

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124965.10 + phase: 0
(1569 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
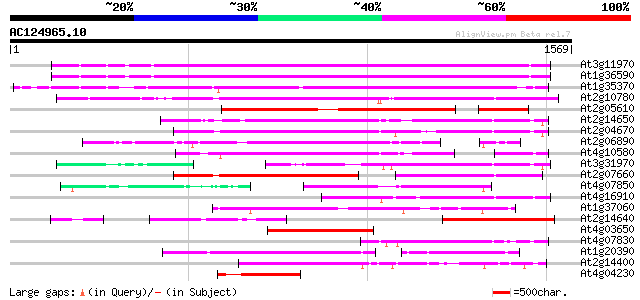
Score E
Sequences producing significant alignments: (bits) Value
At3g11970 hypothetical protein 1095 0.0
At1g36590 hypothetical protein 1087 0.0
At1g35370 hypothetical protein 977 0.0
At2g10780 pseudogene 696 0.0
At2g05610 putative retroelement pol polyprotein 606 e-173
At2g14650 putative retroelement pol polyprotein 604 e-172
At2g04670 putative retroelement pol polyprotein 602 e-172
At2g06890 putative retroelement integrase 488 e-137
At4g10580 putative reverse-transcriptase -like protein 473 e-133
At3g31970 hypothetical protein 408 e-113
At2g07660 putative retroelement pol polyprotein 388 e-107
At4g07850 putative polyprotein 376 e-104
At4g16910 retrotransposon like protein 342 8e-94
At1g37060 Athila retroelment ORF 1, putative 301 3e-81
At2g14640 putative retroelement pol polyprotein 300 3e-81
At4g03650 putative reverse transcriptase 262 1e-69
At4g07830 putative reverse transcriptase 248 3e-65
At1g20390 hypothetical protein 232 1e-60
At2g14400 putative retroelement pol polyprotein 226 6e-59
At4g04230 putative transposon protein 194 3e-49
>At3g11970 hypothetical protein
Length = 1499
Score = 1095 bits (2832), Expect = 0.0
Identities = 582/1410 (41%), Positives = 843/1410 (59%), Gaps = 40/1410 (2%)
Query: 118 KLEFPRFNGKHVLDWIFKAEQYFGYYNTPDAERLIIASVHLEQEVVPWFQMVNRS----R 173
K++FPRF+G + +W+FK E++FG +TP+ ++ +A++H + W Q +S
Sbjct: 111 KIDFPRFDGTRLKEWLFKVEEFFGVDSTPEDMKVKMAAIHFDSHASTWHQSFIQSGVGLE 170
Query: 174 PFQSWNDFTRALELDFGPSIYDCPRASLFKLQQNKSVNEYYLEFTALSNRVYGLSNDALI 233
W + + L+ F D P A L LQ+ + +Y+ +F + RV LS + L+
Sbjct: 171 VLYDWKGYVKLLKERFEDDCDD-PMAELKHLQETDGIIDYHQKFELIKTRV-NLSEEYLV 228
Query: 234 DCFVSGLKDELRRDVMLHTPISIVKAVSLAKLFEEKIAANFNPKQPLKPYTPAQPHQHRA 293
+++GL+ + + V + P ++ + L K +E+ +PK+P
Sbjct: 229 SVYLAGLRTDTQMHVRMFQPQTVRHCLFLGKTYEKA-----HPKKPANTTWSTNRSAPTG 283
Query: 294 PFNPTRNDQNQTIEKAPNLPLLPTPPTRPMSHLQKNPAIKRISPAEMQLRREKGLCYFCD 353
+N + + + N +P K++S EM RR KGLCYFCD
Sbjct: 284 GYNKYQKEGESKTDHYGNKGNFKPVSQQP----------KKMSQQEMSDRRSKGLCYFCD 333
Query: 354 DKFSFSHKCPNKQLMLLELDDDLDPPDTPSQTAVTSDTETQEVAEHHLSLNALKGATGMG 413
+K++ H +K+ L +D D + D + D ++ S+NA+ G G
Sbjct: 334 EKYTPEHYLVHKKTQLFRMDVDEEFEDAREELVNDDDEHMPQI-----SVNAVSGIAGYK 388
Query: 414 VIRFKGYIGPISVSILLDGGSSESFIQPRIVHCLNLPIEHTDKCNVLVGNGQHMKAEGVV 473
+R KG + IL+D GS+ +F+ P L ++ V V +G+ ++ EG V
Sbjct: 389 TMRVKGTYDKKIIFILIDSGSTHNFLDPNTAAKLGCKVDTAGLTRVSVADGRKLRVEGKV 448
Query: 474 RKLSLKVQDIDITVPAYLLPVAGSDVILGAPWLASLGPHVADYSVSKLKFYMDGKFVTLQ 533
S K+Q L+P+ G D++LG WL +LG ++ +++F + + V L
Sbjct: 449 TDFSWKLQTTTFQSDILLIPLQGIDMVLGVQWLETLGRISWEFKKLEMRFKFNNQKVLLH 508
Query: 534 GESDNKPAVSQLNHFRRLQHMNAISELFTIQKIDPAVIED----NWEGMPVNIEPEMATL 589
G + + ++LQ + +Q++ + + N + E + +
Sbjct: 509 GLTSGSVREVKAQKLQKLQEDQVQLAMLCVQEVSESTEGELCTINALTSELGEESVVEEV 568
Query: 590 LHTYREIFQIPKGLPPNREL-SHEILLKEGAQPVKVKPYRYPHSQKEQIEKMVQDMLEEG 648
L+ Y +IF P LPP RE +H+I L EG+ PV +PYRY QK +I+K+V+D+L G
Sbjct: 569 LNEYPDIFIEPTALPPFREKHNHKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNG 628
Query: 649 IIQPSLSPFSSPIILVKKKDGTWRCCTDYRALNAITIKDSFPMPTVDELLDELHGAQYFT 708
+Q S SP++SP++LVKKKDGTWR C DYR LN +T+KDSFP+P +++L+DEL GA F+
Sbjct: 629 TVQASSSPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFS 688
Query: 709 KLDLRSGYHQILLKPEDRQKTAFRTHQGHYEWLVMPFGLTSAPATFQRLMNQLFQPLLRK 768
K+DLR+GYHQ+ + P+D QKTAF+TH GH+E+LVMPFGLT+APATFQ LMN +F+P LRK
Sbjct: 689 KIDLRAGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRK 748
Query: 769 CVLVFFDDILVYSPSWHSHLKHVEIVFQILSQNVLYAKLSKCSFGITEVEYLGHVVSGSG 828
VLVFFDDILVYS S H +H++ VF+++ N L+AKLSKC+F + +VEYLGH +S G
Sbjct: 749 FVLVFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQG 808
Query: 829 VAMDKIKVIAVLEWPTPTNLKQLRGFLGLTGYYRRFIRSYATIAGPLTNLLKKDAFKWDE 888
+ D K+ AV EWP PT LKQLRGFLGL GYYRRF+RS+ IAGPL L K DAF+W
Sbjct: 809 IETDPAKIKAVKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIAGPLHALTKTDAFEWTA 868
Query: 889 STAKAFDTLKQAITTAPVLVLPDFSQPFVLETDASGTGVGAVLSQGGHPIAYFSKKMASR 948
+AF+ LK A+ APVL LP F + FV+ETDA G G+GAVL Q GHP+AY S+++ +
Sbjct: 869 VAQQAFEDLKAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQEGHPLAYISRQLKGK 928
Query: 949 MQLQSAYTREFYAITTALAKFRHYLLGHKFILRTDQKSLKSLLDQSLQTPEQQAWLHKFI 1008
S Y +E A+ A+ K+RHYLL FI++TDQ+SLK LL+Q L TP QQ WL K +
Sbjct: 929 QLHLSIYEKELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPKLL 988
Query: 1009 GFDFQIEYKPGKDNQAADALSR-----VMSLSWSAPEHDFLEQLKKEIGNDSHLQTIVQQ 1063
FD++I+Y+ GK+N ADALSR V+ ++ + E D L+ ++ NDS LQ I+
Sbjct: 989 EFDYEIQYRQGKENVVADALSRVEGSEVLHMAMTVVECDLLKDIQAGYANDSQLQDIITA 1048
Query: 1064 CVNNTMDDINYMVKEGLLYWKHRLVIPMESNLIHQILKEYHDTPIGGHAGVTRTLARVTA 1123
+ + + +L K ++V+P N+ + IL H + +GGH+G T RV
Sbjct: 1049 LQRDPDSKKYFSWSQNILRRKSKIVVPANDNIKNTILLWLHGSGVGGHSGRDVTHQRVKG 1108
Query: 1124 QFYWPNMRQHIQKFIEACVTCQQAKSVNTTYAGLLQPLPIPEQVWDDVTMDFITGLPLSF 1183
FYW M + IQ +I +C TCQQ KS GLLQPLPIP+ +W +V+MDFI GLP+S
Sbjct: 1109 LFYWKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPLPIPDTIWSEVSMDFIEGLPVSG 1168
Query: 1184 GFTVIMVVVDRLSKYAHFMPLKADYSNRTVAEAFVNYVVKLHGMPKSIISDRDKVFTSKF 1243
G TVIMVVVDRLSK AHF+ L YS TVA A+++ V KLHG P SI+SDRD VFTS+F
Sbjct: 1169 GKTVIMVVVDRLSKAAHFIALSHPYSALTVAHAYLDNVFKLHGCPTSIVSDRDVVFTSEF 1228
Query: 1244 WQHLFQMQGTTLSMSTTYHPQTDGQSEAVNKCLEMYLRCFTFKNPKTWFKALTWVELWYN 1303
W+ F +QG L +++ YHPQ+DGQ+E VN+CLE YLRC P+ W K L E WYN
Sbjct: 1229 WREFFTLQGVALKLTSAYHPQSDGQTEVVNRCLETYLRCMCHDRPQLWSKWLALAEYWYN 1288
Query: 1304 TFLHTSLGMSPFKALYGREPPMLTRYSVNNSDPTEVQQQLMDRDKLLVELKENLLRAQQV 1363
T H+S M+PF+ +YG+ PP+ Y S V + L +R+ +L+ LK +L+RAQ
Sbjct: 1289 TNYHSSSRMTPFEIVYGQVPPVHLPYLPGESKVAVVARSLQEREDMLLFLKFHLMRAQHR 1348
Query: 1364 MKSNADKKRKDVSFEVGDQVLVKLQPYRQSSVALRKNNKLGMRYFGPFTVIAKVGVVAYK 1423
MK AD+ R + FE+GD V VKLQPYRQ SV +R N KL +YFGP+ +I + G VAYK
Sbjct: 1349 MKQFADQHRTEREFEIGDYVYVKLQPYRQQSVVMRANQKLSPKYFGPYKIIDRCGEVAYK 1408
Query: 1424 LQLPETARIHPVFHVSQLKIFKG-LTTEPYFPLPLTTMEEGPVIQPEVILNVRNIIRGDR 1482
L LP +++HPVFHVSQLK+ G ++T + P + M++ PE ++ + + R +
Sbjct: 1409 LALPSYSQVHPVFHVSQLKVLVGNVSTTVHLP---SVMQDVFEKVPEKVVERKMVNRQGK 1465
Query: 1483 KVEQLLVKWKDMQNSEATWEDKQEMLDSYP 1512
V ++LVKW + EATWE ++ ++P
Sbjct: 1466 AVTKVLVKWSNEPLEEATWEFLFDLQKTFP 1495
>At1g36590 hypothetical protein
Length = 1499
Score = 1087 bits (2810), Expect = 0.0
Identities = 580/1410 (41%), Positives = 842/1410 (59%), Gaps = 40/1410 (2%)
Query: 118 KLEFPRFNGKHVLDWIFKAEQYFGYYNTPDAERLIIASVHLEQEVVPWFQMVNRS----R 173
K++FPRF+G + +W+FK E++FG +TP+ ++ +A++H + W Q +S
Sbjct: 111 KIDFPRFDGTRLKEWLFKVEEFFGVDSTPEDMKVKMAAIHFDSHASTWHQSFIQSGVGLE 170
Query: 174 PFQSWNDFTRALELDFGPSIYDCPRASLFKLQQNKSVNEYYLEFTALSNRVYGLSNDALI 233
W + + L+ F D P A L LQ+ + +Y+ +F + RV LS + L+
Sbjct: 171 VLYDWKGYVKLLKERFEDDCDD-PMAELKHLQETDGIIDYHQKFELIKTRV-NLSEEYLV 228
Query: 234 DCFVSGLKDELRRDVMLHTPISIVKAVSLAKLFEEKIAANFNPKQPLKPYTPAQPHQHRA 293
+++GL+ + + V + P ++ + L K +E+ + K+P
Sbjct: 229 SVYLAGLRTDTQMHVRMFQPQTVRHCLFLGKTYEKA-----HLKKPANTTWSTNRSAPTG 283
Query: 294 PFNPTRNDQNQTIEKAPNLPLLPTPPTRPMSHLQKNPAIKRISPAEMQLRREKGLCYFCD 353
+N + + + N +P K++S EM RR KGLCYFCD
Sbjct: 284 GYNKYQKEGESKTDHYGNKGNFKPVSQQP----------KKMSQQEMSDRRSKGLCYFCD 333
Query: 354 DKFSFSHKCPNKQLMLLELDDDLDPPDTPSQTAVTSDTETQEVAEHHLSLNALKGATGMG 413
+K++ H +K+ L +D D + D + D ++ S+NA+ G G
Sbjct: 334 EKYTPEHYLVHKKTQLFRMDVDEEFEDAREELVNDDDEHMPQI-----SVNAVSGIAGYK 388
Query: 414 VIRFKGYIGPISVSILLDGGSSESFIQPRIVHCLNLPIEHTDKCNVLVGNGQHMKAEGVV 473
+R KG + IL+D GS+ +F+ P L ++ V V +G+ ++ EG V
Sbjct: 389 TMRVKGTYDKKIIFILIDSGSTHNFLDPNTAAKLGCKVDTAGLTRVSVADGRKLRVEGKV 448
Query: 474 RKLSLKVQDIDITVPAYLLPVAGSDVILGAPWLASLGPHVADYSVSKLKFYMDGKFVTLQ 533
S K+Q L+P+ G D++LG WL +LG ++ +++F + + V L
Sbjct: 449 TDFSWKLQTTTFQSDILLIPLQGIDMVLGVQWLETLGRISWEFKKLEMRFKFNNQKVLLH 508
Query: 534 GESDNKPAVSQLNHFRRLQHMNAISELFTIQKIDPAVIED----NWEGMPVNIEPEMATL 589
G + + ++LQ + +Q++ + + N + E + +
Sbjct: 509 GLTSGSVREVKAQKLQKLQEDQVQLAMLCVQEVSESTEGELCTINALTSELGEESVVEEV 568
Query: 590 LHTYREIFQIPKGLPPNREL-SHEILLKEGAQPVKVKPYRYPHSQKEQIEKMVQDMLEEG 648
L+ Y +IF P LPP RE +H+I L EG+ PV +PYRY QK +I+K+V+D+L G
Sbjct: 569 LNEYPDIFIEPTALPPFREKHNHKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNG 628
Query: 649 IIQPSLSPFSSPIILVKKKDGTWRCCTDYRALNAITIKDSFPMPTVDELLDELHGAQYFT 708
+Q S SP++SP++LVKKKDGTWR C DYR LN +T+KDSFP+P +++L+DEL GA F+
Sbjct: 629 TVQASSSPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFS 688
Query: 709 KLDLRSGYHQILLKPEDRQKTAFRTHQGHYEWLVMPFGLTSAPATFQRLMNQLFQPLLRK 768
K+DLR+GYHQ+ + P+D QKTAF+TH GH+E+LVMPFGLT+APATFQ LMN +F+P LRK
Sbjct: 689 KIDLRAGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRK 748
Query: 769 CVLVFFDDILVYSPSWHSHLKHVEIVFQILSQNVLYAKLSKCSFGITEVEYLGHVVSGSG 828
VLVFFDDILVYS S H +H++ VF+++ N L+AKLSKC+F + +VEYLGH +S G
Sbjct: 749 FVLVFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQG 808
Query: 829 VAMDKIKVIAVLEWPTPTNLKQLRGFLGLTGYYRRFIRSYATIAGPLTNLLKKDAFKWDE 888
+ D K+ AV EWP PT LKQLRGFLGL GYYRRF+RS+ IAGPL L K DAF+W
Sbjct: 809 IETDPAKIKAVKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIAGPLHALTKTDAFEWTA 868
Query: 889 STAKAFDTLKQAITTAPVLVLPDFSQPFVLETDASGTGVGAVLSQGGHPIAYFSKKMASR 948
+AF+ LK A+ APVL LP F + FV+ETDA G G+GAVL Q GHP+AY S+++ +
Sbjct: 869 VAQQAFEDLKAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQEGHPLAYISRQLKGK 928
Query: 949 MQLQSAYTREFYAITTALAKFRHYLLGHKFILRTDQKSLKSLLDQSLQTPEQQAWLHKFI 1008
S Y +E A+ A+ K+RHYLL FI++TDQ+SLK LL+Q L TP QQ WL K +
Sbjct: 929 QLHLSIYEKELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPKLL 988
Query: 1009 GFDFQIEYKPGKDNQAADALSR-----VMSLSWSAPEHDFLEQLKKEIGNDSHLQTIVQQ 1063
FD++I+Y+ GK+N ADALSR V+ ++ + E D L+ ++ NDS LQ I+
Sbjct: 989 EFDYEIQYRQGKENVVADALSRVEGSEVLHMAMTVVECDLLKDIQAGYANDSQLQDIITA 1048
Query: 1064 CVNNTMDDINYMVKEGLLYWKHRLVIPMESNLIHQILKEYHDTPIGGHAGVTRTLARVTA 1123
+ + + +L K ++V+P N+ + IL H + +GGH+G T RV
Sbjct: 1049 LQRDPDSKKYFSWSQNILRRKSKIVVPANDNIKNTILLWLHGSGVGGHSGRDVTHQRVKG 1108
Query: 1124 QFYWPNMRQHIQKFIEACVTCQQAKSVNTTYAGLLQPLPIPEQVWDDVTMDFITGLPLSF 1183
FY M + IQ +I +C TCQQ KS GLLQPLPIP+ +W +V+MDFI GLP+S
Sbjct: 1109 LFYSKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPLPIPDTIWSEVSMDFIEGLPVSG 1168
Query: 1184 GFTVIMVVVDRLSKYAHFMPLKADYSNRTVAEAFVNYVVKLHGMPKSIISDRDKVFTSKF 1243
G TVIMVVVDRLSK AHF+ L YS TVA+A+++ V KLHG P SI+SDRD VFTS+F
Sbjct: 1169 GKTVIMVVVDRLSKAAHFIALSHPYSALTVAQAYLDNVFKLHGCPTSIVSDRDVVFTSEF 1228
Query: 1244 WQHLFQMQGTTLSMSTTYHPQTDGQSEAVNKCLEMYLRCFTFKNPKTWFKALTWVELWYN 1303
W+ F +QG L +++ YHPQ+DGQ+E VN+CLE YLRC P+ W K L E WYN
Sbjct: 1229 WREFFTLQGVALKLTSAYHPQSDGQTEVVNRCLETYLRCMCHDRPQLWSKWLALAEYWYN 1288
Query: 1304 TFLHTSLGMSPFKALYGREPPMLTRYSVNNSDPTEVQQQLMDRDKLLVELKENLLRAQQV 1363
T H+S M+PF+ +YG+ PP+ Y S V + L +R+ +L+ LK +L+RAQ
Sbjct: 1289 TNYHSSSRMTPFEIVYGQVPPVHLPYLPGESKVAVVARSLQEREDMLLFLKFHLMRAQHR 1348
Query: 1364 MKSNADKKRKDVSFEVGDQVLVKLQPYRQSSVALRKNNKLGMRYFGPFTVIAKVGVVAYK 1423
MK AD+ R + FE+GD V VKLQPYRQ SV +R N KL +YFGP+ +I + G VAYK
Sbjct: 1349 MKQFADQHRTEREFEIGDYVYVKLQPYRQQSVVMRANQKLSPKYFGPYKIIDRCGEVAYK 1408
Query: 1424 LQLPETARIHPVFHVSQLKIFKG-LTTEPYFPLPLTTMEEGPVIQPEVILNVRNIIRGDR 1482
L LP +++HPVFHVSQLK+ G ++T + P + M++ PE ++ + + R +
Sbjct: 1409 LALPSYSQVHPVFHVSQLKVLVGNVSTTVHLP---SVMQDVFEKVPEKVVERKMVNRQGK 1465
Query: 1483 KVEQLLVKWKDMQNSEATWEDKQEMLDSYP 1512
V ++LVKW + EATWE ++ ++P
Sbjct: 1466 AVTKVLVKWSNEPLEEATWEFLFDLQKTFP 1495
>At1g35370 hypothetical protein
Length = 1447
Score = 977 bits (2525), Expect = 0.0
Identities = 563/1515 (37%), Positives = 844/1515 (55%), Gaps = 122/1515 (8%)
Query: 12 TELRRQAEAQEKSNVENKARFDRLEAMQTASDLRFNQLSATLERFMQQAPQHNVSHGNTN 71
T+L Q KS +++ A D + AM + R + L+R + ++
Sbjct: 28 TKLEEMVAEQHKSMLKHMA--DMMSAMSRTTAKRVTEGEKVLDRSVPRS----------- 74
Query: 72 SGSSSSVISATVVGSQSTFVGHTQSAVVGKQSTSSMSSQPFQVRHIKLEFPRFNGKHVLD 131
S+SS+ + V Q F + +V ++ + + R K++FPRF+G + +
Sbjct: 75 --STSSMARSGFVEHQRDFQRDFRPEMVRQEVNNQYGNL---TRLGKIDFPRFDGSRINE 129
Query: 132 WIFKAEQYFGYYNTPDAERLIIASVHLEQEVVPWFQMVNRSR----PFQSWNDFTRALEL 187
W+FK E++FG TP+ ++ + ++H + W +S F +W ++ + L+
Sbjct: 130 WLFKVEEFFGVDFTPEEMKVKMVAIHFDSHAATWHHSFIQSGIGLDVFFNWPEYVKLLKD 189
Query: 188 DFGPSIYDCPRASLFKLQQNKSVNEYYLEFTALSNRVYGLSNDALIDCFVSGLKDELRRD 247
F + D P A L KLQ+ + EY+ +F + R+ LS + L+ +++GL+ + +
Sbjct: 190 RFEDACDD-PMAELKKLQETDGIVEYHQQFELIKVRL-NLSEEYLVSVYLAGLRTDTQMH 247
Query: 248 VMLHTPISIVKAVSLAKLFEEKIAANFNPKQPLKPYTPAQPHQHRAPFNPTRNDQNQTIE 307
V + P ++ + L K +E +PK+ + + + + P + + ++
Sbjct: 248 VRMFEPKTVRDCLRLGKYYERA-----HPKKTVSSTWSQKGTRSGGSYRPVKEVEQKS-- 300
Query: 308 KAPNLPLLPTPPTRPMSHLQKNPAIKRISPAEMQLRREKGLCYFCDDKFSFSHKCPNKQL 367
HL GLCYFCD+KF+ H +K+
Sbjct: 301 ----------------DHL--------------------GLCYFCDEKFTPEHYLVHKKT 324
Query: 368 MLLELDDDLDPPDTPSQTAVTSDTETQEVAEHHLSLNALKGATGMGVIRFKGYIGPISVS 427
L +D D + D V SD + ++ +S+NA+ G +G + KG + +
Sbjct: 325 QLFRMDVDEEFEDA---VEVLSDDDHEQKPMPQISVNAVSGISGYKTMGVKGTVDKRDLF 381
Query: 428 ILLDGGSSESFIQPRIVHCLNLPIEHTDKCNVLVGNGQHMKAEGVVRKLSLKVQDIDITV 487
IL+D GS+ +FI + L +E V V +G+ + +G ++ + K+Q
Sbjct: 382 ILIDSGSTHNFIDSTVAAKLGCHVESAGLTKVAVADGRKLNVDGQIKGFTWKLQSTTFQS 441
Query: 488 PAYLLPVAGSDVILGAPWLASLGPHVADYSVSKLKFYMDGKFVTLQGESDNKPAVSQLNH 547
L+P+ G D++LG WL +LG ++ +++F+ + V L G + +
Sbjct: 442 DILLIPLQGVDMVLGVQWLETLGRISWEFKKLEMQFFYKNQRVWLHGIITGSVRDIKAHK 501
Query: 548 FRRLQHMNAISELFTIQKIDPAVIEDNWEGMPVNI-------EPEMATLLHTYREIFQIP 600
++ Q + ++++ V ++ E ++ E + ++ + ++F P
Sbjct: 502 LQKTQADQIQLAMVCVREV---VSDEEQEIGSISALTSDVVEESVVQNIVEEFPDVFAEP 558
Query: 601 KGLPPNREL-SHEILLKEGAQPVKVKPYRYPHSQKEQIEKMVQDMLEEGIIQPSLSPFSS 659
LPP RE H+I L EGA PV +PYRY QK++I+K+VQDM++ G IQ S SPF+S
Sbjct: 559 TDLPPFREKHDHKIKLLEGANPVNQRPYRYVVHQKDEIDKIVQDMIKSGTIQVSSSPFAS 618
Query: 660 PIILVKKKDGTWRCCTDYRALNAITIKDSFPMPTVDELLDELHGAQYFTKLDLRSGYHQI 719
P++LVKKKDGTWR C DY LN +T+KD F +P +++L+DEL G+ F+K+DLR+GYHQ+
Sbjct: 619 PVVLVKKKDGTWRLCVDYTELNGMTVKDRFLIPLIEDLMDELGGSVVFSKIDLRAGYHQV 678
Query: 720 LLKPEDRQKTAFRTHQGHYEWLVMPFGLTSAPATFQRLMNQLFQPLLRKCVLVFFDDILV 779
+ P+D QKTAF+TH GH+E+LVM FGLT+APATFQ LMN +F+ LRK VLVFFDDIL+
Sbjct: 679 RMDPDDIQKTAFKTHNGHFEYLVMLFGLTNAPATFQSLMNSVFRDFLRKFVLVFFDDILI 738
Query: 780 YSPSWHSHLKHVEIVFQILSQNVLYAKLSKCSFGITEVEYLGHVVSGSGVAMDKIKVIAV 839
YS S H +H+ +VF+++ + L+AK SK E+LGH +S + D K+ AV
Sbjct: 739 YSSSIEEHKEHLRLVFEVMRLHKLFAKGSK--------EHLGHFISAREIETDPAKIQAV 790
Query: 840 LEWPTPTNLKQLRGFLGLTGYYRRFIRSYATIAGPLTNLLKKDAFKWDESTAKAFDTLKQ 899
EWPTPT +KQ+RGFLG GYYRRF+R++ IAGPL L K D F W AFDTLK
Sbjct: 791 KEWPTPTTVKQVRGFLGFAGYYRRFVRNFGVIAGPLHALTKTDGFCWSLEAQSAFDTLKA 850
Query: 900 AITTAPVLVLPDFSQPFVLETDASGTGVGAVLSQGGHPIAYFSKKMASRMQLQSAYTREF 959
+ APVL LP F + F++ETDA G G+ AVL Q GHP+AY S+++ + S Y +E
Sbjct: 851 VLCNAPVLALPVFDKQFMVETDACGQGIRAVLMQKGHPLAYISRQLKGKQLHLSIYEKEL 910
Query: 960 YAITTALAKFRHYLLGHKFILRTDQKSLKSLLDQSLQTPEQQAWLHKFIGFDFQIEYKPG 1019
A A+ K+RHYLL FI++TDQ+SLK LL+Q L TP QQ WL K + FD++I+Y+ G
Sbjct: 911 LAFIFAVRKWRHYLLPSHFIIKTDQRSLKYLLEQRLNTPVQQQWLPKLLEFDYEIQYRQG 970
Query: 1020 KDNQAADALSR-----VMSLSWSAPEHDFLEQLKKEIGNDSHLQTIVQQCVNNTMDDINY 1074
K+N ADALSR V+ ++ S E DFL++++ +D L+ I+ + +Y
Sbjct: 971 KENLVADALSRVEGSEVLHMALSIVECDFLKEIQVAYESDGVLKDIISALQQHPDAKKHY 1030
Query: 1075 MVKEGLLYWKHRLVIPMESNLIHQILKEYHDTPIGGHAGVTRTLARVTAQFYWPNMRQHI 1134
+ +L K ++V+P + + +++L+ H + +GG +G + RV + FYW M + I
Sbjct: 1031 SWSQDILRRKSKIVVPNDVEITNKLLQWLHCSGMGGRSGRDASHQRVKSLFYWKGMVKDI 1090
Query: 1135 QKFIEACVTCQQAKSVNTTYAGLLQPLPIPEQVWDDVTMDFITGLPLSFGFTVIMVVVDR 1194
Q FI +C TCQQ KS N Y GLLQPLPIP+++W DV+MDFI GLP S G +VIMVVVDR
Sbjct: 1091 QAFIRSCGTCQQCKSDNAAYPGLLQPLPIPDKIWCDVSMDFIEGLPNSGGKSVIMVVVDR 1150
Query: 1195 LSKYAHFMPLKADYSNRTVAEAFVNYVVKLHGMPKSIISDRDKVFTSKFWQHLFQMQGTT 1254
LSK AHF+ L YS TVA+AF++ V K HG P SI+SDRD +FTS FW+ F++QG
Sbjct: 1151 LSKAAHFVALAHPYSALTVAQAFLDNVYKHHGCPTSIVSDRDVLFTSDFWKEFFKLQGVE 1210
Query: 1255 LSMSTTYHPQTDGQSEAVNKCLEMYLRCFTFKNPKTWFKALTWVELWYNTFLHTSLGMSP 1314
L MS+ YHPQ+DGQ+E VN+CLE YLRC P W K L E WYNT H+S M+P
Sbjct: 1211 LRMSSAYHPQSDGQTEVVNRCLENYLRCMCHARPHLWNKWLPLAEYWYNTNYHSSSQMTP 1270
Query: 1315 FKALYGREPPMLTRYSVNNSDPTEVQQQLMDRDKLLVELKENLLRAQQVMKSNADKKRKD 1374
F+ +YG+ PP+ Y S V + L +R+ +L+ LK +L+RAQ MK AD+ R +
Sbjct: 1271 FELVYGQAPPIHLPYLPGKSKVAVVARSLQERENMLLFLKFHLMRAQHRMKQFADQHRTE 1330
Query: 1375 VSFEVGDQVLVKLQPYRQSSVALRKNNKLGMRYFGPFTVIAKVGVVAYKLQLPETARIHP 1434
+F++GD V VKLQPYRQ SV LR N KL +YFGP+ +I K G V
Sbjct: 1331 RTFDIGDFVYVKLQPYRQQSVVLRVNQKLSPKYFGPYKIIEKCGEV-------------- 1376
Query: 1435 VFHVSQLKIFKGLTTEPYFPLPLTTMEEGPVIQPEVILNVRNIIRGDRKVEQLLVKWKDM 1494
+ +TT P L + E PE IL + + R R +LVKW
Sbjct: 1377 --------MVGNVTTSTQLPSVLPDIFEK---APEYILERKLVKRQGRAATMVLVKWIGE 1425
Query: 1495 QNSEATWE---DKQE 1506
EATW+ D+Q+
Sbjct: 1426 PVEEATWKFLFDRQQ 1440
>At2g10780 pseudogene
Length = 1611
Score = 696 bits (1795), Expect = 0.0
Identities = 475/1467 (32%), Positives = 729/1467 (49%), Gaps = 122/1467 (8%)
Query: 131 DWIFKAEQYFGYYNTPDAERLIIASVHLEQEVVPWFQMVNRSR--PFQSWNDFTRALELD 188
+W + ++ F P+ + IA LE + W+ V + R +S+ DF
Sbjct: 193 EWRSRLKRNFKSTRCPEDYQRDIAVHFLEGDAHNWWLTVEKRRGDEVRSFADFEDEFNKK 252
Query: 189 -FGPSIYDCPRASLFKLQQ-NKSVNEYYLEFTALSNRVYGLS---NDALIDCFVSGLKDE 243
F P +D + L Q N++V EY EF L R G A + F+ GL+ E
Sbjct: 253 YFPPEAWDRLECAYLDLVQGNRTVREYDEEFNRL-RRYVGRELEEEQAQLRRFIRGLRIE 311
Query: 244 LRRDVMLHTPISIVKAVSLAKLFEEKIAANF---NPKQPLKPYTPAQPHQHRAPFNPTRN 300
+R ++ T S+ + V A + EE I K P++ +P + F+ N
Sbjct: 312 IRNHCLVRTFNSVSELVERAAMIEEGIEEERYLNREKAPIRNNQSTKPADKKRKFDKVDN 371
Query: 301 DQN-----QTIEKAPNLPLLPTPPTRPMSHL-QKNPAIK---RISPAEMQ-LRREKGLCY 350
++ + + N K+ AI+ ++ P + + L E C+
Sbjct: 372 TKSDAKTGECVTCGKNHSGTCWKAIGACGRCGSKDHAIQSCPKMEPGQSKVLGEETRTCF 431
Query: 351 FCDDKFSFSHKCPNKQLMLLELDDDLDPPDTPSQTAVTSDTETQEVAEHHLSLNALKGAT 410
+C +CP L + D + + Q VA L+ + A
Sbjct: 432 YCGKTGHLKRECPK-----LTAEKQAGQRDNRGGNGLPPPPKRQAVASRVYELS--EEAN 484
Query: 411 GMGVIRFKGYIGPISVSILLDGGSSESFIQPRIVHCLNLPIEHTDKCNVLVGNGQHMKAE 470
G R + GG + +TD V GQ M
Sbjct: 485 DAGNFR------------AITGGFRKE--------------PNTDYGMVRAAGGQAMYPT 518
Query: 471 GVVRKLSLKVQDIDITVPAYLLPVAGSDVILGAPWLASLGPHVADYSVSKLKFYMDGKFV 530
G+VR +S+ V +++ ++P+ DVILG WL H+ D +++F D +
Sbjct: 519 GLVRGISVVVNGVNMPADLIIVPLKKHDVILGMDWLGKYKAHI-DCHRGRIQFERDEGML 577
Query: 531 TLQG--ESDNKPAVSQLNHFRRL-QHMNAISELFTIQKIDPAVIEDNWEGMPVNIEPEMA 587
QG + +S + R L + A T +++ + E +
Sbjct: 578 KFQGIRTTSGSLVISAIQAERMLGKGCEAYLATITTKEVGASA------------ELKDI 625
Query: 588 TLLHTYREIFQIPKGLPPNRELSHEILLKEGAQPVKVKPYRYPHSQKEQIEKMVQDMLEE 647
+++ + ++F G+PP+R I L+ G P+ PYR ++ +++K ++++L++
Sbjct: 626 PIVNEFSDVFAAVSGVPPDRSDPFTIELEPGTTPISKAPYRMAPAEMAKLKKQLEELLDK 685
Query: 648 GIIQPSLSPFSSPIILVKKKDGTWRCCTDYRALNAITIKDSFPMPTVDELLDELHGAQYF 707
G I+PS SP+ +P++ VKKKDG++R C DYR LN +T+K+ +P+P +DEL+D+L GAQ+F
Sbjct: 686 GFIRPSSSPWGAPVLFVKKKDGSFRLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWF 745
Query: 708 TKLDLRSGYHQILLKPEDRQKTAFRTHQGHYEWLVMPFGLTSAPATFQRLMNQLFQPLLR 767
+K+DL SGYHQI ++P D +KTAFRT H+E++VMPFGLT+APA F ++MN +F+ L
Sbjct: 746 SKIDLASGYHQIPIEPTDVRKTAFRTRYDHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLD 805
Query: 768 KCVLVFFDDILVYSPSWHSHLKHVEIVFQILSQNVLYAKLSKCSFGITEVEYLGHVVSGS 827
+ V++F +DILVYS SW +H +H+ V + L ++ L+AKLSKCSF V +LGHV+S
Sbjct: 806 EFVIIFINDILVYSKSWEAHQEHLRAVLERLREHELFAKLSKCSFWQRSVGFLGHVISDQ 865
Query: 828 GVAMDKIKVIAVLEWPTPTNLKQLRGFLGLTGYYRRFIRSYATIAGPLTNLLKKD-AFKW 886
GV++D K+ ++ EWP P N ++R FLGL GYYRRF+ S+A++A PLT L KD AF W
Sbjct: 866 GVSVDPEKIRSIKEWPRPRNATEIRSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNW 925
Query: 887 DESTAKAFDTLKQAITTAPVLVLPDFSQPFVLETDASGTGVGAVLSQGGHPIAYFSKKMA 946
+ K+F LK +T APVLVLP+ +P+ + TDAS G+G VL Q G IAY S+++
Sbjct: 926 SDECEKSFLELKAMLTNAPVLVLPEEGEPYTVYTDASIVGLGCVLMQKGSVIAYASRQLR 985
Query: 947 SRMQLQSAYTREFYAITTALAKFRHYLLGHKFILRTDQKSLKSLLDQSLQTPEQQAWLHK 1006
+ + E A+ L +R YL G K + TD KSLK + Q Q+ W+
Sbjct: 986 KHEKNYPTHDLEMAAVVFFLKIWRSYLYGAKVQIYTDHKSLKYIFTQPELNLRQRRWMEL 1045
Query: 1007 FIGFDFQIEYKPGKDNQAADALSR----------------------VMSLS--------W 1036
++ I Y PGK NQ ADALSR V +LS
Sbjct: 1046 VADYNLDIAYHPGKANQVADALSRRRSEVEAERSQVDLVNMMGTLHVNALSKEVEPLGLG 1105
Query: 1037 SAPEHDFLEQLKKEIGNDSHLQTIVQQCVNNTMDDINYMVKEGLLYWKHRLVIPMESNLI 1096
+A + D L +++ D ++ Q NN + G + R+ +P + L
Sbjct: 1106 AADQADLLSRIRLAQERDEEIKGWAQ---NNKTE--YQTSNNGTIVVNGRVCVPNDRALK 1160
Query: 1097 HQILKEYHDTPIGGHAGVTRTLARVTAQFYWPNMRQHIQKFIEACVTCQQAKSVNTTYAG 1156
+IL+E H + H G + + ++W M++ + +++ C TCQ K+ + +G
Sbjct: 1161 EEILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWVAKCPTCQLVKAEHQVPSG 1220
Query: 1157 LLQPLPIPEQVWDDVTMDFITGLP--LSFGFTVIMVVVDRLSKYAHFMPLKADYSNRTVA 1214
LLQ LPIPE WD +TMDF+TGLP + + VVVDRL+K AHFM + +A
Sbjct: 1221 LLQNLPIPEWKWDHITMDFVTGLPTGIKSKHNAVWVVVDRLTKSAHFMAISDKDGAEIIA 1280
Query: 1215 EAFVNYVVKLHGMPKSIISDRDKVFTSKFWQHLFQMQGTTLSMSTTYHPQTDGQSEAVNK 1274
E +++ +V+LHG+P SI+SDRD FTSKFW+ + GT +++ST YHPQTD QSE +
Sbjct: 1281 EKYIDEIVRLHGIPVSIVSDRDTRFTSKFWKAFQKALGTRVNLSTAYHPQTDEQSERTIQ 1340
Query: 1275 CLEMYLRCFTFKNPKTWFKALTWVELWYNTFLHTSLGMSPFKALYG---REPPMLTRYSV 1331
LE LR W K L VE YN S+GMSP++ALYG R P T
Sbjct: 1341 TLEDMLRACVLDWGGNWEKYLRLVEFAYNNSFQASIGMSPYEALYGRACRTPLCWTPVGE 1400
Query: 1332 NNSDPTEVQQQLMDRDKLL-VELKENLLRAQQVMKSNADKKRKDVSFEVGDQVLVKLQPY 1390
+ + +R K L ++LKE AQ KS A+K+RK++ F+VGD V +K Y
Sbjct: 1401 RRLFGPTIVDETTERMKFLKIKLKE----AQDRQKSYANKRRKELEFQVGDLVYLKAMTY 1456
Query: 1391 RQSSVALRKNNKLGMRYFGPFTVIAKVGVVAYKLQL-PETARIHPVFHVSQLKIFKGLTT 1449
+ + KL RY GP+ VI +VG VAYKL L P+ H VFHVSQL+
Sbjct: 1457 KGAG-RFTSRKKLSPRYVGPYKVIERVGAVAYKLDLPPKLNAFHNVFHVSQLRKCLSDQE 1515
Query: 1450 EPYFPLPLTTMEEGPVIQPEVILNVRNIIRGDR-KVEQLL-VKWKDMQNSEATWEDKQEM 1507
E +P ++E ++ + + + +G R K LL V W E TWE + +M
Sbjct: 1516 ESVEDIP-PGLKENMTVEAWPVRIMDRMTKGTRGKARDLLKVLWNCRGREEYTWETENKM 1574
Query: 1508 LDSYPNLNLE-DKIVLEGEGNVMSVEG 1533
++P E K L+ + ++G
Sbjct: 1575 KANFPEWFKEMGKDQLDADSRTNPIQG 1601
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 606 bits (1562), Expect = e-173
Identities = 314/660 (47%), Positives = 416/660 (62%), Gaps = 57/660 (8%)
Query: 593 YREIFQIPKGLPPNREL-SHEILLKEGAQPVKVKPYRYPHSQKEQIEKMVQDMLEEGIIQ 651
+ +IF P LPP R H+I L E + PV +PYRY QK +I+K+V DML G IQ
Sbjct: 14 FPDIFVEPTDLPPFRAPHDHKIELLEDSNPVNQRPYRYVVHQKNEIDKIVDDMLASGTIQ 73
Query: 652 PSLSPFSSPIILVKKKDGTWRCCTDYRALNAITIKDSFPMPTVDELLDELHGAQYFTKLD 711
S SP++SP++LVKKKDGTWR C DYR LN +T+KD FP+P +++L+DEL G+ ++K+D
Sbjct: 74 ASSSPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDRFPIPLIEDLMDELGGSNVYSKID 133
Query: 712 LRSGYHQILLKPEDRQKTAFRTHQGHYEWLVMPFGLTSAPATFQRLMNQLFQPLLRKCVL 771
LR+GYHQ+ + P D KTAF+TH GHYE+LVMPFGLT+APA+FQ LMN F+P LRK VL
Sbjct: 134 LRAGYHQVRMDPLDIHKTAFKTHNGHYEYLVMPFGLTNAPASFQSLMNSFFKPFLRKFVL 193
Query: 772 VFFDDILVYSPSWHSHLKHVEIVFQILSQNVLYAKLSKCSFGITEVEYLGHVVSGSGVAM 831
VFFDDIL+YS S H KH+E VF+++ + L+AK+SKC+F + VEYLGH +SG G+A
Sbjct: 194 VFFDDILIYSTSMEEHKKHLEAVFEVMRVHHLFAKMSKCAFAVPRVEYLGHFISGEGIAT 253
Query: 832 DKIKVIAVLEWPTPTNLKQLRGFLGLTGYYRRFIRSYATIAGPLTNLLKKDAFKWDESTA 891
D K+ AV +WP P NLKQL GFLGLTGYYRRF
Sbjct: 254 DPAKIKAVQDWPVPVNLKQLCGFLGLTGYYRRF--------------------------- 286
Query: 892 KAFDTLKQAITTAPVLVLPDFSQPFVLETDASGTGVGAVLSQGGHPIAYFSKKMASRMQL 951
FV+E DA G G+GAVL Q GHP+A+ S+++ +
Sbjct: 287 ------------------------FVVEMDACGHGIGAVLMQEGHPLAFISRQLKGKQLH 322
Query: 952 QSAYTREFYAITTALAKFRHYLLGHKFILRTDQKSLKSLLDQSLQTPEQQAWLHKFIGFD 1011
S Y +E A+ + K+RHYL+ FI++TDQ+SLK LL+Q L TP QQ WL K + FD
Sbjct: 323 LSIYEKELLAVIFVVRKWRHYLISSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPKLLEFD 382
Query: 1012 FQIEYKPGKDNQAADALSR-----VMSLSWSAPEHDFLEQLKKEIGNDSHLQTIVQQCVN 1066
++I+YK GK+N ADALSR V+ ++ S E D L++++ D +Q I+
Sbjct: 383 YEIQYKQGKENLVADALSRVEGSEVLHMALSVVECDLLKEIQAGYVTDGDIQGIITILQQ 442
Query: 1067 NTMDDINYMVKEGLLYWKHRLVIPMESNLIHQILKEYHDTPIGGHAGVTRTLARVTAQFY 1126
+Y +G+L K+++V+P S + IL+ H + +GGH+G T RV FY
Sbjct: 443 QADSKKHYTWSQGVLRRKNKIVVPNNSGIKDTILRWLHCSGMGGHSGKEVTHQRVKGLFY 502
Query: 1127 WPNMRQHIQKFIEACVTCQQAKSVNTTYAGLLQPLPIPEQVWDDVTMDFITGLPLSFGFT 1186
W +M + IQ FI +C TCQQ KS N GLLQPLPIP+++W DV+MDFI GLPLS G T
Sbjct: 503 WKSMVKDIQAFIRSCGTCQQCKSDNAASPGLLQPLPIPDRIWSDVSMDFIDGLPLSNGKT 562
Query: 1187 VIMVVVDRLSKYAHFMPLKADYSNRTVAEAFVNYVVKLHGMPKSIISDRDKVFTSKFWQH 1246
VIMVVVDRLSK AHF+ L YS TVA+A+++ V KLHG P SI+ +V H
Sbjct: 563 VIMVVVDRLSKAAHFIALAHPYSAMTVAQAYLDNVFKLHGCPSSIVVSSRRVLVQHQLSH 622
Score = 136 bits (342), Expect = 1e-31
Identities = 66/138 (47%), Positives = 92/138 (65%)
Query: 1312 MSPFKALYGREPPMLTRYSVNNSDPTEVQQQLMDRDKLLVELKENLLRAQQVMKSNADKK 1371
M+P++A+YG+ PP+ Y S V + + +R+ +++ LK +L+RAQ MK AD+
Sbjct: 625 MTPYEAVYGQPPPLHLPYLPGESKVAVVARSMQERESMILFLKFHLMRAQHRMKQLADQH 684
Query: 1372 RKDVSFEVGDQVLVKLQPYRQSSVALRKNNKLGMRYFGPFTVIAKVGVVAYKLQLPETAR 1431
+ FEVGD V VKLQPYRQ SV +R KL +YFGP+ VI + G VAYKLQLP ++
Sbjct: 685 ITEREFEVGDYVFVKLQPYRQQSVVMRSTQKLSPKYFGPYKVIDRCGEVAYKLQLPANSQ 744
Query: 1432 IHPVFHVSQLKIFKGLTT 1449
+HPVFHVSQL++ G T
Sbjct: 745 VHPVFHVSQLRVLVGTVT 762
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 604 bits (1557), Expect = e-172
Identities = 386/1101 (35%), Positives = 569/1101 (51%), Gaps = 105/1101 (9%)
Query: 421 IGPISVSILLDGGSSESFIQPRIVHCLNLPIEHTDKCN-VLVGNGQHMKAEGVVRKLSLK 479
+G + +L D G+S FI P N+ + ++ V V GQ + G + + ++
Sbjct: 308 VGGVEAHVLFDSGASHCFITPESASRGNIRGDPGEQLGAVKVAGGQFVAVLGRTKGVDIQ 367
Query: 480 VQDIDITVPAYLLPVAGSDVILGAPWLASLGPHVADYSVSKLKFYMDGKFVTLQGESDNK 539
+ + + PV DVILG WL H+ D ++ F + QG +
Sbjct: 368 IAGESMPADLIISPVELYDVILGMDWLDHYRVHL-DCHRGRVSFERPEGSLVYQGV---R 423
Query: 540 PAVSQLNHFRRLQHMNAISELFTIQKIDPAVIEDNWEGMPVNIEPEMATLLHTYREIFQI 599
P L ++A+ I+K A +E + ++FQ
Sbjct: 424 PTSGSLV-------ISAVQAEKMIEKGCEAYLE--------------------FEDVFQS 456
Query: 600 PKGLPPNRELSHEILLKEGAQPVKVKPYRYPHSQKEQIEKMVQDMLEEGIIQPSLSPFSS 659
+GLPP+R I L+ G P+ PYR ++ +++K ++D+L +
Sbjct: 457 LQGLPPSRSDPFTIELEPGTAPLSKAPYRMAPAEMAELKKQLEDLL------------GA 504
Query: 660 PIILVKKKDGTWRCCTDYRALNAITIKDSFPMPTVDELLDELHGAQYFTKLDLRSGYHQI 719
P++ VKKKDG++R C DYR LN +T+K+ +P+P +DELLD+L GA F+K+DL SGYH I
Sbjct: 505 PVLFVKKKDGSFRLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHLI 564
Query: 720 LLKPEDRQKTAFRTHQGHYEWLVMPFGLTSAPATFQRLMNQLFQPLLRKCVLVFFDDILV 779
+ D +KTAFRT GH+E++VMPFGLT+APA F RLMN +FQ +L + V++F DDILV
Sbjct: 565 PIAEADVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEVLDEFVIIFIDDILV 624
Query: 780 YSPSWHSHLKHVEIVFQILSQNVLYAKLSKCSFGITEVEYLGHVVSGSGVAMDKIKVIAV 839
YS S H H+ V + L + L+AKLSKCSF E+ +LGH+VS GV++D K+ A+
Sbjct: 625 YSKSLEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAI 684
Query: 840 LEWPTPTNLKQLRGFLGLTGYYRRFIRSYATIAGPLTNLLKKDA-FKWDESTAKAFDTLK 898
+W TPTN ++R FLGL GYYRRF++ +A++A P+T L KD F W + F +LK
Sbjct: 685 RDWHTPTNATEIRSFLGLAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLK 744
Query: 899 QAITTAPVLVLPDFSQPFVLETDASGTGVGAVLSQGGHPIAYFSKKMASRMQLQSAYTRE 958
+ +T+ PVL LP+ +P+++ TDASG G+G VL Q G IAY S+++ + E
Sbjct: 745 EMLTSTPVLALPEHGEPYMVYTDASGVGLGCVLMQRGKVIAYASRQLRKHEGNYPTHDLE 804
Query: 959 FYAITTALAKFRHYLLGHKFILRTDQKSLKSLLDQSLQTPEQQAWLHKFIGFDFQIEYKP 1018
A+ AL +R YL G + TD KSLK + Q Q+ W+ +D +I Y P
Sbjct: 805 MAAVIFALKIWRSYLYGGNVQVFTDHKSLKYIFTQPELNLRQRQWMELVADYDLEIAYHP 864
Query: 1019 GKDNQAADALSRVMSLSWSAPEHDFLEQLKKEIGNDSHLQTIVQQCVNNTMDDINYMVKE 1078
GK N ADALS A +E L EIG ++ C V
Sbjct: 865 GKANVVADALSHKRV---GAAPGQSVEALVSEIG-------ALRLCAVAREPLGLEAVDR 914
Query: 1079 GLLYWKHRLVIPMESNLIHQILKEYHDTPIGGHAGVTRTLARVTAQFYWPNMRQHIQKFI 1138
L + RL + LI Y + + ++
Sbjct: 915 ADLLTRVRLAQEKDEGLI---------------------------AAYKAEGSEDVANWV 947
Query: 1139 EACVTCQQAKSVNTTYAGLLQPLPIPEQVWDDVTMDFITGLPLSFGFTVIMVVVDRLSKY 1198
C CQ K+ + G+LQ LPIPE WD +T+DF+ GLP+S I V+VDRL+K
Sbjct: 948 AECDVCQLVKAEHQVPGGMLQSLPIPEWKWDFITIDFVVGLPVSRTKDAIWVIVDRLTKS 1007
Query: 1199 AHFMPLKADYSNRTVAEAFVNYVVKLHGMPKSIISDRDKVFTSKFWQHLFQMQGTTLSMS 1258
AHF+ ++ +A+ +V+ +VKLHG+P SI+SDRD FTS FW+ GT + MS
Sbjct: 1008 AHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTSAFWRAFQAEMGTKVQMS 1067
Query: 1259 TTYHPQTDGQSEAVNKCLEMYLRCFTFKNPKTWFKALTWVELWYNTFLHTSLGMSPFKAL 1318
T YHPQT GQSE + LE LR W L+ VE YN S+GM+PF+AL
Sbjct: 1068 TAYHPQTYGQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYPASIGMAPFEAL 1127
Query: 1319 YG---REPPMLTRYSVNNSDPTEVQQQLMDRDKLLVELKENLLRAQQVMKSNADKKRKDV 1375
Y R P LT+ + + Q+ +R ++ LK N+ AQ +S ADK+R+++
Sbjct: 1128 YERPCRTPLCLTQVGERSIYGADYVQETTERIRV---LKLNMKEAQDRQRSYADKRRREL 1184
Query: 1376 SFEVGDQVLVKLQPYRQSSVALRKNNKLGMRYFGPFTVIAKVGVVAYKLQLPETAR-IHP 1434
FEVGD+V +K+ R + ++ KL RY GPF ++ +VG VAY+L+LP+ R H
Sbjct: 1185 EFEVGDRVYLKMAMLRGPNRSI-SETKLSPRYMGPFRIVERVGPVAYRLELPDVMRAFHK 1243
Query: 1435 VFHVSQLKIFKGLTTEPYFPLPLTTMEEGPVIQPEVILNVRNIIRGDRKVEQL------L 1488
VFHVS L+ E +P +QP + L R + +R++++L L
Sbjct: 1244 VFHVSMLRKCLHKDDEVLAKIP-------EDLQPNMTLEARPVRVLERRIKELRRKKIPL 1296
Query: 1489 VK--WKDMQNSEATWEDKQEM 1507
+K W ++ TWE + M
Sbjct: 1297 IKVLWDCDGVTKETWEPEARM 1317
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 602 bits (1552), Expect = e-172
Identities = 371/1093 (33%), Positives = 564/1093 (50%), Gaps = 102/1093 (9%)
Query: 458 NVLVGNGQHMKAEGVVRKLSLKVQDIDITVPAYLLPVAGSDVILGAPWLASLGPHVADYS 517
+V V GQ + G + + +++ + + PV DVILG WL H+ D
Sbjct: 364 HVKVAGGQFLAVLGRAKGVDIQIAGESMPADLIISPVELYDVILGMDWLDHYRVHL-DCH 422
Query: 518 VSKLKFYM-DGKFVTLQGESDNKPAVSQLNHFRRLQHMNAISELFTI---QKIDPAVIED 573
++ F +G+ V + V +++ + L TI + + + D
Sbjct: 423 RGRVSFERPEGRLVYQGVRPTSGSLVISAVQAKKMIEKGCEAYLVTISMPESLGQVAVSD 482
Query: 574 NWEGMPVNIEPEMATLLHTYREIFQIPKGLPPNRELSHEILLKEGAQPVKVKPYRYPHSQ 633
++ + ++FQ +GLPP+R I L+ G P+ PYR ++
Sbjct: 483 -------------IRVIQEFEDVFQSLQGLPPSRSDPFTIELEPGTAPLSKAPYRMAPAE 529
Query: 634 KEQIEKMVQDMLEEGIIQPSLSPFSSPIILVKKKDGTWRCCTDYRALNAITIKDSFPMPT 693
+++K ++D+L +G I+PS SP+ +P++ VKKKDG++R C DYR LN +T+K+ +P+P
Sbjct: 530 MTELKKQLEDLLGKGFIRPSTSPWGAPVLFVKKKDGSFRLCIDYRGLNWVTVKNKYPLPR 589
Query: 694 VDELLDELHGAQYFTKLDLRSGYHQILLKPEDRQKTAFRTHQGHYEWLVMPFGLTSAPAT 753
+DELLD+L GA F+K+DL SGYHQI + D +KTAFRT GH+E++VMPF LT+APA
Sbjct: 590 IDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAFRTRYGHFEFVVMPFALTNAPAA 649
Query: 754 FQRLMNQLFQPLLRKCVLVFFDDILVYSPSWHSHLKHVEIVFQILSQNVLYAKLSKCSFG 813
F RLMN +FQ L + V++F DDILVYS S H H+ V + L + L+AKLSKCSF
Sbjct: 650 FMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEHEVHLRRVMEKLREQKLFAKLSKCSFW 709
Query: 814 ITEVEYLGHVVSGSGVAMDKIKVIAVLEWPTPTNLKQLRGFLGLTGYYRRFIRSYATIAG 873
E+ +LGH+VS GV++D K+ A+ +WP PTN ++R FL LTGYYRRF++ +A++A
Sbjct: 710 QREIGFLGHIVSAEGVSVDPEKIEAIRDWPRPTNATEIRSFLRLTGYYRRFVKGFASMAQ 769
Query: 874 PLTNLLKKDA-FKWDESTAKAFDTLKQAITTAPVLVLPDFSQPFVLETDASGTGVGAVLS 932
P+T L KD F W + F +LK+ +T+ PVL LP+ QP+++ TDAS G+G VL
Sbjct: 770 PMTKLTGKDVPFVWSPECEEGFVSLKEMLTSTPVLALPEHGQPYMVYTDASRVGLGCVLM 829
Query: 933 QGGHPIAYFSKKMASRMQLQSAYTREFYAITTALAKFRHYLLGHKFILRTDQKSLKSLLD 992
Q G IAY S+++ + E + AL +R YL G K + TD KSLK + +
Sbjct: 830 QRGKVIAYASRQLRKHEGNYPTHDLEMAVVIFALKIWRSYLYGGKVQVFTDHKSLKYIFN 889
Query: 993 QSLQTPEQQAWLHKFIGFDFQIEYKPGKDNQAADALSRVMSLSWSAPEHDFLEQLKKEIG 1052
Q Q W+ +D +I Y PGK N ADALSR A +E L EIG
Sbjct: 890 QPELNLRQMRWMELVADYDLEIAYHPGKANVVADALSRKRV---GAAPGQSVEALVSEIG 946
Query: 1053 NDSHLQTIVQQCVNNTMDDINYMVK--------------------------EGLLYWKHR 1086
+ +D + + + G ++ R
Sbjct: 947 ALRLCVVAREPLGLEAVDRADLLTRARLAQEKDEGLIAASKAEGSEYQFAANGTIFVYGR 1006
Query: 1087 LVIPMESNLIHQILKEYHDTPIGGHAGVTRTLARVTAQFYWPNMRQHIQKFIEACVTCQQ 1146
+ +P + L +IL E H + H G T+ + + W M++ + ++ C CQ
Sbjct: 1007 VCVPKDEELRREILSEAHASMFSIHPGATKMYRDLKRYYQWVGMKRDVANWVAECDVCQL 1066
Query: 1147 AKSVNTTYAGLLQPLPIPEQVWDDVTMDFITGLPLSFGFTVIMVVVDRLSKYAHFMPLKA 1206
K+ + +P+ +W V++DRL+K AHF+ ++
Sbjct: 1067 VKAEH----------QVPDAIW---------------------VIMDRLTKSAHFLAIRK 1095
Query: 1207 DYSNRTVAEAFVNYVVKLHGMPKSIISDRDKVFTSKFWQHLFQMQGTTLSMSTTYHPQTD 1266
+A+ +V+ +VKLHG+P SI+SDRD FT FW+ GT + MST YHPQTD
Sbjct: 1096 TDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTFAFWRAFQAKMGTKVQMSTAYHPQTD 1155
Query: 1267 GQSEAVNKCLEMYLRCFTFKNPKTWFKALTWVELWYNTFLHTSLGMSPFKALYGR---EP 1323
GQSE + LE LR W L+ VE YN S+GM+PF+ALYGR P
Sbjct: 1156 GQSERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYQASIGMAPFEALYGRPCWTP 1215
Query: 1324 PMLTRYSVNNSDPTEVQQQLMDRDKLLVELKENLLRAQQVMKSNADKKRKDVSFEVGDQV 1383
T+ + + Q+ +R ++ LK N+ AQ +S ADK+R+++ FEVGD+V
Sbjct: 1216 LRWTQVEERSIYGADYVQETTERIRV---LKLNMKEAQARQRSYADKRRRELEFEVGDRV 1272
Query: 1384 LVKLQPYRQSSVALRKNNKLGMRYFGPFTVIAKVGVVAYKLQLPETAR-IHPVFHVSQLK 1442
+K+ R + ++ + KL RY GPF ++ +VG VAY+L+LP+ R H VFHV L+
Sbjct: 1273 YLKMAMLRGPNRSILE-TKLSPRYMGPFRIVERVGPVAYRLELPDVMRAFHKVFHVLMLR 1331
Query: 1443 IFKGLTTEPYFPLPLTTMEEGPVIQPEVILNVRNIIRGDRKVEQL------LVK--WKDM 1494
E +P +QP + L R + +R++++L L+K W
Sbjct: 1332 KCLHKDDEVLVKIP-------EDLQPNMTLEARPVRVLERRIKELRRKKIPLIKVLWDCD 1384
Query: 1495 QNSEATWEDKQEM 1507
+E TWE + M
Sbjct: 1385 GVTEETWEPEARM 1397
>At2g06890 putative retroelement integrase
Length = 1215
Score = 488 bits (1256), Expect = e-137
Identities = 335/1035 (32%), Positives = 519/1035 (49%), Gaps = 97/1035 (9%)
Query: 205 QQNKSVNEYYLEFTALSNRVYGLSND--ALIDCFVSGLKDELRRDVMLHTPISIVKAVSL 262
Q N+SV EYY E L R +S D A + F+ L +++ + + I + +
Sbjct: 27 QGNRSVEEYYKEMETLMLRA-DISEDREATLSRFLGDLNRDIQDRLETQYYVQIEEMLHK 85
Query: 263 AKLFEEKIAANFNPKQPLKPYTPAQPHQHRAPFNPTRNDQNQTIEKAPNLPLLPTPPTRP 322
A LFE+++ + + T A+P R T + N+ I + P ++P
Sbjct: 86 AILFEQQVKRKSSSRSSYGSGTIAKPTYQRE--ERTSSYHNKPI-------VSPRAESKP 136
Query: 323 MSHLQKNPAIKRISPAEMQLRREKGLCYFCDDKFSFSHKCPNKQLMLLELDDDLDP---- 378
+ +Q + IS + ++ R CY C K ++++CPNK++M+L + +++P
Sbjct: 137 YAAVQDHKGKAEISTSRVRDVR----CYKCQGKGHYANECPNKRVMILLDNGEIEPEEEI 192
Query: 379 PDTPSQTAVTSDTETQE---VAEHHLSLNALKGATGM--GVIRFKGYIGPISVSILLDGG 433
PD+PS + Q VA LS+ + + ++ S+++DGG
Sbjct: 193 PDSPSSLKENEELPAQGELLVARRTLSVQTKTDEQEQRKNLFHTRCHVHGKVCSLIIDGG 252
Query: 434 SSESFIQPRIVHCLNLP-IEHTDKCNVLVGNGQHMKAEGVVRKLSLKVQDIDITVPAYLL 492
S + +V L L + + K V K + VV + K +D + +L
Sbjct: 253 SCTNVASETMVKKLGLKWLNDSGKMRV--------KNQVVVPIVIGKYED---EILCDVL 301
Query: 493 PVAGSDVILGAPWLASL----------------GPHVADYSVSKLKFYMDGKFVTLQGES 536
P+ ++LG PW + G S++ + Y D + +
Sbjct: 302 PMEAGHILLGRPWQSDRKVMHDGFTNRHSFEFKGGKTILVSMTPHEVYQD----QIHLKQ 357
Query: 537 DNKPAVSQLNHFRRLQHMNAI--SELFTIQKIDPAVIEDNWEGMPVNIEPEMATLLHTYR 594
+ V Q N F + + + S+ + + + PV + EM +LL Y+
Sbjct: 358 KKEQVVKQPNFFAKSGEVKSAYSSKQPMLLFVFKEALTSLTNFAPV-LPSEMTSLLQDYK 416
Query: 595 EIF--QIPKGLPPNRELSHEILLKEGAQPVKVKPYR-YPHSQKEQIEKMVQDMLEEGIIQ 651
++F PKGLPP R + H+I GA YR P KE
Sbjct: 417 DVFPEDNPKGLPPIRGIEHQIDFVPGASLPNRPAYRTNPVETKE---------------- 460
Query: 652 PSLSPFSSPIILVKKKDGTWRCCTDYRALNAITIKDSFPMPTVDELLDELHGAQYFTKLD 711
L ++KDG+WR C D RA+N +T+K P+P +D++LDELHG+ F+K+D
Sbjct: 461 -----------LQRQKDGSWRMCFDCRAINNVTVKYCHPIPRLDDMLDELHGSSIFSKID 509
Query: 712 LRSGYHQILLKPEDRQKTAFRTHQGHYEWLVMPFGLTSAPATFQRLMNQLFQPLLRKCVL 771
L+SGYHQI + D KTAF+T G YEWLVMPFGLT AP+TF RLMN + + + V+
Sbjct: 510 LKSGYHQIRMNEGDEWKTAFKTKHGLYEWLVMPFGLTHAPSTFMRLMNHVLRAFIGIFVI 569
Query: 772 VFFDDILVYSPSWHSHLKHVEIVFQILSQNVLYAKLSKCSFGITEVEYLGHVVSGSGVAM 831
V+FDDILVYS S H++H++ V +L + LYA L KC+F + +LG VVS GV +
Sbjct: 570 VYFDDILVYSESLREHIEHLDSVLNVLRKEELYANLKKCTFCTDNLVFLGFVVSADGVKV 629
Query: 832 DKIKVIAVLEWPTPTNLKQLRGFLGLTGYYRRFIRSYATIAGPLTNLLKKD-AFKWDEST 890
D+ KV A+ +WP+P + ++R F GL G+YRRF + ++TI PLT ++KKD FKW+++
Sbjct: 630 DEEKVKAIRDWPSPKTVGEVRSFHGLAGFYRRFFKDFSTIVAPLTEVMKKDVGFKWEKAQ 689
Query: 891 AKAFDTLKQAITTAPVLVLPDFSQPFVLETDASGTGVGAVLSQGGHPIAYFSKKMASRMQ 950
+AF +LK +T APVL+L +F + F +E DASG G+GAVL Q IA+FS+K+
Sbjct: 690 EEAFQSLKDKLTNAPVLILSEFLKTFEIECDASGIGIGAVLMQDQKLIAFFSEKLGGATL 749
Query: 951 LQSAYTREFYAITTALAKFRHYLLGHKFILRTDQKSLKSLLDQSLQTPEQQAWLHKFIGF 1010
Y +E YA+ AL +++HYL F++ TD +SLK L Q W+ F
Sbjct: 750 NYPTYDKELYALVRALQRWQHYLWPKVFVIHTDHESLKHLKGQQKLNKRHARWVEFIETF 809
Query: 1011 DFQIEYKPGKDNQAADALS-RVMSLSWSAPEHDFLEQLKKEIGNDSHLQTIVQQCVNNTM 1069
+ I+YK GKDN ADALS R LS + EQ+K+ D Q + + C
Sbjct: 810 AYVIKYKKGKDNVVADALSQRYTLLSTLNVKLMGFEQIKEVYETDHDFQEVYKAC--EKF 867
Query: 1070 DDINYMVKEGLLYWKHRLVIPMESNLIHQILKEYHDTPIGGHAGVTRTLARVTAQFYWPN 1129
Y ++ L++++RL +P +L ++E H + GH G+ +TL +T F WP+
Sbjct: 868 ASGRYFRQDKFLFYENRLCVP-NCSLRDLFVREAHGGGLMGHFGIAKTLEVMTEHFRWPH 926
Query: 1130 MRQHIQKFIEACVTCQQAKSVNTTYAGLLQPLPIPEQVWDDVTMDFITGLPLSFGFTVIM 1189
M+ +++ C TC+QAKS GL PLPIP+ W+D++MDF+ GLP + G I
Sbjct: 927 MKCDVKRICGRCNTCKQAKS-KIQPNGLYTPLPIPKHPWNDISMDFVMGLPRT-GKDSIF 984
Query: 1190 VVVDRLSKYAHFMPL 1204
VV F P+
Sbjct: 985 VVYSPFQIVYGFNPI 999
Score = 54.3 bits (129), Expect = 5e-07
Identities = 38/125 (30%), Positives = 61/125 (48%), Gaps = 18/125 (14%)
Query: 1313 SPFKALYGREP---------PMLTRYSVNNSDPTEVQQQLMDRDKLLVELKENLLRAQQV 1363
SPF+ +YG P P+ R S++ E+ QQ+ + + +E K L Q
Sbjct: 988 SPFQIVYGFNPISPFDLIPLPLSERVSIDGKKKAELVQQIHENARRNIEEKTKLYAKQ-- 1045
Query: 1364 MKSNADKKRKDVSFEVGDQVLVKLQPYRQSSVALRKNNKLGMRYFGPFTVIAKVGVVAYK 1423
A+K R++ FEVGD V + L R+ ++ +KL R GPF +I ++ AY+
Sbjct: 1046 ----ANKGRREQIFEVGDMVWIHL---RKERFPAQRKSKLMPRIDGPFKIIKRINDNAYQ 1098
Query: 1424 LQLPE 1428
L L +
Sbjct: 1099 LDLQD 1103
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 473 bits (1217), Expect = e-133
Identities = 281/792 (35%), Positives = 422/792 (52%), Gaps = 60/792 (7%)
Query: 464 GQHMKAEGVVRKLSLKVQDIDITVPAYLLPVAGSDVILGAPWLASLGPHVADYSVSKLKF 523
GQ + G + + +++ + + PV DVILG WL H+ D+ ++ F
Sbjct: 344 GQFLAVLGRAKGVDIQIAGESMPADLIISPVELYDVILGMDWLDYYRVHL-DWHRGRVFF 402
Query: 524 YMDGKFVTLQGESDNKPAVSQLNHFRRLQHMNAISELFTIQKIDPA-VIEDNWEGMPVNI 582
+ QG + IS I + +IE E V I
Sbjct: 403 ERPEGRLVYQG-------------------VRPISGSLVISAVQAEKMIEKGCEAYLVTI 443
Query: 583 E-PEMA--------TLLHTYREIFQIPKGLPPNRELSHEILLKEGAQPVKVKPYRYPHSQ 633
PE ++ ++++FQ +GLPP++ I L+ G P+ PYR ++
Sbjct: 444 SMPESVGQVAVSDIRVVQEFQDVFQSLQGLPPSQSDPFTIELEPGTAPLSKAPYRMAPAE 503
Query: 634 KEQIEKMVQDMLEEGIIQPSLSPFSSPIILVKKKDGTWRCCTDYRALNAITIKDSFPMPT 693
+++K ++D+L +G I+PS SP+ +P++ VKKKDG++R C DYR LN +T+K+ +P+P
Sbjct: 504 MAELKKQLKDLLGKGFIRPSTSPWGAPVLFVKKKDGSFRLCIDYRELNRVTVKNRYPLPR 563
Query: 694 VDELLDELHGAQYFTKLDLRSGYHQILLKPEDRQKTAFRTHQGHYEWLVMPFGLTSAPAT 753
+DELLD+L GA F+K+DL SGYHQI + D +KTAFRT GH+E++VMPFGLT+APA
Sbjct: 564 IDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAFRTRYGHFEFVVMPFGLTNAPAV 623
Query: 754 FQRLMNQLFQPLLRKCVLVFFDDILVYSPSWHSHLKHVEIVFQILSQNVLYAKLSKCSFG 813
F RLMN +FQ L + V++F DDILVYS S H+ V + L + L+AKLSKCSF
Sbjct: 624 FMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEEQEVHLRRVMEKLREQKLFAKLSKCSFW 683
Query: 814 ITEVEYLGHVVSGSGVAMDKIKVIAVLEWPTPTNLKQLRGFLGLTGYYRRFIRSYATIAG 873
E+ +LGH+VS GV++D K+ A+ +WP PTN ++R FLG GYYRRF++ +A++A
Sbjct: 684 QREMGFLGHIVSAEGVSVDPEKIEAIRDWPRPTNATEIRSFLGWAGYYRRFVKGFASMAQ 743
Query: 874 PLTNLLKKDA-FKWDESTAKAFDTLKQAITTAPVLVLPDFSQPFVLETDASGTGVGAVLS 932
P+T L KD F W + + F +LK+ +T+ PVL LP+ QP+++ TDAS G+G VL
Sbjct: 744 PMTKLTGKDVPFVWSQECEEGFVSLKEMLTSTPVLALPEHGQPYMVYTDASRVGLGCVLM 803
Query: 933 QGGHPIAYFSKKMASRMQLQSAYTREFYAITTALAKFRHYLLGHKFILRTDQKSLKSLLD 992
Q G IAY S+++ + E A+ AL +R YL G K + TD KSLK +
Sbjct: 804 QHGKVIAYASRQLMKHEGNYPTHDLEMAAVIFALKIWRSYLYGGKVQVFTDHKSLKYIFT 863
Query: 993 QSLQTPEQQAWLHKFIGFDFQIEYKPGKDNQAADALSRVMSLSWSAPEHDFLEQLKKEIG 1052
Q Q+ W+ +D +I Y PGK N DALSR A +E L EIG
Sbjct: 864 QPELNLRQRRWMELVADYDLEIAYHPGKANVVVDALSRKRV---GAALGQSVEVLVSEIG 920
Query: 1053 NDSHLQTIVQQCVNNTMDDINYMVKEGLLYWKHRLVIPMESNLIHQILKEYHDTPIGGHA 1112
+ +D + + + L K + + +++ LK Y
Sbjct: 921 ALRLCAVAREPLGLEAVDRADLLTRVRLAQKKDE---GLRATKMYRDLKRY--------- 968
Query: 1113 GVTRTLARVTAQFYWPNMRQHIQKFIEACVTCQQAKSVNTTYAGLLQPLPIPEQVWDDVT 1172
+ W M+ + ++ C CQ K+ + G+LQ LPIPE WD +T
Sbjct: 969 ------------YQWVGMKMDVANWVAECDVCQLVKAEHQVLGGMLQSLPIPEWKWDFIT 1016
Query: 1173 MDFITGLPLSFGFTVIMVVVDRLSKYAHFMPLKADYSNRTVAEAFVNYVVKLHGMPKSI- 1231
MD + GL +S I V+VDRL+K AHF+ ++ +A+ FV+ +VKLHG+P ++
Sbjct: 1017 MDLVVGLRVSRTKDAIWVIVDRLTKSAHFLAIRKTDGAAVLAKKFVSEIVKLHGVPLNMK 1076
Query: 1232 -ISDRDKVFTSK 1242
DR + + K
Sbjct: 1077 EAQDRQRSYADK 1088
Score = 80.9 bits (198), Expect = 5e-15
Identities = 56/158 (35%), Positives = 83/158 (52%), Gaps = 11/158 (6%)
Query: 1356 NLLRAQQVMKSNADKKRKDVSFEVGDQVLVKLQPYRQSSVALRKNNKLGMRYFGPFTVIA 1415
N+ AQ +S ADK+R+++ FEVGD+V +K+ R + ++ + KL RY GPF ++
Sbjct: 1074 NMKEAQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSISET-KLSPRYMGPFKIVE 1132
Query: 1416 KVGVVAYKLQLPETAR-IHPVFHVSQL-----KIFKGLTTEPYFPLPLTTMEEGPVIQPE 1469
+V VAY+L+LP+ R H VFHVS L K + L P P T+E PV
Sbjct: 1133 RVEPVAYRLELPDVMRAFHKVFHVSMLRKCLHKDDEALAKIPEDLQPNMTLEARPV---- 1188
Query: 1470 VILNVRNIIRGDRKVEQLLVKWKDMQNSEATWEDKQEM 1507
+L R +K+ + V W +E TWE + M
Sbjct: 1189 RVLERRIKELRQKKIPLIKVLWDCDGVTEETWEPEARM 1226
>At3g31970 hypothetical protein
Length = 1329
Score = 408 bits (1049), Expect = e-113
Identities = 278/828 (33%), Positives = 410/828 (48%), Gaps = 110/828 (13%)
Query: 716 YHQILLKPEDRQKTAFRTHQGHYEWLVMPFGLTSAPATFQRLMNQLFQPLLRKCVLVFFD 775
+ I + + +KTAFRT GH+E++VMPFGLT+ P F RLMN +FQ L + V++F D
Sbjct: 570 FEDIPIAEANVRKTAFRTRYGHFEFVVMPFGLTNIPTAFMRLMNSVFQEFLDEFVIIFID 629
Query: 776 DILVYSPSWHSHLKHVEIVFQILSQNVLYAKLSKCSFGITEVEYLGHVVSGSGVAMDKIK 835
DILVYS S H ++ S+ ++ E G V GV++D K
Sbjct: 630 DILVYSKSPEEH--------EVQSEE-------------SDGEAAGAEV---GVSVDLEK 665
Query: 836 VIAVLEWPTPTNLKQLRGFLGLTGYYRRFIRSYATIAGPLTNLLKKDA-FKWDESTAKAF 894
+ A+ +WP PTN ++R FLGL GYY+RF++ +A++A P+T L KD F W + F
Sbjct: 666 IEAIRDWPRPTNATEIRSFLGLAGYYKRFVKVFASMAQPMTKLTGKDVPFVWSPECDEGF 725
Query: 895 DTLKQAITTAPVLVLPDFSQPFVLETDASGTGVGAVLSQGGHPIAYFSKKMASRMQLQSA 954
+LK+ +T+ PVL LP+ +P+++ TDAS G+ VL Q G
Sbjct: 726 MSLKEMLTSTPVLALPEHGEPYMVYTDASRVGLDCVLMQRGKVF---------------- 769
Query: 955 YTREFYAITTALAKFRHYLLGHKFILRTDQKSLKSLLDQSLQTPEQQAWLHKFIGFDFQI 1014
TD KSLK + Q Q+ W+ +D +I
Sbjct: 770 ---------------------------TDHKSLKYIFTQPELNLRQRRWMELVADYDLEI 802
Query: 1015 EYKPGKDNQAADALSRVM---SLSWSAPEHDFL-------EQLKKEIGNDSHLQTIVQQC 1064
Y GK N ADALSR S+ E L E L E + + L T V+
Sbjct: 803 AYHSGKANVVADALSRKRVGGSVEALVSEIGALRLCVMAQEPLGLEAVDRADLLTRVRLA 862
Query: 1065 VN--------NTMDDINYM-VKEGLLYWKHRLVIPMESNLIHQILKEYHDTPIGGHAGVT 1115
+ D Y G + R+ +P + L +IL E H + H T
Sbjct: 863 QEKDEGLIAASKADGSEYQFAANGTILVHGRVCVPKDEELRREILSEAHASMFSIHPRAT 922
Query: 1116 RTLARVTAQFYWPNMRQHIQKFIEACVTCQQAKSVNTTYAGLLQPLPIPEQVWDDVTMDF 1175
+ + + W M++ + ++ C CQ K+ + GLLQ LPI E WD +TMDF
Sbjct: 923 KMYRDLKRYYQWVGMKRDVANWVTECDVCQLVKAEHQVPGGLLQSLPILEWKWDFITMDF 982
Query: 1176 ITGLPLSFGFTVIMVVVDRLSKYAHFMPLKADYSNRTVAEAFVNYVVKLHGMPKSIISDR 1235
+ GLP+S I V+VDRL+K AHF+ ++ +A+ +V+ +V+LHG+P SI+SDR
Sbjct: 983 VVGLPVSRTKDAIWVIVDRLTKSAHFLAIRKTDGAVLLAKKYVSEIVELHGVPVSIVSDR 1042
Query: 1236 DKVFTSKFWQHLFQMQGTTLSMSTTYHPQTDGQSEAVNKCLEMYLRCFTFKNPKTWFKAL 1295
D FTS FW+ GT + MST YHPQTDGQSE + LE LR W L
Sbjct: 1043 DSKFTSAFWRAFQGEMGTKVQMSTAYHPQTDGQSERTIQTLEDMLRMCVLDRGGHWADHL 1102
Query: 1296 TWVELWYNTFLHTSLGMSPFKALYG---REPPMLTRYSVNNSDPTEVQQQLMDRDKLLVE 1352
+ VE YN S+ M+PF+ALYG R P T+ + + + +R ++
Sbjct: 1103 SLVEFAYNNSYQASIRMAPFEALYGRPCRTPLCWTQVGERSIYGADYVLETTERIRV--- 1159
Query: 1353 LKENLLRAQQVMKSNADKKRKDVSFEVGDQVLVKLQPYRQSSVALRKNNKLGMRYFGPFT 1412
LK N+ AQ +S ADK+R+++ FEVGD+V +K+ R + ++ KL RY GPF
Sbjct: 1160 LKLNMKEAQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSI-SETKLTPRYMGPFR 1218
Query: 1413 VIAKVGVVAYKLQLPETAR-IHPVFHVSQLKIFKGLTTEPYFPLPLTTMEEGPVIQPEVI 1471
++ +VG VAY+L+LP+ R H VFHVS L+ E + +E+ +QP +
Sbjct: 1219 IVERVGPVAYRLELPDVMRAFHKVFHVSMLRKCLHKDDEVLAKI----LED---LQPNMT 1271
Query: 1472 LNVRNIIRGDRKVEQL------LVK--WKDMQNSEATWEDKQEMLDSY 1511
L R + +R++++L L+K W +E TWE + M S+
Sbjct: 1272 LEARPVRILERRIKELRRKKIPLIKVLWNCDGVTEETWEPEARMKASF 1319
Score = 52.0 bits (123), Expect = 3e-06
Identities = 88/391 (22%), Positives = 136/391 (34%), Gaps = 69/391 (17%)
Query: 132 WIFKAEQYFGYYNTPDAERLIIASVHLEQEVVPWFQMVN--RSRPFQSWNDFTRALELDF 189
W + E FG P R+ +A LE + W++ V R + SW DF +
Sbjct: 170 WKSRVEHNFGSSRCPAEYRVDLAVHFLEGDAHLWWRSVTARRRQAHMSWADFVAEFNAKY 229
Query: 190 GPS-IYDCPRASLFKLQQN-KSVNEYYLEFTALSNRVY---GLSND-ALIDCFVSGLKDE 243
P D A +L Q +SV EY EF L VY G+ +D A + F+ GL+ +
Sbjct: 230 FPQEALDRMEARFLELTQGERSVREYEREFNRLL--VYAGRGMQDDQAQMRRFLRGLRPD 287
Query: 244 LRRDVMLHTPISIVKAVSLAKLFEEKIAANFNPKQPLKPYTPAQPHQHRAPFNPTRNDQN 303
LR + + V A EE + Q
Sbjct: 288 LRVRCRVLQYATKAALVETAAEVEEDL-------------------------------QR 316
Query: 304 QTIEKAPNLPLLPTPPTRPMSHLQKNPAIKRISPAEMQLRREKGLCYFCDDKFSFSHKCP 363
Q + +P + +P Q+ K PA+ Q R+E+G CP
Sbjct: 317 QVVGVSPAV--------KPKKTQQQVAPSKGGKPAQGQKRKERG---------HLRPNCP 359
Query: 364 NKQLMLLELDDDLDPPDTPSQTAVTSDTETQEVAEHHLSLNALKGATGMGVIRFKGYIGP 423
Q M + + Q AV + Q+ + + A I
Sbjct: 360 KLQRMAVAV----------VQPAVQHGAQVQQRVQQLAHIAAAPQGYTTREIGSTSKRAI 409
Query: 424 ISVSILLDGGSSESFIQPRIVHCLNLPIEHTDKCN-VLVGNGQHMKAEGVVRKLSLKVQD 482
+L D G+S FI P N+ + ++ V V GQ + G + + +++
Sbjct: 410 TEAHVLFDSGASHCFITPESASRGNIRGDPGEQLGAVKVAGGQFLAVLGRAKGVDIQIAG 469
Query: 483 IDITVPAYLLPVAGSDVILGAPWLASLGPHV 513
+ V + PV DVILG WL H+
Sbjct: 470 ESMPVDLIISPVELYDVILGMDWLDHYRVHL 500
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 388 bits (996), Expect = e-107
Identities = 203/520 (39%), Positives = 315/520 (60%), Gaps = 17/520 (3%)
Query: 459 VLVGNGQHMKAEGVVRKLSLKVQDIDITVPAYLLPVAGSDVILGAPWLASLGPHVADYSV 518
V GQ M +VR +S+ V +++ ++P+ DVILG WL H+ D
Sbjct: 2 VRAAGGQAMYPTRLVRGISVVVNGVNMPADLIIVPLKKHDVILGMDWLGKYKGHI-DCHR 60
Query: 519 SKLKFYMDGKFVTLQG--ESDNKPAVSQLNHFRRL-QHMNAISELFTIQKIDPAVIEDNW 575
+++F D + QG + +S + R L + A T +++ +
Sbjct: 61 ERIQFERDEGMLKFQGIRTTSGSLVISAIQAERMLGKGCEAYLATITTKEVGASA----- 115
Query: 576 EGMPVNIEPEMATLLHTYREIFQIPKGLPPNRELSHEILLKEGAQPVKVKPYRYPHSQKE 635
E + +++ + ++F G+PP+R I L+ G P+ PYR ++
Sbjct: 116 -------ELKDILIVNEFSDVFAAVSGVPPDRSDPFTIELEPGTTPISKAPYRMAPAEMA 168
Query: 636 QIEKMVQDMLEEGIIQPSLSPFSSPIILVKKKDGTWRCCTDYRALNAITIKDSFPMPTVD 695
+++K ++++L +G I+PS SP+ +P++ VKKKDG++R C DYR LN +T+K+ +P+P +D
Sbjct: 169 ELKKQLEELLAKGFIRPSSSPWGAPVLFVKKKDGSFRLCIDYRGLNKVTVKNKYPLPRID 228
Query: 696 ELLDELHGAQYFTKLDLRSGYHQILLKPEDRQKTAFRTHQGHYEWLVMPFGLTSAPATFQ 755
EL+D+L GAQ+F+K+DL SGYHQI ++P D +KTAFRT GH+E++VMPFGLT+APA F
Sbjct: 229 ELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAFRTRYGHFEFVVMPFGLTNAPAAFM 288
Query: 756 RLMNQLFQPLLRKCVLVFFDDILVYSPSWHSHLKHVEIVFQILSQNVLYAKLSKCSFGIT 815
++MN +F+ L + V++F DDILV+S SW +H +H+ V + L ++ L+AKLSK SF
Sbjct: 289 KMMNGVFRDFLDEFVIIFIDDILVHSKSWEAHQEHLRAVLERLREHELFAKLSKFSFWQR 348
Query: 816 EVEYLGHVVSGSGVAMDKIKVIAVLEWPTPTNLKQLRGFLGLTGYYRRFIRSYATIAGPL 875
V +LGHV+S GV++D K+ ++ EWP P N ++R FLGL GYYRRF+ S+A++A PL
Sbjct: 349 SVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATEIRSFLGLAGYYRRFVMSFASMAQPL 408
Query: 876 TNLLKKD-AFKWDESTAKAFDTLKQAITTAPVLVLPDFSQPFVLETDASGTGVGAVLSQG 934
T L KD AF W + K+F LK + APVLVLP+ +P+ + TDAS G+G VL Q
Sbjct: 409 TRLTGKDTAFNWSDECEKSFLELKAMLINAPVLVLPEEGEPYTVYTDASIVGLGCVLMQK 468
Query: 935 GHPIAYFSKKMASRMQLQSAYTREFYAITTALAKFRHYLL 974
G IAY S+++ + + E A+ AL +R YL+
Sbjct: 469 GSVIAYASRQLRKHEKNYPTHDLEMAAVVFALKIWRSYLI 508
Score = 254 bits (649), Expect = 3e-67
Identities = 153/419 (36%), Positives = 225/419 (53%), Gaps = 13/419 (3%)
Query: 1079 GLLYWKHRLVIPMESNLIHQILKEYHDTPIGGHAGVTRTLARVTAQFYWPNMRQHIQKFI 1138
G + R+ +P + L +IL+E H + H G + + ++W MR+ + +++
Sbjct: 535 GTIVVNGRVCVPNDRALKEEILREAHQSKFSIHPGSNKMYRDLKRYYHWVGMRKDVARWV 594
Query: 1139 EACVTCQQAKSVNTTYAGLLQPLPIPEQVWDDVTMDFITGLP--LSFGFTVIMVVVDRLS 1196
C TCQ K+ + +GLLQ LPI E WD +TMDF+T LP + + VVVDRL+
Sbjct: 595 AKCPTCQLVKAEHQVPSGLLQNLPISEWKWDHITMDFVTRLPTGIKSKHNAVWVVVDRLT 654
Query: 1197 KYAHFMPLKADYSNRTVAEAFVNYVVKLHGMPKSIISDRDKVFTSKFWQHLFQMQGTTLS 1256
K AHFM + +AE +++ +++LHG+P SI+SDRD FTSKFW + GT ++
Sbjct: 655 KSAHFMAISDKDGAEIIAEKYIDEIMRLHGIPVSIVSDRDTRFTSKFWNAFQKALGTRVN 714
Query: 1257 MSTTYHPQTDGQSEAVNKCLEMYLRCFTFKNPKTWFKALTWVELWYNTFLHTSLGMSPFK 1316
+ST YHPQTDGQSE + LE LR W K L +E YN S+GMSP++
Sbjct: 715 LSTAYHPQTDGQSERTIQTLEDMLRACVLDWGGNWEKYLRLIEFAYNNSFQASIGMSPYE 774
Query: 1317 ALYG---REPPMLTRYSVNNSDPTEVQQQLMDRDKLL-VELKENLLRAQQVMKSNADKKR 1372
ALYG R P T + + +R K L ++LKE AQ KS A+K+R
Sbjct: 775 ALYGRACRTPLCWTPVGERRLFGPTIVDETTERMKFLKIKLKE----AQDRQKSYANKRR 830
Query: 1373 KDVSFEVGDQVLVKLQPYRQSSVALRKNNKLGMRYFGPFTVIAKVGVVAYKLQLPETARI 1432
K++ F+VGD V +K Y+ + KL RY GP+ VI +VG VAYKL LP +
Sbjct: 831 KELEFQVGDLVYLKAMTYKGAG-RFTSRKKLSPRYVGPYKVIERVGAVAYKLDLPPKLNV 889
Query: 1433 -HPVFHVSQLKIFKGLTTEPYFPLPLTTMEEGPVIQPEVILNVRNIIRGDRKVEQLLVK 1490
H VFHVSQL+ + E +P ++E ++ + + + +G R + L+K
Sbjct: 890 FHNVFHVSQLRKYLSDQEESVEDIP-PGLKENMTVEAWPVRIMDRMSKGTRGKSRDLLK 947
>At4g07850 putative polyprotein
Length = 1138
Score = 376 bits (965), Expect = e-104
Identities = 213/535 (39%), Positives = 297/535 (54%), Gaps = 58/535 (10%)
Query: 823 VVSGSGVAMDKIKVIAVLEWPTPTNLKQLRGFLGLTGYYRRFIRSYATIAGPLTNLLKKD 882
V S GV +D+ KV A+ EWP+P ++ ++R F GL G+YRRF+R ++T+A PLT ++KK+
Sbjct: 531 VASTDGVKVDEEKVKAIREWPSPKSVGKVRSFHGLAGFYRRFVRDFSTLAAPLTEVIKKN 590
Query: 883 A-FKWDESTAKAFDTLKQAITTAPVLVLPDFSQPFVLETDASGTGVGAVLSQGGHPIAYF 941
FKW+++ AF LK+ +T APVL LPDF + F +E DA G G+GAVL Q PIAYF
Sbjct: 591 VGFKWEQAPEDAFQALKEKLTHAPVLSLPDFLKTFEIECDAPGVGIGAVLMQDKKPIAYF 650
Query: 942 SKKMASRMQLQSAYTREFYAITTALAKFRHYLLGHKFILRTDQKSLKSLLDQSLQTPEQQ 1001
S+K+ Y +E YA+ AL ++HYL +F++ TD +SLK L Q
Sbjct: 651 SEKLGGATLNYPTYDKELYALVRALQTWQHYLWPKEFVIHTDHESLKHLKGQQKLNKRHA 710
Query: 1002 AWLHKFIGFDFQIEYKPGKDNQAADALSRVMSLSWSAPEHDFLEQLKKEIGNDSHLQTIV 1061
W+ F + I+YK GKDN ADALSR H
Sbjct: 711 RWVEFIETFPYVIKYKKGKDNVVADALSR---------RH-------------------- 741
Query: 1062 QQCVNNTMDDINYMVKEGLLYWKHRLVIPMESNLIHQILKEYHDTPIGGHAGVTRTLARV 1121
EG L++ +RL IP S+L ++E H + GH GV++TL +
Sbjct: 742 ----------------EGFLFYDNRLCIP-NSSLRELFIREAHGGGLMGHFGVSKTLKVM 784
Query: 1122 TAQFYWPNMRQHIQKFIEACVTCQQAKSVNTTYAGLLQPLPIPEQVWDDVTMDFITGLPL 1181
F+WP+M++ +++ E C TC+QAK+ + + GL PLPIP W+D++MDF+ GLP
Sbjct: 785 QDHFHWPHMKRDVERMCERCTTCKQAKAKSQPH-GLCTPLPIPLHPWNDISMDFVVGLPR 843
Query: 1182 S-FGFTVIMVVVDRLSKYAHFMPLKADYSNRTVAEAFVNYVVKLHGMPKSIISDRDKVFT 1240
+ G I VVVDR SK AHF+P +A F VV+LHGMPK+I+SDRD F
Sbjct: 844 TRTGKDSIFVVVDRFSKMAHFIPCHKTDDAMHIANLFFREVVRLHGMPKTIVSDRDTKFL 903
Query: 1241 SKFWQHLFQMQGTTLSMSTTYHPQTDGQSEAVNKCLEMYLRCFTFKNPKTWFKALTWVEL 1300
S FW+ L+ GT L STT HPQTDGQ+E VN+ L LR KN KTW L VE
Sbjct: 904 SYFWKTLWSKLGTKLLFSTTCHPQTDGQTEVVNRTLSTLLRALIKKNLKTWEDCLPHVEF 963
Query: 1301 WYNTFLHTSLGMSPFKALYGREP---------PMLTRYSVNNSDPTEVQQQLMDR 1346
YN +H++ SPF+ +YG P P+++ + P + + +LM R
Sbjct: 964 AYNHSVHSATKFSPFQIVYGFNPITPLDLMPLPLISIHLRKERFPNKRKSKLMPR 1018
Score = 68.2 bits (165), Expect = 4e-11
Identities = 112/552 (20%), Positives = 208/552 (37%), Gaps = 130/552 (23%)
Query: 142 YYNTPDAERLIIASVHLEQEVVPWFQMVNRSR------PFQSWNDFTRALELDFGPSIYD 195
++ +A ++ +A+ + W+ V ++ ++WN ++ F PS Y
Sbjct: 87 FHEYTEANKVKVAATEFYDYALSWWDQVVTTKRRLGDDSIETWNQLKNIMKRRFVPSHYH 146
Query: 196 CPRASLFK--LQQNKSVNEYYLEFTALSNRVYGLSN-DALIDCFVSGL-KDELRRDVMLH 251
+ +Q N++V EY+ E L R +A + F+ GL +D L R ++H
Sbjct: 147 RELHQRLRNLVQGNRTVEEYFKEMETLMLRADVQEECEATMSRFMGGLNRDILDRFEVIH 206
Query: 252 TPISIVKAVSLAKLFEEKIAANFNPKQPLKP-YTPAQPHQHRAPFNPTRNDQNQTIEKAP 310
++ + A +FE++I ++ KP Y ++P R + + + ++
Sbjct: 207 YE-NLEELFHKAVMFEKQIK-----RRSAKPSYNSSKPSYQREEKSGFQKEYKPFVK--- 257
Query: 311 NLPLLPTPPTRPMSHLQKNPAIKRISPA--EMQLRREKGL-CYFCDDKFSFSHKCPNKQL 367
P ++ IS E ++ R + L C+ C ++ +C NK++
Sbjct: 258 -------------------PKVEEISSKGKEKEVTRTRDLKCFKCHGLGHYASECSNKRI 298
Query: 368 MLLELDDDLDPPDTPSQTAVTSDTETQEVAEHHLSLNALKGATGMG----VIRFKGYIGP 423
M++ +++ D + + + E+ L+ L A + + I
Sbjct: 299 MIIRDSGEVESEDEKPEESDVEEAPKGELLVTMRVLSVLNKAEEQAQRENLFHTRCLIKG 358
Query: 424 ISVSILLDGGSSESFIQPRIVHCLNL---PIEHTDKCNVLVGNGQHMKAEGVVRKLSLKV 480
S+++DGGS + +V L L P K L +G+ V L++
Sbjct: 359 KVCSLIIDGGSCTNVASETMVQKLGLEEFPHPKPYKLQWLNESGEMAVTRQVQVPLAIGK 418
Query: 481 QDIDITVPAYLLPVAGSDVILGAPWLASLGPHVADYSVSKLKFYMDGKFVTLQGESDNKP 540
+ +I +LP+ S V+LG PW +SD K
Sbjct: 419 YEDEILCD--ILPLEASHVLLGRPW-----------------------------QSDRK- 446
Query: 541 AVSQLNHFRRLQHMNAISELFTIQKIDPAVIEDNWEGMPVNIEPEMATLLHTYREIFQIP 600
Q + P E+N EG+P + +I +P
Sbjct: 447 ---------------------DYQDVFP---EENPEGLP--------PIRGIEHQIDFVP 474
Query: 601 KGLPPNRELSHEILLKEGAQPVKVKPYRYPHSQKEQIEKMVQDMLEEGIIQPSLSPFSSP 660
PNR PV+ K ++EK V +++E G I S+SP + P
Sbjct: 475 GASLPNRPAYR-------TNPVETK----------ELEKQVNELMERGHICESMSPCAVP 517
Query: 661 IILVKKKDGTWR 672
++LV KKDG+WR
Sbjct: 518 VLLVPKKDGSWR 529
>At4g16910 retrotransposon like protein
Length = 687
Score = 342 bits (878), Expect = 8e-94
Identities = 234/685 (34%), Positives = 337/685 (49%), Gaps = 65/685 (9%)
Query: 871 IAGPLTNLLKKD-AFKWDESTAKAFDTLKQAITTAPVLVLPDFSQPFVLETDASGTGVGA 929
+A P+T L KD AF W E ++F LK +T APVLVLP+ +P+ + TDAS G+G
Sbjct: 1 MAQPMTQLTGKDTAFNWSEECERSFLELKAMLTNAPVLVLPEEGEPYTVYTDASIVGLGC 60
Query: 930 VLSQGGHPIAYFSKKMASRMQLQSAYTREFYAITTALAKFRHYLLGHKFILRTDQKSLKS 989
VL Q G IAY S+++ + E A+ AL +R YL G K + TD KSLK
Sbjct: 61 VLIQKGSVIAYASRQLRKHEKNYPTNDLEMAAVVFALKIWRSYLYGAKVQIFTDHKSLKY 120
Query: 990 LLDQSLQTPEQQAWLHKFIGFDFQIEYKPGKDNQAADALSRVMSLSWS------------ 1037
+ Q Q+ W+ +D I Y PGK NQ DALSR+ ++ +
Sbjct: 121 IFTQPELNLRQRRWIKLVADYDLNIAYHPGKANQVVDALSRLRTVVEAERNQVNLVNMMG 180
Query: 1038 ------------------APEHDFLEQLKKEIGNDSHLQTIVQQCVNNTMDDINYMVKEG 1079
A + D L +++ D ++ Q N G
Sbjct: 181 TLHLNALSKEVEPLGLRAANQADLLSRIRSAQERDEEIKGWAQNNKTEYQSSNN-----G 235
Query: 1080 LLYWKHRLVIPMESNLIHQILKEYHDTPIGGHAGVTRTLARVTAQFYWPNMRQHIQKFIE 1139
+ R+ P + L +ILKE H + H G + + ++W M++ + +++
Sbjct: 236 TIVVNGRVCGPNDKALKEEILKEAHQSKFSIHPGSNKMYRDLKRYYHWVGMKKDVARWV- 294
Query: 1140 ACVTCQQAKSVNTTYAGLLQPLPIPEQVWDDVTMDFITGLP--LSFGFTVIMVVVDRLSK 1197
AK + +G+LQ LPIPE WD + MDF+TGLP + + VVVDRL+K
Sbjct: 295 -------AKEEHQVPSGMLQNLPIPEWKWDHIMMDFVTGLPTGIKSKHNAVWVVVDRLTK 347
Query: 1198 YAHFMPLKADYSNRTVAEAFVNYVVKLHGMPKSIISDRDKVFTSKFWQHLFQMQGTTLSM 1257
AHFM + + +AE +++ +V+LHG+P SI+SDRD FTSKFW+ ++ GT +++
Sbjct: 348 SAHFMAISDKDAAEIIAEKYIDEIVRLHGIPVSIVSDRDTRFTSKFWKPFQKVLGTRVNL 407
Query: 1258 STTYHPQTDGQSEAVNKCLEMYLRCFTFKNPKTWFKALTWVELWYNTFLHTSLGMSPFKA 1317
ST YHPQTDGQSE + LE LR W K L VE YN S+GMSP++A
Sbjct: 408 STAYHPQTDGQSERTIQTLEDMLRACVLDWGGNWEKYLRLVEFAYNNSFQASIGMSPYEA 467
Query: 1318 LY---GREPPMLTRYSVNN-SDPTEVQQQLMDRDKLLVELKENLLRAQQVMKSNADKKRK 1373
LY GR P T P V + L ++LKE AQ KS A+K+RK
Sbjct: 468 LYGRAGRTPLCWTPVGERRLFGPAVVDETTKKMKFLKIKLKE----AQDRQKSYANKRRK 523
Query: 1374 DVSFEVGDQVLVKLQPYRQSSVALRKNNKLGMRYFGPFTVIAKVGVVAYKLQL-PETARI 1432
++ F+VGD V +K Y+ + KL RY GP+ VI +VG VAYKL L P+
Sbjct: 524 ELEFQVGDLVYLKAMTYKGAG-RFTSRKKLRPRYVGPYKVIERVGAVAYKLDLPPKLDAF 582
Query: 1433 HPVFHVSQLKIFKGLTTEPYFPLP-----LTTMEEGPVIQPEVILNVRNIIRGDRKVEQL 1487
H VFHVSQL+ E +P T+E PV ++ ++ RG + ++ L
Sbjct: 583 HNVFHVSQLRKCLSEQEESMEDVPPGLKENMTVEAWPV---RIMDQMKKGTRG-KSMDLL 638
Query: 1488 LVKWKDMQNSEATWEDKQEMLDSYP 1512
+ W E TWE + +M ++P
Sbjct: 639 KILWNCGGREEYTWETETKMKANFP 663
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 301 bits (770), Expect = 3e-81
Identities = 259/910 (28%), Positives = 407/910 (44%), Gaps = 129/910 (14%)
Query: 568 PAVIEDNWEGMPVNIEPEMATLLHTYREIFQIP----KGLPPNRELSHEILLKEGAQPVK 623
P ++ D VN+ + T L YR+ KG+ P +H I L+ +
Sbjct: 846 PVIVNDELTADQVNL---LITELMKYRKAIGYSLDDIKGISPTL-CTHRIHLENESYSSI 901
Query: 624 VKPYRYPHSQKEQIEKMVQDMLEEGIIQP-SLSPFSSPIILVKKKDGTW----------- 671
R + KE ++K + +L+ G+I P S S + SP+ V KK G
Sbjct: 902 EPQRRLNPNLKEVVKKEILKLLDAGVIYPISDSTWVSPVHCVPKKGGMTVVKNSKDELIP 961
Query: 672 -------RCCTDYRALNAITIKDSFPMPTVDELLDELHGAQYFTKLDLRSGYHQILLKPE 724
R C +YR LN + K+ FP+P +D +L+ L Y+ LD SG+ QI + P
Sbjct: 962 TRTITGHRMCIEYRKLNVASRKEHFPLPFIDHMLERLANHPYYCFLDSYSGFFQIPIHPN 1021
Query: 725 DRQKTAFRTHQGHYEWLVMPFGLTSAPATFQRLMNQLFQPLLRKCVLVFFDDILVYSPSW 784
D+ KT F G + + MPFGL +APATFQR M +F L+ + V VF DD VY S+
Sbjct: 1022 DQGKTTFTCPYGTFAYKRMPFGLCNAPATFQRCMTSIFSDLIEEMVEVFMDDFSVYGSSF 1081
Query: 785 HSHLKHVEIVFQILSQNVLYAKLSKCSFGITEVEYLGHVVSGSGVAMDKIKVIAVLEWPT 844
S L ++ V + + L KC F + E LG +S G+ +DK K+ +++
Sbjct: 1082 SSCLLNLCRVLKRCEETNLVLNWEKCHFMVREGIVLGRKISEEGIEVDKAKIDVMMQLQP 1141
Query: 845 PTNLKQLRGFLGLTGYYRRFIRSYATIAGPLTNLLKKDA-FKWDESTAKAFDTLKQAITT 903
P +K +R FLG G+YR FI+ ++ +A PLT LL K+ F +D+ AF +K+A+ T
Sbjct: 1142 PKTVKDIRSFLGHAGFYRIFIKDFSKLARPLTRLLCKETEFAFDDECLTAFKLIKEALIT 1201
Query: 904 APVLVLPDFSQPFVLETDASGTGVGAVLSQGGHPIAYFSKKMASRMQLQSAYT-REFYAI 962
AP++ P++ PF + T Q++ A T +E A+
Sbjct: 1202 APIVQAPNWDFPFEIIT-------------------------MDDAQVRYATTEKELLAV 1236
Query: 963 TTALAKFRHYLLGHKFILRTDQKSLKSLLDQSLQTPEQQAWLHKFIGFDFQIEYKPGKDN 1022
A KFR YL+G K + TD +L+ + + P W+ FD +I K G +N
Sbjct: 1237 VFAFEKFRSYLVGSKVTIYTDHAALRHIYAKKDTKPRLLRWILLLQEFDMEIVDKKGIEN 1296
Query: 1023 QAADALSRV-----MSLSWSAPEHDF--LEQL--KKEIGNDSHLQTIV--QQCVNNTMDD 1071
AD LSR+ + + S PE ++QL KK H+ +V ++ N + +
Sbjct: 1297 GVADHLSRMRIEDEVLIDDSMPEEQLMAIQQLNEKKLPWYADHVNYLVSGEEPPNLSSYE 1356
Query: 1072 INYMVKE-GLLYWKHRLVIPMESNLIHQ----------ILKEYHDTPIGGHAGVTRTLAR 1120
K+ YW + + + I++ IL H GGH +T+++
Sbjct: 1357 KKKFFKDINHFYWDEPYLYTLCKDKIYRTCVSEDEIEGILLHCHGFAYGGHFATFKTMSK 1416
Query: 1121 V-TAQFYWPNMRQHIQKFIEACVTCQQAKSVNTTYAGLLQPLPIPEQVWDDVTMDFITGL 1179
+ A F+WP+M + Q+FI C + + VW +DF+
Sbjct: 1417 ILQAGFWWPSMFKDAQEFISKCDSFENF------------------DVWG---IDFMGPF 1455
Query: 1180 PLSFGFTVIMVVVDRLSKYAHFMPLKADYSN--RTVAEAFVNYVVKLHGMPKSIISDRDK 1237
P S+G I+V +D +SK+ + A ++N R V + F + G+P+ +ISD K
Sbjct: 1456 PSSYGNKYILVAIDYVSKWVEAI---ASHTNDARVVLKLFKTIIFPRFGVPRIVISDGGK 1512
Query: 1238 VFTSKFWQHLFQMQGTTLSMSTTYHPQTDGQSEAVNKCLEMYLRCFTFKNPKTWFKALTW 1297
F +K +++L + G +T GQ E N+ ++ L K W L
Sbjct: 1513 HFINKGFENLLKKHGV--------KHKTSGQVEISNREIKAILEKTVGSTRKDWSAKLND 1564
Query: 1298 VELWYNTFLHTSLGMSPFKALYGR--------EPPMLTRYSVNNSDPTEVQQ----QLMD 1345
Y T T +G +PF LYG+ E + + N D ++ QL D
Sbjct: 1565 TLWAYRTAFKTPIGTTPFNLLYGKSCHLPVELEYKAMWAVKLLNFDIKTAEEKRLIQLND 1624
Query: 1346 RDKLLVELKENLLRAQQVMKSNADKKRKDVSFEVGDQVLVKLQPYRQSSVALRKNNKLGM 1405
+K+ +E E+ ++ KS DKK F+VGDQVL+ R KL
Sbjct: 1625 LNKIRLEAYESSKIYKERTKSFHDKKIVSRDFKVGDQVLLFNSRLRLFP------GKLKS 1678
Query: 1406 RYFGPFTVIA 1415
R+ GPF+V A
Sbjct: 1679 RWSGPFSVTA 1688
>At2g14640 putative retroelement pol polyprotein
Length = 945
Score = 300 bits (769), Expect = 3e-81
Identities = 149/315 (47%), Positives = 206/315 (65%), Gaps = 2/315 (0%)
Query: 1210 NRTVAEAFVNYVVKLHGMPKSIISDRDKVFTSKFWQHLFQMQGTTLSMSTTYHPQTDGQS 1269
+R+VA FV++VVKLHG+P+SI+SD D +F S FWQ +++ T L MST YHPQTDGQ+
Sbjct: 631 SRSVAAKFVSHVVKLHGIPRSIVSDCDPIFMSLFWQEFWKLSRTKLWMSTAYHPQTDGQT 690
Query: 1270 EAVNKCLEMYLRCFTFKNPKTWFKALTWVELWYNTFLHTSLGMSPFKALYGREPPMLTRY 1329
E VN+C+E +LRCF +PK W + W E WYNT H S GM+PF+ALYGR P + Y
Sbjct: 691 EVVNRCIEQFLRCFVHYHPKQWSSFIPWAEYWYNTTFHASTGMTPFQALYGRPPSPIPAY 750
Query: 1330 SVNNSDPTEVQQQLMDRDKLLVELKENLLRAQQVMKSNADKKRKDVSFEVGDQVLVKLQP 1389
+ + E+ +Q+ RD+LL ELK++L+ A MK AD + +DVSF+VGD VL+++QP
Sbjct: 751 ELGSVVCGELNEQMAARDELLAELKQHLVTANNCMKQQADSRLRDVSFQVGDWVLLRIQP 810
Query: 1390 YRQSSVALRKNNKLGMRYFGPFTVIAKVGVVAYKLQLPETARIHPVFHVSQLKIFKGLTT 1449
YRQ ++ R + KL R++GPF V +K G VAY+L LPE RIHPVFHVS LK + G
Sbjct: 811 YRQKTLFRRSSQKLSHRFYGPFQVASKHGEVAYRLTLPEGTRIHPVFHVSLLKPWVGDGE 870
Query: 1450 EPYFPLPLTTMEEGPVIQPEVILNVRNIIRGDRKVEQLLVKWKDMQNSEATWEDKQEMLD 1509
LP +QP +L VR + ++V LLV+W+ + +ATWE+ ++
Sbjct: 871 PDMGQLPPLRNNGELKLQPTAVLEVRWRSQDKKRVADLLVQWEGLHIEDATWEEYDQLAA 930
Query: 1510 SYPN--LNLEDKIVL 1522
S+P LNLEDK+ L
Sbjct: 931 SFPEFVLNLEDKVRL 945
Score = 212 bits (540), Expect = 1e-54
Identities = 131/385 (34%), Positives = 195/385 (50%), Gaps = 39/385 (10%)
Query: 390 DTETQEVAEHHLSLNALKGATGMGVIRFKGYIGPISVSILLDGGSSESFIQPRIVHCLNL 449
+ T+ + ++L AL+G IR + I + L++ GS+ +FI + V +NL
Sbjct: 293 EESTEAEGKPEITLYALEGVDTTSTIRVRATIHRNRLIALINSGSTHNFIGEKAVRGMNL 352
Query: 450 PIEHTDKCNVLVGNGQHMKAEGVVRKLSLKVQDIDITVPAYLLPVAGSDVILGAPWLASL 509
T V V NG + + + + + V Y LP+ G D+ +G WL++L
Sbjct: 353 KATTTKPFTVRVVNGMPLVCRSRYEAIPVVMGGVVFPVTLYALPLMGLDLAMGVQWLSTL 412
Query: 510 GPHVADYSVSKLKFYMDGKFVTLQGESDNKPAVSQLNHFRRLQHMNAISELFTIQKIDPA 569
GP + ++ L+F+ G V L G KP R ++H + I
Sbjct: 413 GPTLCNWKEQTLQFHWAGDEVRLMGI---KPT-----GLRGVEHKTITKKARMGHTIFAI 464
Query: 570 VIEDNWEGMPVNIEPEMATLLHTYREIFQIPKGLPPNRELSHEILLKEGAQPVKVKPYRY 629
+ N P + E+ L+ + +F+ P LP R + H I LK+G PV V+PYRY
Sbjct: 465 TMAHN-SSDPNTPDGEIKKLIEEFAGLFETPTQLPSERPIVHRIALKKGTDPVNVRPYRY 523
Query: 630 PHSQKEQIEKMVQDMLEEGIIQPSLSPFSSPIILVKKKDGTWRCCTDYRALNAITIKDSF 689
QK+++ + GII+PS SPF SP++L + F
Sbjct: 524 AFYQKDEMSRA-------GIIRPSSSPFLSPVLL-----------------------ERF 553
Query: 690 PMPTVDELLDELHGAQYFTKLDLRSGYHQILLKPEDRQKTAFRTHQGHYEWLVMPFGLTS 749
P+PTVD++LDEL+GA YFTKLDL +GY Q+ + D K AF+TH GHYE+LVMPFGL +
Sbjct: 554 PIPTVDDMLDELNGAVYFTKLDLTAGYQQVRMHSPDIPKIAFQTHNGHYEYLVMPFGLCN 613
Query: 750 APATFQRLMNQLFQPLLRKCVLVFF 774
AP+TFQ LMN++F PLL + V F
Sbjct: 614 APSTFQALMNEIFWPLLSRSVAAKF 638
Score = 64.7 bits (156), Expect = 4e-10
Identities = 39/148 (26%), Positives = 66/148 (44%), Gaps = 28/148 (18%)
Query: 115 RHIKLEFPRFNGKHVLDWIFKAEQYFGYYNTPDAERLIIASVHLEQEVVPWFQMVNRSRP 174
R KL+FPRF+G + W+ KA+QYF Y+ P + + + HL+ W+Q R
Sbjct: 102 RPSKLDFPRFHGGDLTKWLSKAKQYFEYHEGPVEQAVRFTTFHLDGVANGWWQATLRRSA 161
Query: 175 FQSWNDFTRALELDFGPSIYDCPRASLFKLQQNKSVNEYYLEFTALSNRVYGLSNDALID 234
+ Q S++EY EF L N+VYG + + L+
Sbjct: 162 ----------------------------TIGQTGSLSEYQHEFEWLQNKVYGWTQEVLVG 193
Query: 235 CFVSGLKDELRRDVMLHTPISIVKAVSL 262
F++GL + + + P S+ + ++L
Sbjct: 194 AFMNGLYYSISNGIRMFQPRSLREVINL 221
>At4g03650 putative reverse transcriptase
Length = 839
Score = 262 bits (669), Expect = 1e-69
Identities = 135/298 (45%), Positives = 186/298 (62%), Gaps = 1/298 (0%)
Query: 720 LLKPEDRQKTAFRTHQGHYEWLVMPFGLTSAPATFQRLMNQLFQPLLRKCVLVFFDDILV 779
LL D +KTAFRT GH+E++VMPFGLT+APA F RLMN +FQ L + V++F DDILV
Sbjct: 515 LLAEADVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILV 574
Query: 780 YSPSWHSHLKHVEIVFQILSQNVLYAKLSKCSFGITEVEYLGHVVSGSGVAMDKIKVIAV 839
YS S H H+ V + L + L+AKLSKCSF E+ +LGH+VS GV++D K+ A+
Sbjct: 575 YSKSPEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAI 634
Query: 840 LEWPTPTNLKQLRGFLGLTGYYRRFIRSYATIAGPLTNLLKKDA-FKWDESTAKAFDTLK 898
+WP PTN ++R FLGL GYYRRFI+ +A++A P+T L KD F W + F +LK
Sbjct: 635 RDWPRPTNATEIRSFLGLAGYYRRFIKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLK 694
Query: 899 QAITTAPVLVLPDFSQPFVLETDASGTGVGAVLSQGGHPIAYFSKKMASRMQLQSAYTRE 958
+ +T+ PVL LP+ +P+++ TDASG G+G L Q G IAY S+++ + E
Sbjct: 695 EMLTSTPVLALPEHGEPYMVYTDASGVGLGCALMQRGKVIAYASRQLRKHEGNYPTHDLE 754
Query: 959 FYAITTALAKFRHYLLGHKFILRTDQKSLKSLLDQSLQTPEQQAWLHKFIGFDFQIEY 1016
A+ AL +R YL G K + TD KSLK + Q Q+ W+ +D +I Y
Sbjct: 755 MAAVIFALKIWRSYLYGGKVQVFTDHKSLKYIFTQPELNLRQRRWMELVADYDLEIAY 812
Score = 40.4 bits (93), Expect = 0.008
Identities = 46/219 (21%), Positives = 87/219 (39%), Gaps = 40/219 (18%)
Query: 445 HCL-NLPIEHTDKCNVLVGNGQHMKAEGVVRKLSLKV----QDIDITVPAYLLP------ 493
HC+ +LP E + N+ G+ + A V L V + +DI + LP
Sbjct: 332 HCIASLPPESASRGNIRGDPGEQLGAVKVAGGQFLAVLGGAKGVDIQIAGESLPADLIIS 391
Query: 494 -VAGSDVILGAPWLASLGPHVADYSVSKLKFYMDGKFVTLQGESDNKPAVSQLNHFRRLQ 552
V DVILG WL H+ D ++ F + + + + + + +
Sbjct: 392 HVELYDVILGMDWLDHYRVHL-DCHRGRVSFERPEGSAGRENDREGLRGLPGDDIYAGVC 450
Query: 553 HMNAISELFTIQKIDPAVIEDNWEGMPVNIEPEMATLLHTYREIFQIPKGLPPNRELSHE 612
A+S++ +Q+ ++ +FQ +GLPP+R
Sbjct: 451 GQVAVSDIRVVQE---------------------------FQYVFQSLQGLPPSRSDPFT 483
Query: 613 ILLKEGAQPVKVKPYRYPHSQKEQIEKMVQDMLEEGIIQ 651
I L+ P+ PYR ++ +++K ++D+L E ++
Sbjct: 484 IELEPKTAPLSKAPYRMAPAKMAELKKQLEDLLAEADVR 522
Score = 40.4 bits (93), Expect = 0.008
Identities = 19/25 (76%), Positives = 21/25 (84%), Gaps = 1/25 (4%)
Query: 1246 HLFQMQGTTLSMSTTYHPQTDGQSE 1270
HL +M GT + MSTTYHPQTDGQSE
Sbjct: 813 HLAEM-GTKVQMSTTYHPQTDGQSE 836
>At4g07830 putative reverse transcriptase
Length = 611
Score = 248 bits (632), Expect = 3e-65
Identities = 183/563 (32%), Positives = 261/563 (45%), Gaps = 71/563 (12%)
Query: 982 TDQKSLKSLLDQSLQTPEQQAWLHKFIGFDFQIEYKPGKDNQAADALSRVMSLSWSAPEH 1041
TD KSLK + Q + W+ +D +I Y PGK N DALSR A
Sbjct: 33 TDHKSLKYIFTQLELNLRLRRWMKLVADYDLEIAYHPGKANVVTDALSRKRV---GAAPG 89
Query: 1042 DFLEQLKKEIG-----------------NDSHLQTIVQQCVNNTMDDINYMVKEGLLYW- 1083
+E L EIG + + L + VQ I EG Y
Sbjct: 90 QSVETLVIEIGALRLCAVAREPLGLEAVDQTDLLSRVQLAQEKDEGLIAASKAEGFEYQF 149
Query: 1084 --------KHRLVIPMESNLIHQILKEYHDTPIGGHAGVTRTLARVTAQFYWPNMRQHIQ 1135
R+ +P + L +IL E H + H G T+ + + W M++ +
Sbjct: 150 AANGTILVHGRVCVPKDKELRQEILSEAHASMFSIHPGATKMYRDLKRYYQWVGMKRDVG 209
Query: 1136 KFIEACVTCQQAKSVNTTYAGLLQPLPIPEQVWDDVTMDFITGLPLSFGFTVIMVVVDRL 1195
++E C CQ K + LLQ LPIPE WD +TMDF+ GLP+S I V+VDRL
Sbjct: 210 NWVEECDVCQLVKIEHQVSGSLLQSLPIPEWKWDFITMDFVVGLPVSRTKDAIWVIVDRL 269
Query: 1196 SKYAHFMPLKADYSNRTVAEAFVNYVVKLHGMPKSIISDRDKVFTSKFWQHLFQMQGTTL 1255
+K AHF+ ++ +A+ +V+ +VKLHG+P SI+SDRD FTS FW+ GT +
Sbjct: 270 TKSAHFLAIRKTDGAAVLAKKYVSEIVKLHGVPVSIVSDRDSKFTSAFWRAFQAEMGTKV 329
Query: 1256 SMSTTYHPQTDGQSEAVNKCLEMYLRCFTFKNPKTWFKALTWVELWYNTFLHTSLGMSPF 1315
MST YHPQTDGQSE + LE LR W L+ VE YN S+GM+PF
Sbjct: 330 QMSTAYHPQTDGQSERTIQTLEDMLRMCVLDWRGHWADHLSLVEFAYNNSYQASIGMAPF 389
Query: 1316 KALYGREPPML--TRYSVNNSDPTEVQQQLMDRDKLLVELKENLLRAQQVMKSNADKKRK 1373
+ LYGR L T+ + + Q++ +R ++ LK N+ AQ +S ADK+RK
Sbjct: 390 EVLYGRPCRTLCWTQVGERSIYGADYVQEITERIRV---LKLNMKEAQNRQRSYADKRRK 446
Query: 1374 DVSFEVGDQVLVKLQPYRQSSVALRKNNKLGMRYFGPFTVIAKVGVVAYKLQLPETAR-I 1432
++ FEVGD V Q R +VG VA++L+L + R
Sbjct: 447 ELEFEVGDSV-------SQDGHVARSEQ--------------RVGPVAFRLELSDVMRAF 485
Query: 1433 HPVFHVSQLKIFKGLTTEPYFPLPLTTMEEGPVIQPEVILNVRNIIRGDRKVEQLLVK-- 1490
H VFHVS L+ E +P +QP + L R + +R++++L K
Sbjct: 486 HKVFHVSMLRKCLHKDDEVLAKIP-------EDLQPNMTLEARPVRVLERRIKELRRKKI 538
Query: 1491 --WKDMQN----SEATWEDKQEM 1507
K ++N +E TWE + +
Sbjct: 539 PLIKVLRNCDGVTEETWEPEARL 561
>At1g20390 hypothetical protein
Length = 1791
Score = 232 bits (591), Expect = 1e-60
Identities = 172/609 (28%), Positives = 289/609 (47%), Gaps = 22/609 (3%)
Query: 428 ILLDGGSSESFIQPRIVHCLNLPIEHTDKCNV-LVG-NGQHMKAEGVVRKLSLKVQDIDI 485
+L+D GSS I ++ +N+ + L G +G + G + KL + V +
Sbjct: 585 VLIDTGSSVDLIFKDVLTAMNITDRQIKPVSKPLAGFDGDFVMTIGTI-KLPIFVGGLIA 643
Query: 486 TVPAYLL-PVAGSDVILGAPWLASLGPHVADYSVSKLKFYMDGKFVTLQGESDNKPAVSQ 544
V ++ A +VILG PW+ + + Y +KF TL+ + K
Sbjct: 644 WVKFVVIGKPAVYNVILGTPWIHQMQAIPSTYHQC-VKFPTHNGIFTLRAPKEAKTPSRS 702
Query: 545 LNHFRRLQHMNAISELFTIQKIDPAVIEDNWEGMPVNIEPEMATLLHTYREIF----QIP 600
+ +E+ I + DP + +I E+ LL + F +
Sbjct: 703 YEESELCR-----TEMVNIDESDPTRCVGVGAEISPSIRLELIALLKRNSKTFAWSIEDM 757
Query: 601 KGLPPNRELSHEILLKEGAQPVKVKPYRYPHSQKEQIEKMVQDMLEEG-IIQPSLSPFSS 659
KG+ P +HE+ + +PVK K + + + + V+ +L+ G II+ + +
Sbjct: 758 KGIDP-AITAHELNVDPTFKPVKQKRRKLGPERARAVNEEVEKLLKAGQIIEVKYPEWLA 816
Query: 660 PIILVKKKDGTWRCCTDYRALNAITIKDSFPMPTVDELLDELHGAQYFTKLDLRSGYHQI 719
++VKKK+G WR C DY LN KDS+P+P +D L++ G + +D SGY+QI
Sbjct: 817 NPVVVKKKNGKWRVCVDYTDLNKACPKDSYPLPHIDRLVEATSGNGLLSFMDAFSGYNQI 876
Query: 720 LLKPEDRQKTAFRTHQGHYEWLVMPFGLTSAPATFQRLMNQLFQPLLRKCVLVFFDDILV 779
L+ +D++KT+F T +G Y + VM FGL +A AT+QR +N++ + + V V+ DD+LV
Sbjct: 877 LMHKDDQEKTSFVTDRGTYCYKVMSFGLKNAGATYQRFVNKMLADQIGRTVEVYIDDMLV 936
Query: 780 YSPSWHSHLKHVEIVFQILSQNVLYAKLSKCSFGITEVEYLGHVVSGSGVAMDKIKVIAV 839
S H++H+ F +L+ + +KC+FG+T E+LG+VV+ G+ + ++ A+
Sbjct: 937 KSLKPEDHVEHLSKCFDVLNTYGMKLNPTKCTFGVTSGEFLGYVVTKRGIEANPKQIRAI 996
Query: 840 LEWPTPTNLKQLRGFLGLTGYYRRFIRSYATIAGPLTNLLKKDA-FKWDESTAKAFDTLK 898
LE P+P N ++++ G RFI P NLLK+ A F WD+ + +AF+ LK
Sbjct: 997 LELPSPRNAREVQRLTGRIAALNRFISRSTDKCLPFYNLLKRRAQFDWDKDSEEAFEKLK 1056
Query: 899 QAITTAPVLVLPDFSQPFVLETDASGTGVGAVLSQGG----HPIAYFSKKMASRMQLQSA 954
++T P+LV P+ + L S V +VL + PI Y SK +
Sbjct: 1057 DYLSTPPILVKPEVGETLYLYIAVSDHAVSSVLVREDRGEQRPIFYTSKSLVEAETRYPV 1116
Query: 955 YTREFYAITTALAKFRHYLLGHKFILRTDQKSLKSLLDQSLQTPEQQAWLHKFIGFDFQI 1014
+ A+ T+ K R Y H + TDQ L+ L Q+ W + +D
Sbjct: 1117 IEKAALAVVTSARKLRPYFQSHTIAVLTDQ-PLRVALHSPSQSGRMTKWAVELSEYDIDF 1175
Query: 1015 EYKPGKDNQ 1023
+P +Q
Sbjct: 1176 RPRPAMKSQ 1184
Score = 88.2 bits (217), Expect = 3e-17
Identities = 92/344 (26%), Positives = 144/344 (41%), Gaps = 24/344 (6%)
Query: 1096 IHQILKEYHDTPIGGHAGVTRTLARVTAQ--FYWPNMRQHIQKFIEACVTCQQAKSVNTT 1153
I++I+KE H+ G H+G R LA + FYWP M + F C CQ+
Sbjct: 1435 INEIMKETHEGAAGNHSG-GRALALKLKKLGFYWPTMISDCKTFTAKCEQCQRHAPTIHQ 1493
Query: 1154 YAGLLQP--LPIPEQVWDDVTMDFITGLPLSFGFTVIMVVVDRLSKYAHFMPLKADYSNR 1211
LL+ P P W MD + +P S I+V+ D +K+ +N
Sbjct: 1494 PTELLRAGVAPYPFMRW---AMDIVGPMPASRQKRFILVMTDYFTKWVEAESYATIRAN- 1549
Query: 1212 TVAEAFVNYVVKLHGMPKSIISDRDKVFTSKFWQHLFQMQGTTLSMSTTYHPQTDGQSEA 1271
V +++ HG+P II+D F S +++ L+ ST +PQ +GQ+EA
Sbjct: 1550 DVQNFVWKFIICRHGLPYEIITDNGSQFISLSFENFCASWKIRLNKSTPRYPQGNGQAEA 1609
Query: 1272 VNKCLEMYLRCFTFKNPKTWFKALTWVELWYNTFLHTSLGMSPFKALYGRE---PPMLTR 1328
NK + L+ + W L V Y T ++ +PF YG E P +
Sbjct: 1610 TNKTILSGLKKRLDEKKGAWADELDGVLWSYRTTPRSATDQTPFAHAYGMEAMAPAEVGY 1669
Query: 1329 YSVNNS----DPTEVQQQLMDRDKLLVELKENLL----RAQQVMKSNADKKRKDVSFEVG 1380
S+ S +P + ++DR L E++ L Q + ++K + F+VG
Sbjct: 1670 SSLRRSMMVKNPELNDRMMLDRLDDLEEIRNAALCRIQNYQLAAAKHYNQKVHNRHFDVG 1729
Query: 1381 DQVLVKLQPYRQSSVALRKNNKLGMRYFGPFTVIAKVGVVAYKL 1424
D VL K+ + A KLG + G + V V Y+L
Sbjct: 1730 DLVLRKV----FENTAEINAGKLGANWEGSYQVSKIVRPGDYEL 1769
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 226 bits (577), Expect = 6e-59
Identities = 228/938 (24%), Positives = 393/938 (41%), Gaps = 153/938 (16%)
Query: 641 VQDMLEEGIIQPSLSP-FSSPIILVKKKDGTWRCCTDYRALNAITIKDSFPMPTVDELLD 699
V +L+ G I+ P + + ++VKKK+G R C D+ LN KDSFP+P +D L++
Sbjct: 484 VDKLLKIGSIREVQYPDWLANTVVVKKKNGKDRVCIDFTDLNKACPKDSFPLPHIDRLVE 543
Query: 700 ELHGAQYFTKLDLRSGYHQILLKPEDRQKTAFRTHQGHYEWLVMPFGLTSAPATFQRLMN 759
G + + +D SGY+QI++ PED++KT F T +G Y + VMPFGL +A AT+ RL+N
Sbjct: 544 STAGNELLSFMDAFSGYNQIMMNPEDQEKTLFITDRGIYCYKVMPFGLRNAGATYPRLVN 603
Query: 760 QLFQPLLRKCVLVFFDDILVYSPSWHSHLKHVEIVFQILSQNVLYAKLSKCSFGITEVEY 819
++F + K + V+ DD+L+ S H+KH+E F IL+Q + +KC+FG+ E+
Sbjct: 604 KMFSEHVGKTMEVYIDDMLIKSLKKEDHVKHLEECFAILNQYQMKLNPAKCTFGVPSGEF 663
Query: 820 LGHVVSGSGVAMDKIKVIAVLEWPTPTNLKQLRGFLGLTGYYRRFIRSYATIAGPLTNLL 879
LG++V+ G+ + ++ A L P+P N K+++ G RFI + P +L
Sbjct: 664 LGYIVTKRGIEANPNQINAFLNMPSPKNFKEVQRLTGRIAALNRFISRSTDKSLPFYQIL 723
Query: 880 K-KDAFKWDESTAKAFDTLKQAITTAPVLVLPDFSQPFVLETDASGTGVGAVLSQGGH-- 936
K F WDE +AF LK +T+ PVL P+ + L S V VL +
Sbjct: 724 KGNKEFLWDEKCEEAFGQLKAYLTSPPVLFKPELDEKLYLYVSVSNHAVSGVLIREDRGE 783
Query: 937 --PIAY-----FSKKMASRMQLQSAYTREFYAITTALAKFRHYLLGHKFILRTD------ 983
PI Y F+ ++ AY + KF + D
Sbjct: 784 QKPIYYIIVNQFNGDYEAKDSRMEAYLHVVKDLAKNFRKFELIRIPRGQNTTADALAALA 843
Query: 984 -----------------QKSLKSLLDQSLQTPEQQAWLHKFIGFDFQIEYKPGKDNQAAD 1026
++S+K + + T + A + D +E P K ++ +
Sbjct: 844 STSDPEVNSIIPVECISERSIKEEKEAFVVTRSRAAARDR---GDTAVELPPVKRRKSKE 900
Query: 1027 ALSRVMSLSWSAPEHDFLEQLKKEIGNDSHLQTIVQQCVNNTM---------DDINYMVK 1077
A ++ A + + +L +E+ +D+ ++ + V+ + + +N + +
Sbjct: 901 A-----TVPKPAVQENIGIELIEEVISDAKIEAETEPWVDEDILAPNEHPEPEALNSLYR 955
Query: 1078 EGLLYWKHRLVIPMESNLIHQILKEYHDTPIGGHAGVTRTLARVTAQ-FYWPNMRQHIQK 1136
G+ ++ + ++ ++ E H+ G H+ ++ +YWP M K
Sbjct: 956 RGV---SDPYLLSIFGPVVEIVMSEVHEGLCGSHSSGRAMAFKIKKMCYYWPTMITDCVK 1012
Query: 1137 FIEACVTCQQAKSVNTTYAGLLQPL--PIPEQVWDDVTMDFITGLPLSF-GFTVIMVVVD 1193
+ + C CQ + + L + P P W +MD I L S G ++V+ D
Sbjct: 1013 YAQRCKRCQLHAPLIHQPSELFSSISAPYPFMRW---SMDIIGPLHRSTRGVQYLLVLTD 1069
Query: 1194 RLSKYAHFMPLKADYSNRTVAEA-FVNYVVKLHGMPKSIISDRDKVFTSKFWQHLFQMQG 1252
SK+ AEA +N+ +K
Sbjct: 1070 YFSKWIE-------------AEAILLNWGIK----------------------------- 1087
Query: 1253 TTLSMSTTYHPQTDGQSEAVNKCLEMYLRCFTFKNPKTWFKALTWVELWYNTFLHTSLGM 1312
+S ST +PQ +GQ+EA NK + L+ W+ L V Y T S G
Sbjct: 1088 --VSYSTLRYPQGNGQAEAANKTILSNLKKRLNHLKGGWYDELQPVLWAYRTTPRRSTGE 1145
Query: 1313 SPFKALYGRE---PPML-------TRYSVNNSDPTEVQQQLMD-----RDKLLVELKENL 1357
+PF +YG + P L T +N + + + +D RD+ L++++
Sbjct: 1146 TPFSLVYGMKAVVPAELNVPGLRRTEAPLNEKENSAMLDDSLDTINERRDQALIQIQNYQ 1205
Query: 1358 LRAQQVMKSNADKKRKDVSFEVGDQVLVKLQPYRQSSVALRKNNKLGMRYFGPFTVIAKV 1417
A + S K K F VGD VL ++ ++ A KLG+ + GP+ V V
Sbjct: 1206 HAAARYYNS----KVKSRPFFVGDYVLKRVFDNKKEEGA----GKLGINWEGPYIVTEVV 1257
Query: 1418 GVVAYKLQLPETARIHPVFHV--------------SQLKIFKGLTTEPYFPLPLTTMEEG 1463
Y+L+ E + ++V S+ G+ E +F L +++EG
Sbjct: 1258 RNGVYRLKDLEDRPVQRPWNVNGHHTIRDASKLCDSEFDPISGVRVEAFFVLSRESLDEG 1317
Query: 1464 PVIQPEVILNVRNIIRGDRKVEQLLVKWKDMQNSEATW 1501
R+++ RKV + D+ + W
Sbjct: 1318 H----------RDVLVSARKVPDKCTRSSDLGSKVTKW 1345
>At4g04230 putative transposon protein
Length = 315
Score = 194 bits (494), Expect = 3e-49
Identities = 101/233 (43%), Positives = 143/233 (61%), Gaps = 41/233 (17%)
Query: 582 IEPEMATLLHTYREIF--QIPKGLPPNRELSHEILLKEGAQPVKVKPYRYPHSQKEQIEK 639
+ E+ +LH Y+++F +IP GLPP
Sbjct: 99 LSAEVQVVLHWYKDLFPEEIPPGLPP---------------------------------- 124
Query: 640 MVQDMLEEGIIQPSLSPFSSPIILVKKKDGTWRCCTDYRALNAITIKDSFPMPTVDELLD 699
+ +G I+ SLSP + P++LV KKDGTWR C D RA+N ITIK P+P + ++LD
Sbjct: 125 -----IHKGYIRESLSPCAVPVLLVPKKDGTWRMCLDCRAINNITIKYRHPIPRLYDMLD 179
Query: 700 ELHGAQYFTKLDLRSGYHQILLKPEDRQKTAFRTHQGHYEWLVMPFGLTSAPATFQRLMN 759
EL GA F+K+DLRSGYHQ+ ++ D KTAF+T QG YE LVMPFGLT+AP+TF RLMN
Sbjct: 180 ELSGAIIFSKVDLRSGYHQVRMREGDEWKTAFKTKQGLYECLVMPFGLTNAPSTFMRLMN 239
Query: 760 QLFQPLLRKCVLVFFDDILVYSPSWHSHLKHVEIVFQILSQNVLYAKLSKCSF 812
Q+ + + K V+V+FDDIL+Y+ S+ H++ +E++ + L + LYA L KC+F
Sbjct: 240 QVLRSFIGKFVVVYFDDILIYNKSYSDHIQPLELILKTLRKEGLYANLKKCTF 292
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,275,864
Number of Sequences: 26719
Number of extensions: 1550172
Number of successful extensions: 4656
Number of sequences better than 10.0: 105
Number of HSP's better than 10.0 without gapping: 71
Number of HSP's successfully gapped in prelim test: 34
Number of HSP's that attempted gapping in prelim test: 4289
Number of HSP's gapped (non-prelim): 217
length of query: 1569
length of database: 11,318,596
effective HSP length: 112
effective length of query: 1457
effective length of database: 8,326,068
effective search space: 12131081076
effective search space used: 12131081076
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC124965.10


