKIKKFLAQSFSIKDLGNLSYILGIEVSRSK 1035
F+Y + + VLVYVDD++I GN+ + K K++L + FS+KDLG L Y LGIEVSR
Sbjct: 817 FSYTRNGIELRVLVYVDDLLICGNDGYMLQKFKEYLGRCFSMKDLGKLKYFLGIEVSRGS 876
Query: 1036 KGIFLCQRKYTLDILSDSGMTGCRPSDFPMEQHLRLRPNDGTPLSDPTVYRRHVGRLLYL 1095
+GIFL QRKY LDI++DSG GCRP+ P+EQ+ L +DG L+D YRR VGRLLYL
Sbjct: 877 EGIFLSQRKYALDIITDSGNLGCRPALTPLEQNHHLATDDGPLLADAKPYRRLVGRLLYL 936
Query: 1096 TVTRPDIQYAVNTLSQFMQSPYSSHFDAATQVLRYLKGSVGKGLFLSASSSINLVGYADS 1155
TRP++ Y+V+ LSQFMQ+P +H AA +++R+LKGS G+G+ L+A + + L Y DS
Sbjct: 937 LHTRPELSYSVHVLSQFMQTPREAHLAAAMRIVRFLKGSPGQGILLNADTELQLDVYCDS 996
Query: 1156 DWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPTISRSSAEAEYRSLATLSSELQWLKYLL 1215
D+ CP TRRS + Y +LG +PISWKTKKQ T+S SSAEAEYR+++ E++WL+ LL
Sbjct: 997 DFQTCPKTRRSLSAYVVLLGGSPISWKTKKQDTVSHSSAEAEYRAMSVALREIKWLRKLL 1056
Query: 1216 SDLGIDHPQPITIYCDSQAAIHIAENPVFHERTKHIEIDCHFVREKIKSGLIAPSYIRSS 1275
+L NPVFHERTKHIE DCH VR+ ++ G+I ++R++
Sbjct: 1057 KELA---------------------NPVFHERTKHIESDCHSVRDAVRDGIITTHHVRTN 1095
Query: 1276 DQLADIFTKPLGGDAYKRILGKLGVIEISIP 1306
+QLADIFTK LG + ++ KLG+ ++ P
Sbjct: 1096 EQLADIFTKALGRSQFLYLMSKLGIQDLHTP 1126
Score = 108 bits (271), Expect = 2e-23
Identities = 53/111 (47%), Positives = 74/111 (65%), Gaps = 1/111 (0%)
Query: 546 FKQTYLFYFGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNM 605
F+ + F GE VLTAAYLIN+ PT + SP+++L S P+Y LRVFG C+
Sbjct: 263 FQASLPIQFWGEAVLTAAYLINRTPTSLHNGLSPYEILHNSKPNYEHLRVFGSACYVHRA 322
Query: 606 NI-QHKFDERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPY 655
+ + KF ERS+ +F+GYPF QKG++++DM+ ++ VSRDV F E VFPY
Sbjct: 323 SRDKDKFGERSRLCVFIGYPFAQKGWKVFDMEKKEFLVSRDVVFREDVFPY 373
>At4g14460 retrovirus-related like polyprotein
Length = 1489
Score = 552 bits (1422), Expect = e-157
Identities = 300/793 (37%), Positives = 448/793 (55%), Gaps = 91/793 (11%)
Query: 546 FKQTYLFYFGGECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAK-N 604
F+ + +CV TA +LIN+LP+P+L KSP++++L P YS L+ FGCLCF N
Sbjct: 750 FQSNIPMQYWSDCVTTAVFLINRLPSPLLNNKSPYELILNKQPDYSLLKNFGCLCFVSTN 809
Query: 605 MNIQHKFDERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQD------- 657
+ + KF R++ +F+GYP KGY++ D+++ + VSR+V F E VFP++
Sbjct: 810 AHERTKFTPRARACVFLGYPSGYKGYKVLDLESHSVTVSRNVVFKEHVFPFKTSELLNKA 869
Query: 658 -----------------IQSPPFNNAISINTQILDNEFDDLFTGSPTH--PNIPPENSHN 698
+++ P + S+ D+ D S + P+I P +S+
Sbjct: 870 VDMFPNSILPLPAPLHFVETMPLIDEDSLIPTTTDSRTADNHASSSSSALPSIIPPSSNT 929
Query: 699 DNSNDTIVTISTPEDDASSNPPSFSAESLSNNPPSTEMNPPNHRHSQRIRNPPIHLKDYI 758
+ + + ++ PS+ +E + PS PP + PIH I
Sbjct: 930 ETQDIDSNAVPITRSKRTTRAPSYLSEYHCSLVPSISTLPPTDS------SIPIHPLPEI 983
Query: 759 CKINNVTSKINFPLENYLSLSNLSNSHRAFLINIIENKEPKSYSQAMKSVEWRDAMAKEI 818
++ +P+ +S + ++++ EPK++SQAMKS +W +E+
Sbjct: 984 FTASSPKKTTPYPISTVVSYDKYTPLCQSYIFAYNTETEPKTFSQAMKSEKWIRVAVEEL 1043
Query: 819 QALESNNTWILCPLPEGKSAIGCKWIYKIKYHSDGSIDRYKARLVAKGYSQVQGIDYHDT 878
QA+E N TW + LP K+ +GCKW++ IKY+ DG+++RYKARLVA+G++Q +GID+ DT
Sbjct: 1044 QAMELNKTWSVESLPPDKNVVGCKWVFTIKYNPDGTVERYKARLVAQGFTQQEGIDFLDT 1103
Query: 879 FAPVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFLQGDLSEEVYMKLPPGFSHKKKPC--- 935
F+PVAKL + +++L +AAI W L Q DV++AFL GDL EE++M LP G++
Sbjct: 1104 FSPVAKLTSAKMMLGLAAITGWTLTQMDVSDAFLHGDLDEEIFMSLPQGYTPPAGTILPP 1163
Query: 936 --VCKLNKSIYGLKQASRQWFSKFSTTLIQKGFRQSISDYSLFTYNCDQTTIFVLVYVDD 993
VC+L KSIYGLKQASRQW+ +F + LVY+DD
Sbjct: 1164 NPVCRLLKSIYGLKQASRQWYKRF---------------------------VAALVYIDD 1196
Query: 994 IIITGNNENAISKIKKFLAQSFSIKDLGNLSYILGIEVSRSKKGIFLCQRKYTLDILSDS 1053
I+I NN+ + +K L F IKDLG + LG+
Sbjct: 1197 IMIASNNDAEVENLKALLRSEFKIKDLGPARFFLGL------------------------ 1232
Query: 1054 GMTGCRPSDFPMEQHLRLRPNDGTPLSDPTVYRRHVGRLLYLTVTRPDIQYAVNTLSQFM 1113
GC+PS PM+ L L + GTPL +PT YR+ +GRLLYLT+TRPDI YAV+ LSQF+
Sbjct: 1233 --LGCKPSSIPMDPTLHLVRDMGTPLPNPTAYRKLIGRLLYLTITRPDITYAVHQLSQFI 1290
Query: 1114 QSPYSSHFDAATQVLRYLKGSVGKGLFLSASSSINLVGYADSDWAGCPTTRRSTTGYFTM 1173
+P H AA +VLRY+K + G+GL SA I L G++D+DWA C TRRS +G+
Sbjct: 1291 SAPSDIHLQAAHKVLRYIKANPGQGLMYSADYEICLNGFSDADWAACKDTRRSISGFCIY 1350
Query: 1174 LGSNPISWKTKKQPTISRSSAEAEYRSLATLSSELQWLKYLLSDLGIDHPQPITIYCDSQ 1233
LG++ ISWK+KKQ SRSS E+EYRS+A + E+ WL+ LL DL I P ++CD++
Sbjct: 1351 LGTSLISWKSKKQAVASRSSTESEYRSMAQATCEIIWLQQLLKDLHIPLTCPAKLFCDNK 1410
Query: 1234 AAIHIAENPVFHERTKHIEIDCHFVREKIKSGLIAPSYIRSSDQLADIFTKPLGGDAYKR 1293
+A+H + NPVFHERTKHIEIDCH VR++IK+G + ++ + +Q ADI TK L +
Sbjct: 1411 SALHSSLNPVFHERTKHIEIDCHTVRDQIKAGNLKALHVPTENQHADILTKALHPGPFHH 1470
Query: 1294 ILGKLGVIEISIP 1306
+L ++ + + +P
Sbjct: 1471 LLRQMSLSSLFLP 1483
Score = 202 bits (514), Expect = 1e-51
Identities = 99/277 (35%), Positives = 163/277 (58%), Gaps = 19/277 (6%)
Query: 53 DPCVIHHSDNPSTVLVTP-LLTGDNYGSWSRAVTMALRAKNKFGFVDGSLTIPKKKE-DI 110
+P +H +D+ +LV+ L T ++ SW R++ MAL +NK GF++G++T P + D
Sbjct: 32 NPYYLHSADHAGLILVSDRLTTASDFHSWRRSILMALNVRNKLGFINGTITKPPEDHRDF 91
Query: 111 LTWQRCNDLVASWILNSVSTEIRPSILYAETAEQIWSDLQDRFSQSNAPKIYQLKQSISA 170
W RCND+V++W++NSV +I S+LY T + IW++L RF Q +AP+I+ ++Q +S
Sbjct: 92 GAWSRCNDIVSTWLMNSVDKKIGQSLLYIATVQGIWNNLLSRFKQDDAPRIFDIEQKLSK 151
Query: 171 LKQEGMYVSLYFTQLKSLWDELNSIALVNPCICGN-----AKSILDQQNQDRAMEFLQGV 225
++Q M +S Y+T L +LW+E + + C CG A Q + R +FL+ +
Sbjct: 152 IEQGSMDISTYYTALLTLWEEHRNYVELPVCTCGRCECDAAVKWEHLQQRSRVTKFLKEL 211
Query: 226 HDRFSAVRSQILLMDPFPSIQRIYNIVRQEEKQQEINFRPVPAVESAALQTSK------- 278
++ F R IL++ P P+I+ +N+V Q+E+Q+ N +P+ V+S A Q +
Sbjct: 212 NEGFDQTRRHILMLKPIPTIKEAFNMVTQDERQR--NVKPLTRVDSVAFQNTSMINEDEN 269
Query: 279 ---APYRTPGKRQRPFCEHCNRHGHTITTCYQIHGFP 312
A Y T Q+P C HC + GHTI CY++HG+P
Sbjct: 270 AYVAAYNTVRPNQKPICTHCGKVGHTIQKCYKVHGYP 306
Score = 95.1 bits (235), Expect = 2e-19
Identities = 56/151 (37%), Positives = 78/151 (51%), Gaps = 25/151 (16%)
Query: 368 SWIIDSGASNHICTSLSLFSSYYPVNNQISVQQPDGSQALVKHIGTINCSPSLILTNVYH 427
+WIIDSGAS+H+C+ L++F V+ GT++ + LIL NV H
Sbjct: 481 AWIIDSGASSHVCSDLAMFRELKSVS------------------GTVHITQKLILHNVLH 522
Query: 428 VPTFKFNLMSVTQLTESLNCDAIFSSSGCVFQDQATKKMIGRGSARNELYYLNQDLVSNK 487
VP FKFNLMSV+ L ++++C A F C+ Q+ + MIGRG + LY L + S
Sbjct: 523 VPDFKFNLMSVSSLVKTISCSAHFYVDCCLIQELSQGLMIGRGRLYHNLYILETENTSPS 582
Query: 488 HDKDHYCLSHPLSSLFSSKHCNKFDLWHLRL 518
S P + LF+ N LWH RL
Sbjct: 583 -------TSTPAACLFTGSVLNDGHLWHQRL 606
>At1g53810
Length = 1522
Score = 533 bits (1373), Expect = e-151
Identities = 306/791 (38%), Positives = 444/791 (55%), Gaps = 56/791 (7%)
Query: 557 ECVLTAAYLINKLPTPIL-KFKSPHQVLLGSPPSYSSLRVFGCLCFAKNMNIQH-KFDER 614
E TA ++IN LPT L +SP+Q L G P YS+LRVFGC C+ + KFD R
Sbjct: 647 ESFFTANFVINLLPTSSLDNNESPYQKLYGKAPEYSALRVFGCACYPTLRDYASTKFDPR 706
Query: 615 SKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFPYQDIQSPPFNNAISINTQIL 674
S +F+GY KGYR T +IY+SR V F E P++ I S T +L
Sbjct: 707 SLKCVFLGYNEKYKGYRCLYPPTGRIYISRHVVFDENTHPFESIYSHLHPQD---KTPLL 763
Query: 675 DNEFDDLFTGSPTHPN--------IP-PE------------------NSHNDNS--NDTI 705
+ F +PT P+ IP PE N+ +D S N+TI
Sbjct: 764 EAWFKSFHHVTPTQPDQSRYPVSSIPQPETTDLSAAPASVAAETAGPNASDDTSQDNETI 823
Query: 706 VTIS-TPE-----DDASSNPPSFSAESLSNNPPSTEMNPPNHRHSQRIRNPPIHLKDYIC 759
+S +PE D AS S + S++P +P + I+ P +
Sbjct: 824 SVVSGSPERTTGLDSASIGDSYHSPTADSSHPSPARSSPASSPQGSPIQMAPA--QQVQA 881
Query: 760 KINNVTSKINFPLENYLSLSNLSNSHRAFLINIIENKEPKSYSQAMKSVEWRDAMAKEIQ 819
+ N + + E + N L + + EPK+ ++A+K W +AM +E+
Sbjct: 882 PVTNEHAMVTRGKEGI----SKPNKRYVLLTHKVSIPEPKTVTEALKHPGWNNAMQEEMG 937
Query: 820 ALESNNTWILCPLPEGKSAIGCKWIYKIKYHSDGSIDRYKARLVAKGYSQVQGIDYHDTF 879
+ TW L P + +G W+++ K H+DGS+D+ KARLVAKG+ Q +GIDY +T+
Sbjct: 938 NCKETETWTLVPYSPNMNVLGSMWVFRTKLHADGSLDKLKARLVAKGFKQEEGIDYLETY 997
Query: 880 APVAKLVTVRLLLSIAAIKNWPLYQFDVNNAFLQGDLSEEVYMKLPPGFSHKKKPC-VCK 938
+PV + TVRL+L +A + W L Q DV NAFL GDL+E VYM+ P GF K KP VC
Sbjct: 998 SPVVRTPTVRLILHVATVLKWELKQMDVKNAFLHGDLTETVYMRQPAGFVDKSKPDHVCL 1057
Query: 939 LNKSIYGLKQASRQWFSKFSTTLIQKGFRQSISDYSLFTYNCDQTTIFVLVYVDDIIITG 998
L+KS+YGLKQ+ R WF +FS L++ GF S+ D SLF Y+ + I +L+YVDD++ITG
Sbjct: 1058 LHKSLYGLKQSPRAWFDRFSNFLLEFGFICSLFDPSLFVYSSNNDVILLLLYVDDMVITG 1117
Query: 999 NNENAISKIKKFLAQSFSIKDLGNLSYILGIEVSRSKKGIFLCQRKYTLDILSDSGMTGC 1058
NN +++ + L + F +KD+G + Y LGI++ G+F+ Q+KY D+L + M C
Sbjct: 1118 NNSQSLTHLLAALNKEFRMKDMGQVHYFLGIQIQTYDGGLFMSQQKYAEDLLITASMANC 1177
Query: 1059 RPSDFPMEQHLRLRPNDGTPLSDPTVYRRHVGRLLYLTVTRPDIQYAVNTLSQFMQSPYS 1118
P P+ L N SDPT +R G+L YLT+TRPDIQ+AVN + Q M P
Sbjct: 1178 SPMPTPLPLQLDRVSNQDEVFSDPTYFRSLAGKLQYLTLTRPDIQFAVNFVCQKMHQPSV 1237
Query: 1119 SHFDAATQVLRYLKGSVGKGLFLSASSS---------INLVGYADSDWAGCPTTRRSTTG 1169
S F+ ++LRY+KG+V G+ +++SS +L Y+DSD+A C TRRS G
Sbjct: 1238 SDFNLLKRILRYIKGTVSMGIQYNSNSSSVVSAYESDYDLSAYSDSDYANCKETRRSVGG 1297
Query: 1170 YFTMLGSNPISWKTKKQPTISRSSAEAEYRSLATLSSELQWLKYLLSDLGIDHPQPITIY 1229
Y T +G N ISW +KKQPT+SRSS EAEYRSL+ +SE++W+ +L ++G+ P ++
Sbjct: 1298 YCTFMGQNIISWSSKKQPTVSRSSTEAEYRSLSETASEIKWMSSILREIGVSLPDTPELF 1357
Query: 1230 CDSQAAIHIAENPVFHERTKHIEIDCHFVREKIKSGLIAPSYIRSSDQLADIFTKPLGGD 1289
CD+ +A+++ NP FH RTKH ++D H++RE++ + +I QLADIFTK L +
Sbjct: 1358 CDNLSAVYLTANPAFHARTKHFDVDHHYIRERVALKTLVVKHIPGHLQLADIFTKSLPFE 1417
Query: 1290 AYKRILGKLGV 1300
A+ R+ KLGV
Sbjct: 1418 AFTRLRFKLGV 1428
Score = 93.6 bits (231), Expect = 7e-19
Identities = 99/504 (19%), Positives = 184/504 (35%), Gaps = 107/504 (21%)
Query: 68 VTPLLTGDNYGSWSRAVTMALRAKNKFGFVDGSLTIPKKKEDIL--------------TW 113
VT L NY W L + GFV GS++ P + + TW
Sbjct: 15 VTVTLNQQNYILWKSQFESFLSGQGLLGFVTGSISAPAQTRSVTHNNVTSEEPNPEFYTW 74
Query: 114 QRCNDLVASWILNSVSTEIRPSILYAETAEQIWSDLQDRFSQSNAPKIYQLKQSISALKQ 173
+ + +V SW+L S + +I ++ T+ Q+W L + F++ ++ ++++L++ + L++
Sbjct: 75 HQTDQVVKSWLLGSFAEDILSVVVNCFTSHQVWLTLANHFNRVSSSRLFELQRRLQTLEK 134
Query: 174 EGMYVSLYFTQLKSLWDELNSIALVNP--------------------CICGNAKSILDQQ 213
+ + ++ LK + D+L S+ P N+
Sbjct: 135 KDNTMEVFLKDLKHICDQLASVGSPVPEKMKIFSALNGLGREYEPIKTTIENSVDSNPSL 194
Query: 214 NQDRAMEFLQGVHDRFSAVRSQILLMDPFPSIQRIYNIVRQEEKQQEINFRPVPAVESAA 273
+ D L+G DR Q + +P S +N+ + N R S
Sbjct: 195 SLDEVASKLRGYDDRL-----QSYVTEPTISPHVAFNVTHSDSGYYHNNNRGKGRSNSG- 248
Query: 274 LQTSKAPYRTPGK---------------RQRPFCEHCNRHGHTITTCYQIHGFPSNPSKP 318
+ K+ + T G+ C+ C + GH C+
Sbjct: 249 --SGKSSFSTRGRGFHQQISPTSGSQAGNSGLVCQICGKAGHHALKCW------------ 294
Query: 319 QKKTETSPSTSANQLSSAQYHKLLTLLAKEDNVGSSVNLAGTAFTCIPFSWIIDSGASNH 378
++ ++ H+ L + + + G WI DS AS H
Sbjct: 295 ------------HRFDNSYQHEDLPMALATMRITDVTDHHG-------HEWIPDSAASAH 335
Query: 379 ICTSLSLFSSYYPVNNQISVQQPDGSQALVKHIGT---INCSPSLILTNVYHVPTFKFNL 435
+ + + P + S+ DG+ + H G+ + S + L V P +L
Sbjct: 336 VTNNRHVLQQSQPYHGSDSIMVADGNFLPITHTGSGSIASSSGKIPLKEVLVCPDIVKSL 395
Query: 436 MSVTQLTESLNCDAIFSSSGCVFQDQATKKMIGRGSARNELYYLNQDLVSNKHDKDHYCL 495
+SV++LT C F + D+ATKK++ G R+ LY L +
Sbjct: 396 LSVSKLTSDYPCSVEFDADSVRINDKATKKLLVMGRNRDGLYSLEEP------------- 442
Query: 496 SHPLSSLFSSK-HCNKFDLWHLRL 518
L L+S++ + ++WH RL
Sbjct: 443 --KLQVLYSTRQNSASSEVWHRRL 464
>At4g23160 putative protein
Length = 1240
Score = 531 bits (1369), Expect = e-151
Identities = 265/553 (47%), Positives = 372/553 (66%), Gaps = 7/553 (1%)
Query: 714 DASSNPPSFSAESLSNNPPSTEMNPPNHRHSQRIRNPPIHLKDYICKINNVTSKINFPLE 773
DA ++ S S + + + ++ P+ S R P +L+DY C ++V S +
Sbjct: 4 DADASTSSSSIDIMPSANIQNDVPEPSVHTSHRRTRKPAYLQDYYC--HSVASLTIHDIS 61
Query: 774 NYLSLSNLSNSHRAFLINIIENKEPKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLP 833
+LS +S + +FL+ I + KEP +Y++A + + W AM EI A+E+ +TW +C LP
Sbjct: 62 QFLSYEKVSPLYHSFLVCIAKAKEPSTYNEAKEFLVWCGAMDDEIGAMETTHTWEICTLP 121
Query: 834 EGKSAIGCKWIYKIKYHSDGSIDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLS 893
K IGCKW+YKIKY+SDG+I+RYKARLVAKGY+Q +GID+ +TF+PV KL +V+L+L+
Sbjct: 122 PNKKPIGCKWVYKIKYNSDGTIERYKARLVAKGYTQQEGIDFIETFSPVCKLTSVKLILA 181
Query: 894 IAAIKNWPLYQFDVNNAFLQGDLSEEVYMKLPPGFSHKKKP-----CVCKLNKSIYGLKQ 948
I+AI N+ L+Q D++NAFL GDL EE+YMKLPPG++ ++ VC L KSIYGLKQ
Sbjct: 182 ISAIYNFTLHQLDISNAFLNGDLDEEIYMKLPPGYAARQGDSLPPNAVCYLKKSIYGLKQ 241
Query: 949 ASRQWFSKFSTTLIQKGFRQSISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENAISKIK 1008
ASRQWF KFS TLI GF QS SD++ F + VLVYVDDIII NN+ A+ ++K
Sbjct: 242 ASRQWFLKFSVTLIGFGFVQSHSDHTYFLKITATLFLCVLVYVDDIIICSNNDAAVDELK 301
Query: 1009 KFLAQSFSIKDLGNLSYILGIEVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDFPMEQH 1068
L F ++DLG L Y LG+E++RS GI +CQRKY LD+L ++G+ GC+PS PM+
Sbjct: 302 SQLKSCFKLRDLGPLKYFLGLEIARSAAGINICQRKYALDLLDETGLLGCKPSSVPMDPS 361
Query: 1069 LRLRPNDGTPLSDPTVYRRHVGRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDAATQVL 1128
+ + G D YRR +GRL+YL +TR DI +AVN LSQF ++P +H A ++L
Sbjct: 362 VTFSAHSGGDFVDAKAYRRLIGRLMYLQITRLDISFAVNKLSQFSEAPRLAHQQAVMKIL 421
Query: 1129 RYLKGSVGKGLFLSASSSINLVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPT 1188
Y+KG+VG+GLF S+ + + L ++D+ + C TRRST GY LG++ ISWK+KKQ
Sbjct: 422 HYIKGTVGQGLFYSSQAEMQLQVFSDASFQSCKDTRRSTNGYCMFLGTSLISWKSKKQQV 481
Query: 1189 ISRSSAEAEYRSLATLSSELQWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVFHERT 1248
+S+SSAEAEYR+L+ + E+ WL +L + +P ++CD+ AAIHIA N VFHERT
Sbjct: 482 VSKSSAEAEYRALSFATDEMMWLAQFFRELQLPLSKPTLLFCDNTAAIHIATNAVFHERT 541
Query: 1249 KHIEIDCHFVREK 1261
KHIE DCH VRE+
Sbjct: 542 KHIESDCHSVRER 554
>At4g27210 putative protein
Length = 1318
Score = 521 bits (1343), Expect = e-148
Identities = 299/774 (38%), Positives = 431/774 (55%), Gaps = 38/774 (4%)
Query: 554 FGGECVLTAAYLINKLPTPILKFK-SPHQVLLGSPPSYSSLRVFGCLCFAKNMNI-QHKF 611
F E TA +LIN LPT LK SP++ L P Y+SLR FG CF + ++KF
Sbjct: 455 FWVEAFFTANFLINLLPTSALKESISPYEKLYDKKPDYTSLRSFGSACFPTLRDYAENKF 514
Query: 612 DERSKPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFP----YQDIQSPPFNNAI 667
+ S +F+GY KGYR T ++Y+SR V F E+V+P Y+ + P +
Sbjct: 515 NPCSLKCVFLGYNEKYKGYRCLYPPTGRLYISRHVIFDESVYPFSHTYKHLHPQPRTPLL 574
Query: 668 SI----------NTQILDNEFDDLFTGS----------PTHPNIPPENSHNDNSNDTIVT 707
+ +T + LFT + P P + P +S + SN I T
Sbjct: 575 AAWLRSSDSPAPSTSTSPSSRSPLFTSADFPPLPQRKTPLLPTLVPISSVSHASN--ITT 632
Query: 708 ISTPEDDASSNPPSFSAESLSNNPPSTEMNPPNHRHSQRIRNPPIHLKDYICKINNVTSK 767
+P+ D S F + S+ ++ S++ + Q+ + +H ++ + ++
Sbjct: 633 QQSPDFD-SERTTDFDSASIGDSSHSSQAGSDSEETIQQA-SVNVHQTHASTNVHPMVTR 690
Query: 768 INFPLENYLSLSNLSNSHRAFLINIIENKEPKSYSQAMKSVEWRDAMAKEIQALESNNTW 827
+ N FL + + EPK+ + A+K W AM +EI TW
Sbjct: 691 AKVGISK-------PNPRYVFLSHKVSYPEPKTVTAALKHPGWTGAMTEEIGNCSETQTW 743
Query: 828 ILCPLPEGKSAIGCKWIYKIKYHSDGSIDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVT 887
L P +G KW+++ K H+DG++++ KAR+VAKG+ Q +GIDY +T++PV + T
Sbjct: 744 SLVPYKSDMHVLGSKWVFRTKLHADGTLNKLKARIVAKGFLQEEGIDYLETYSPVVRTPT 803
Query: 888 VRLLLSIAAIKNWPLYQFDVNNAFLQGDLSEEVYMKLPPGFSHKKKPC-VCKLNKSIYGL 946
VRL+L +A NW + Q DV NAFL GDL E VYM P GF KP VC L+KSIYGL
Sbjct: 804 VRLVLHLATALNWDIKQMDVKNAFLHGDLKETVYMTQPAGFVDPSKPDHVCLLHKSIYGL 863
Query: 947 KQASRQWFSKFSTTLIQKGFRQSISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENAISK 1006
KQ+ R WF KFST L++ GF S SD SLF Y + I +L+YVDD++ITGN+ ++
Sbjct: 864 KQSPRAWFDKFSTFLLEFGFFCSKSDPSLFIYAHNNNLILLLLYVDDMVITGNSSQTLTS 923
Query: 1007 IKKFLAQSFSIKDLGNLSYILGIEVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDFPME 1066
+ L + F + D+G L Y LGI+V R + G+F+ Q+KY D+L + M C P P+
Sbjct: 924 LLAALNKEFRMTDMGQLHYFLGIQVQRQQNGLFMSQQKYAEDLLIAASMEHCTPLPTPLP 983
Query: 1067 QHLRLRPNDGTPLSDPTVYRRHVGRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDAATQ 1126
L P+ SDPT +R G+L YLT+TRPDIQ+AVN + Q M P S F +
Sbjct: 984 VQLDRVPHQEELFSDPTYFRSIAGKLQYLTLTRPDIQFAVNFVCQKMHQPTISDFHLLKR 1043
Query: 1127 VLRYLKGSVGKGLFLSASSSINLVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQ 1186
+LRY+KG++ G+ S S L Y+DSDW C TRRS G T +G+N +SW +KK
Sbjct: 1044 ILRYIKGTITMGISYSRDSPTLLQAYSDSDWGNCKQTRRSVGGLCTFMGTNLVSWSSKKH 1103
Query: 1187 PTISRSSAEAEYRSLATLSSELQWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVFHE 1246
PT+SRSS EAEY+SL+ +SE+ WL LL +L I P ++CD+ +A+++ NP FH
Sbjct: 1104 PTVSRSSTEAEYKSLSDAASEILWLSTLLRELRIPLPDTPELFCDNLSAVYLTANPAFHA 1163
Query: 1247 RTKHIEIDCHFVREKIKSGLIAPSYIRSSDQLADIFTKPLGGDAYKRILGKLGV 1300
RTKH +ID HFVRE++ + +I S+Q+ADIFTK L +A+ + GKLGV
Sbjct: 1164 RTKHFDIDFHFVRERVALKALVVKHIPGSEQIADIFTKSLPYEAFIHLRGKLGV 1217
Score = 63.5 bits (153), Expect = 7e-10
Identities = 43/154 (27%), Positives = 71/154 (45%), Gaps = 18/154 (11%)
Query: 369 WIIDSGASNHICTSLSLFSSYYPVNNQISVQQPDGSQALVKHIGTINCSPS---LILTNV 425
W+ DS A+ H+ S P + ++ DG+ + H G+ N + S + LT+V
Sbjct: 177 WLPDSAATAHVTNSPRSLQQSQPYHGTDAIMVDDGNYLPITHTGSTNLASSSGTVPLTDV 236
Query: 426 YHVPTFKFNLMSVTQLTESLNCDAIFSSSGCVFQDQATKKMIGRGSARNELYYLNQDLVS 485
P+ +L+S+++LT+ C F G D+ATKK++ GS R+ LY L D
Sbjct: 237 LVCPSITKSLLSMSKLTQDFPCTVEFEYDGVRVNDKATKKLLLMGSNRDGLYCLKDD--- 293
Query: 486 NKHDKDHYCLSHPLSSLFSSKHCNKFD-LWHLRL 518
+ FS++ + D +WH RL
Sbjct: 294 -----------KQFQAFFSTRQRSASDEVWHRRL 316
>At1g47650 hypothetical protein
Length = 1409
Score = 514 bits (1324), Expect = e-145
Identities = 257/507 (50%), Positives = 358/507 (69%), Gaps = 15/507 (2%)
Query: 784 SHRAFLI-NIIENKE-------PKSYSQAMKSVEWRDAMAKEIQALESNNTWILCPLPEG 835
SH F+ N++ ++E PKS S ++K ++ EI A+E+ +TW + LP G
Sbjct: 710 SHTVFISRNVVFHEEVFPLAVDPKSES-SLKLFTPMVPVSSEIGAMETTHTWEITTLPAG 768
Query: 836 KSAIGCKWIYKIKYHSDGSIDRYKARLVAKGYSQVQGIDYHDTFAPVAKLVTVRLLLSIA 895
K A+GCKW++ +K+ +DGS++RYKARLVAKGY+Q +G+DY DTF+PVAK+ T++LLL I+
Sbjct: 769 KKAVGCKWVFSLKFLADGSLERYKARLVAKGYTQKEGLDYTDTFSPVAKMTTIKLLLKIS 828
Query: 896 AIKNWPLYQFDVNNAFLQGDLSEEVYMKLPPGFSHKK-----KPCVCKLNKSIYGLKQAS 950
A K W L Q DV+NAFL G+L EE+YMKLP G++ +K VC+L +SIYGLKQAS
Sbjct: 829 ASKKWFLKQLDVSNAFLNGELEEEIYMKLPEGYAARKGITLPPNSVCRLKRSIYGLKQAS 888
Query: 951 RQWFSKFSTTLIQKGFRQSISDYSLFTYNCDQTTIFVLVYVDDIIITGNNENAISKIKKF 1010
RQWF KFS +L+ GF+++ D++LF D + VLVYVDDI I +E A ++ +
Sbjct: 889 RQWFKKFSASLLDLGFKKTHGDHTLFIKEYDGEFVIVLVYVDDIAIASTSEGAAIQLTQD 948
Query: 1011 LAQSFSIKDLGNLSYILGIEVSRSKKGIFLCQRKYTLDILSDSGMTGCRPSDFPMEQHLR 1070
L Q F ++DLG+L Y LG+E++R++ GI +CQRKY L++L+ +GM C+P PM +++
Sbjct: 949 LQQRFKLRDLGDLKYFLGLEIARTEAGISICQRKYALELLASTGMVNCKPVSVPMIPNVK 1008
Query: 1071 LRPNDGTPLSDPTVYRRHVGRLLYLTVTRPDIQYAVNTLSQFMQSPYSSHFDAATQVLRY 1130
+ DG L D YRR VG+L+YLT+TR DI +AVN L QF +P +SH AA +VL+Y
Sbjct: 1009 MMKTDGDLLDDREQYRRIVGKLMYLTITRLDITFAVNKLCQFSSAPRTSHLQAAYRVLQY 1068
Query: 1131 LKGSVGKGLFLSASSSINLVGYADSDWAGCPTTRRSTTGYFTMLGSNPISWKTKKQPTIS 1190
+KGSVG+GLF SAS+ + L G+ADSDWA CP +RRSTTG+ +G + ISW++KKQ +S
Sbjct: 1069 IKGSVGQGLFYSASADLTLKGFADSDWASCPDSRRSTTGFSMFVGDSLISWRSKKQHVVS 1128
Query: 1191 RSSAEAEYRSLATLSSELQWLKYLLSDLGIDHPQPITIYCDSQAAIHIAENPVFHERTKH 1250
RSSAEAEYR+LA ++ EL WL LL L + P+ +Y DS AI+IA NPVFHERTKH
Sbjct: 1129 RSSAEAEYRALALVTCELVWLHTLLMSLKASYLVPV-LYSDSTTAIYIATNPVFHERTKH 1187
Query: 1251 IEIDCHFVREKIKSGLIAPSYIRSSDQ 1277
IE+DCH VREK+ +G + ++R+ DQ
Sbjct: 1188 IELDCHTVREKLDNGELKLLHVRTEDQ 1214
Score = 220 bits (560), Expect = 5e-57
Identities = 138/455 (30%), Positives = 216/455 (47%), Gaps = 54/455 (11%)
Query: 30 PPSENSPLSSISSPITVHSTTNP--DPCVIHHSDNPSTVLVTPLLTGDNYGSWSRAVTMA 87
PP S +S S I + +P P +H +D+P +++ L NYG W+ A+ ++
Sbjct: 38 PPVHKSNVSGASRVIAPPESVDPTQSPFFLHSADHPGLNIISHRLDETNYGDWNVAMLIS 97
Query: 88 LRAKNKFGFVDGSLTIPKKKE-DILTWQRCNDLVASWILNSVSTEIRPSILYAETAEQIW 146
L AKNK GF+DG+L P + + + W RC +V SW+LNSVS +I SIL A IW
Sbjct: 98 LDAKNKSGFIDGTLPRPLESDKNFRLWSRCKSMVKSWLLNSVSPQIYRSILRMNDASDIW 157
Query: 147 SDLQDRFSQSNAPKIYQLKQSISALKQEGMYVSLYFTQLKSLWDELNSI-ALVNPCICGN 205
DL RF+ +N P+ Y L Q I +Q + +S Y+T+LK+LWD+L S L +PCICG
Sbjct: 158 RDLHSRFNVTNLPRTYNLTQEIQDFRQGTLSLSEYYTRLKTLWDQLESTEELDDPCICGK 217
Query: 206 AKSILDQQNQDRAMEFLQGVHDRFSAVRSQILLMDPFPSIQRIYNIVRQEEKQQEINFRP 265
A + + Q + ++ L G++D ++ +R QI+ PS+ +Y+I+ Q+ QQ +
Sbjct: 218 AMRLQQKAEQAKIVKLLAGLNDSYAIIRRQIIAKKALPSLAEVYHILDQDNSQQGFSNVV 277
Query: 266 VPAVESAALQTSKAPYRTPGKRQRPFCEHCNRHGHTITTCYQIHGFPSNPSKPQKKTETS 325
P AA Q S+A T E GH CY+ HGFP + K + S
Sbjct: 278 AP---PAAFQVSEAMMATNNDVNMLCSEWV---GHIAERCYKKHGFPLGFTPKGKGGDKS 331
Query: 326 PSTSANQLSSAQYHKLLTLLAKEDNVGSSVNLAGTAFTCIPFSWIIDSGASNHICTSLSL 385
+ + + K L L + +V S+VNL
Sbjct: 332 QVIDSGAIHHVSHDKSL-FLNLDTSVVSAVNL---------------------------- 362
Query: 386 FSSYYPVNNQISVQQPDGSQALVKHIGTINCSPSLILTNVYHVPTFKFNLMSVTQLTESL 445
P G + +GT+ + ++L N+ +P F+ NL+S++ LT+ +
Sbjct: 363 ---------------PAGPTVRISGVGTLRLNDDILLKNILFIPEFRLNLISISSLTDDI 407
Query: 446 NCDAIFSSSGCVFQDQATKKMIGRGSARNELYYLN 480
IF + C QD +M+G+G LY L+
Sbjct: 408 GSRVIFDKTSCEIQDPIKARMLGQGRRIANLYVLD 442
Score = 90.9 bits (224), Expect = 4e-18
Identities = 43/99 (43%), Positives = 65/99 (65%), Gaps = 13/99 (13%)
Query: 556 GECVLTAAYLINKLPTPILKFKSPHQVLLGSPPSYSSLRVFGCLCFAKNMNIQHKFDERS 615
G+CVLTA +LIN+ P+ +L K+P+++L G+ P Y FAK +HKF RS
Sbjct: 642 GDCVLTAVFLINRTPSQLLSNKTPYEILTGTVPFY----------FAKK---RHKFQPRS 688
Query: 616 KPGIFVGYPFNQKGYRIYDMKTRQIYVSRDVQFHETVFP 654
K +F+GYP KGY++ D+++ +++SR+V FHE VFP
Sbjct: 689 KACVFLGYPAGYKGYKLMDLESHTVFISRNVVFHEEVFP 727
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.134 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,438,994
Number of Sequences: 26719
Number of extensions: 1360242
Number of successful extensions: 5018
Number of sequences better than 10.0: 159
Number of HSP's better than 10.0 without gapping: 130
Number of HSP's successfully gapped in prelim test: 29
Number of HSP's that attempted gapping in prelim test: 4112
Number of HSP's gapped (non-prelim): 443
length of query: 1309
length of database: 11,318,596
effective HSP length: 111
effective length of query: 1198
effective length of database: 8,352,787
effective search space: 10006638826
effective search space used: 10006638826
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 66 (30.0 bits)
Medicago: description of AC124966.5


