

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124963.7 + phase: 0
(512 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
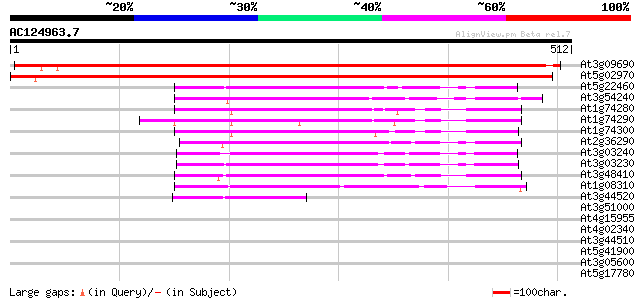
Score E
Sequences producing significant alignments: (bits) Value
At3g09690 unknown protein 604 e-173
At5g02970 unknown protein 598 e-171
At5g22460 putative protein 151 9e-37
At3g54240 putative protein (fragment) 136 2e-32
At1g74280 unknown protein 136 3e-32
At1g74290 unknown protein 131 8e-31
At1g74300 unknown protein 130 2e-30
At2g36290 unknown protein 126 3e-29
At3g03240 hypothetical protein 121 1e-27
At3g03230 hypothetical protein 115 6e-26
At3g48410 unknown protein 115 8e-26
At1g08310 unknown protein 111 1e-24
At3g44520 putative protein 97 2e-20
At3g51000 epoxide hydrolase-like protein 40 0.003
At4g15955 unknown protein 40 0.004
At4g02340 unknown protein 39 0.009
At3g44510 putative protein 38 0.012
At5g41900 unknown protein 37 0.026
At3g05600 putative epoxide hydrolase 36 0.044
At5g17780 putative protein 34 0.22
>At3g09690 unknown protein
Length = 527
Score = 604 bits (1558), Expect = e-173
Identities = 288/506 (56%), Positives = 376/506 (73%), Gaps = 13/506 (2%)
Query: 5 RGTTWWSEELASLMENSLPESST----TTSFEDVRKSSSSEE---VEVGESLKEHAVGFL 57
RG W +ELASLM+ L + T E KSS E E ESLK+ GF+
Sbjct: 6 RGVPTWQDELASLMDGGLQYDGSPIDLTADTESRSKSSGFESGSGSESVESLKDQVTGFM 65
Query: 58 MAWCEILMELGRGCRDILQQNFFNEDSYLVQKLRGPCSKLSKRLSFLNDFLPEDRDPLLA 117
+W E+LM+L GC+D++QQ +DS++V+KLR P +K+SK+LSFLN++LPEDRDP+ A
Sbjct: 66 KSWGEMLMDLAIGCKDVVQQMVVTDDSFVVRKLRKPAAKVSKKLSFLNEYLPEDRDPVHA 125
Query: 118 WSIVFTVFLLAFAAISVDSNHQTSTKAAM-VRMHPPIASRILLPDGRYMAYQDQGVPPGR 176
W ++F VFLLA A+S S+H S +R+HP ASR+ LPDGRY+AYQ+ GV R
Sbjct: 126 WPVIFFVFLLALTALSFSSDHDRSVPLLKKIRLHPTSASRVQLPDGRYLAYQELGVSADR 185
Query: 177 ARFSLVAPHSFLSSRLAGIPGVKASLLEDYGVRLVTYDLPGFGESDPHPSRNFNSSAMDM 236
AR SL+ PHSFLSSRLAGIPGVK SLL+DYGVRLV+YDLPGFGESDPH +RN +SSA DM
Sbjct: 186 ARHSLIVPHSFLSSRLAGIPGVKESLLKDYGVRLVSYDLPGFGESDPHRARNLSSSASDM 245
Query: 237 LHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIPERIAGAAMLAPMVSPYESHMTKDEMKR 296
+ L A+ + DKFW+L +SSG +HAWA+++Y P++IAG AM+APM++PYE MTK+EM +
Sbjct: 246 IDLAAALGIVDKFWLLGYSSGSVHAWAAMRYFPDQIAGVAMVAPMINPYEPSMTKEEMAK 305
Query: 297 TWEKWLPRRKYMYSLAYRFPKLLSFFYRKSFLPEKHERIDKQFSLSLGKKDEILVDEPAF 356
TWE+W +RK+MY LA R+P LL F YR+SFL E +DK S+SLG+KD+++ +P F
Sbjct: 306 TWEQWQRKRKFMYFLARRWPSLLPFSYRRSFLSGNLEPLDKWMSVSLGEKDKLVTADPVF 365
Query: 357 EEYWQRDLEESVRQGNLKPFIEEALLQVSRWDFNIEELHVHKKCQTGGLLLWLKSMYGQA 416
E+ +QR++EESVRQG KPF+EEA LQVS W F++ E H+ KKC+T G+L WL SMY ++
Sbjct: 366 EDLYQRNVEESVRQGTAKPFVEEAALQVSNWGFSLPEFHMQKKCRTNGVLSWLMSMYSES 425
Query: 417 ECELAGYLGRIHIWQGLDDRMVPPSMTEYIERVLPEAVIHKLPNEGHFSYFFFCDECHKQ 476
ECEL G+ IHIWQG+DDR+ PPS+T+YI RV+PEA +H+LPNEGHFSYF+ CDECH Q
Sbjct: 426 ECELIGFRKPIHIWQGMDDRVTPPSVTDYISRVIPEATVHRLPNEGHFSYFYLCDECHTQ 485
Query: 477 IFSTLFGTPQGPFERQEETMLEKNTE 502
IFS +FG P+GP E +LE+ TE
Sbjct: 486 IFSAIFGEPKGPVE-----LLEQRTE 506
>At5g02970 unknown protein
Length = 514
Score = 598 bits (1541), Expect = e-171
Identities = 283/501 (56%), Positives = 373/501 (73%), Gaps = 6/501 (1%)
Query: 1 MAEARGTTWWSEELASLMENSL-----PESSTTTSFEDVRKSSSSEEVEVGESLKEHAVG 55
M E RG W EELASL++ L P T + S+ E E+LK+ G
Sbjct: 1 MEEIRGVPTWQEELASLVDAGLRYDGAPIDLTAATKRSGFVSADGSGSEPKETLKDQVTG 60
Query: 56 FLMAWCEILMELGRGCRDILQQNFFNEDSYLVQKLRGPCSKLSKRLSFLNDFLPEDRDPL 115
F+ +W E+L+EL +GC+DI+QQ +DS+LV+KLR P +K+SK+LSFLN+FLPEDRDP+
Sbjct: 61 FMKSWGEMLLELAKGCKDIVQQTVVTDDSFLVRKLRKPAAKVSKKLSFLNEFLPEDRDPI 120
Query: 116 LAWSIVFTVFLLAFAAISVD-SNHQTSTKAAMVRMHPPIASRILLPDGRYMAYQDQGVPP 174
AW ++F VFLLA AA+S N + T +R+HP A+R+ LPDGRY+AYQ+ GV
Sbjct: 121 HAWPVIFFVFLLALAALSFSPENDRPVTVITKLRLHPTGATRVQLPDGRYIAYQELGVSA 180
Query: 175 GRARFSLVAPHSFLSSRLAGIPGVKASLLEDYGVRLVTYDLPGFGESDPHPSRNFNSSAM 234
RAR+SLV PHSFLSSRLAGIPGVK SLL +YGVRLV+YDLPGFGESDPH RN +SSA
Sbjct: 181 ERARYSLVMPHSFLSSRLAGIPGVKKSLLVEYGVRLVSYDLPGFGESDPHRGRNLSSSAS 240
Query: 235 DMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIPERIAGAAMLAPMVSPYESHMTKDEM 294
DM++L A+ + +KFW+L +S+G IH WA +KY PE+IAGAAM+AP+++PYE M K+E+
Sbjct: 241 DMINLAAAIGIDEKFWLLGYSTGSIHTWAGMKYFPEKIAGAAMVAPVINPYEPSMVKEEV 300
Query: 295 KRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSFLPEKHERIDKQFSLSLGKKDEILVDEP 354
+TWE+WL +RK+MY LA RFP LL FFYR+SFL +++D+ +LSLG+KD++L+ +P
Sbjct: 301 VKTWEQWLTKRKFMYFLARRFPILLPFFYRRSFLSGNLDQLDQWMALSLGEKDKLLIKDP 360
Query: 355 AFEEYWQRDLEESVRQGNLKPFIEEALLQVSRWDFNIEELHVHKKCQTGGLLLWLKSMYG 414
F+E +QR++EESVRQG KPF+EEA+LQVS W F + E KKC T G+L WL SMY
Sbjct: 361 TFQEVYQRNVEESVRQGITKPFVEEAVLQVSNWGFTLSEFRTQKKCATNGVLSWLMSMYS 420
Query: 415 QAECELAGYLGRIHIWQGLDDRMVPPSMTEYIERVLPEAVIHKLPNEGHFSYFFFCDECH 474
+AECEL G+ IHIWQG++DR+ PPSM++YI R++PEA +HK+ NEGHFS+F+FCDECH
Sbjct: 421 EAECELIGFRKPIHIWQGMEDRVAPPSMSDYISRMIPEATVHKIRNEGHFSFFYFCDECH 480
Query: 475 KQIFSTLFGTPQGPFERQEET 495
+QIF LFG P+G ER +ET
Sbjct: 481 RQIFYALFGEPKGQLERVKET 501
>At5g22460 putative protein
Length = 340
Score = 151 bits (381), Expect = 9e-37
Identities = 91/314 (28%), Positives = 159/314 (49%), Gaps = 28/314 (8%)
Query: 151 PPIAS-RILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPGVKASLLEDYGVR 209
PP+ S RI L DGRY+AY++ GV A + ++ H F SS+ P + ++E+ G+
Sbjct: 35 PPVTSPRIKLSDGRYLAYRESGVDRDNANYKIIVVHGFNSSKDTEFP-IPKDVIEELGIY 93
Query: 210 LVTYDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIP 269
V YD G+GESDPHPSR S A D+ L D + + KF+VL S G ++ LKYIP
Sbjct: 94 FVFYDRAGYGESDPHPSRTVKSEAYDIQELADKLKIGPKFYVLGISLGAYSVYSCLKYIP 153
Query: 270 ERIAGAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSFLP 329
R+AGA ++ P V+ + + + ++++ + E + ++ + +A+ P LL ++ + P
Sbjct: 154 HRLAGAVLMVPFVNYWWTKVPQEKLSKALELMPKKDQWTFKVAHYVPWLLYWWLTQKLFP 213
Query: 330 EKHERIDKQFSLSLGKKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRWDF 389
S KD +++ + E + LE+ +QG+ + + + + W+F
Sbjct: 214 SSSMVTGNNALCS--DKDLVVIKKKM--ENPRPGLEKVRQQGDHECLHRDMIAGFATWEF 269
Query: 390 NIEELHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIERV 449
+ E L++ + + E G +H+WQG++DR++P + YI
Sbjct: 270 DPTE---------------LENPFAEGE-------GSVHVWQGMEDRIIPYEINRYISEK 307
Query: 450 LPEAVIHKLPNEGH 463
LP H++ GH
Sbjct: 308 LPWIKYHEVLGYGH 321
>At3g54240 putative protein (fragment)
Length = 342
Score = 136 bits (343), Expect = 2e-32
Identities = 95/339 (28%), Positives = 156/339 (45%), Gaps = 32/339 (9%)
Query: 151 PPI-ASRILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPG--VKASLLEDYG 207
PPI A RI L DGRY+AY++ GV A F +V H+F + R + V+ LE G
Sbjct: 31 PPITAPRIRLSDGRYLAYEEHGVSRQNATFKIVFIHAFSTFRRDAVIANRVRPGFLEKNG 90
Query: 208 VRLVTYDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKY 267
+ +V+YD PG+GESDPH SRN + A D+ L D + + KF+V+ +S G W LKY
Sbjct: 91 IYVVSYDRPGYGESDPHSSRNEKTLAHDVEQLADQLQLGSKFYVVGYSMGGQAVWGVLKY 150
Query: 268 IPERIAGAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSF 327
IP R+AGA +L P+ + + W K ++ + + P LL ++ +
Sbjct: 151 IPHRLAGATLLCPVTNSWWPSFPDSLTWELWNKQSKSERFSMLITHHTPWLLYWWNNQKL 210
Query: 328 LPEKHERIDKQFSLSLGKKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRW 387
+ + +D L+ + A ++ ++ +QG + + ++ +W
Sbjct: 211 F--STTAVMQSSPNMFSPQDLALLPKLAARVSYK---NQTTQQGTHESLDRDLIVGFGKW 265
Query: 388 DFNIEELHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIE 447
F+ + +++ + + E G +H+WQG DDR+VP + I
Sbjct: 266 SFD---------------PMKIENPFPKGE-------GSVHMWQGDDDRLVPIQLQRIIA 303
Query: 448 RVLPEAVIHKLPNEGHFSYFFFCDECHKQIFSTLFGTPQ 486
+ L H++P GH F D + I L PQ
Sbjct: 304 QKLTWIKYHEIPGAGHI--FPMADGMAETILKELLPIPQ 340
>At1g74280 unknown protein
Length = 372
Score = 136 bits (342), Expect = 3e-32
Identities = 94/326 (28%), Positives = 157/326 (47%), Gaps = 41/326 (12%)
Query: 151 PPI-ASRILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPGVKAS--LLEDYG 207
PPI A RI L DGRY+AY++ G+P +A +V H R + S L+E+ G
Sbjct: 55 PPITAPRIKLQDGRYLAYKEHGLPREKANRKIVFIHGSDCCRHDAVFATLLSPDLVEELG 114
Query: 208 VRLVTYDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKY 267
V +V++D PG+ ESDPHPSR S D+ L D +++ KF+VL +S G AW LKY
Sbjct: 115 VYMVSFDRPGYCESDPHPSRTPRSLVSDIEELADQLSLGSKFYVLGYSMGGQAAWGCLKY 174
Query: 268 IPERIAGAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSF 327
IP R+AG ++AP+V+ Y ++ + + R + +A+ P L+ ++ + +
Sbjct: 175 IPHRLAGVTLVAPVVNYYWKNLPLNVSTEGFNFQQKRDQLAVRVAHYTPWLIYWWNTQKW 234
Query: 328 LPEKHERIDKQFSLSLGKKDEILVD------EPAFEEYWQRDLEESVRQGNLKPFIEEAL 381
P ++ SL L + D+ ++ +P + E Q+ + ES+ + + +
Sbjct: 235 FPGS-SIANRDHSL-LAQPDKDIISKLGSSRKPHWAEVRQQGIHESINR--------DMI 284
Query: 382 LQVSRWDFNIEELHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPS 441
+ W+F +L E G +H+WQG +D +VP
Sbjct: 285 VGFGNWEFGPLDL----------------------ENPFLNKEGSVHLWQGDEDMLVPAK 322
Query: 442 MTEYIERVLPEAVIHKLPNEGHFSYF 467
+ Y+ LP H++P GHF ++
Sbjct: 323 LQRYLAHQLPWVHYHEVPRSGHFFHY 348
>At1g74290 unknown protein
Length = 371
Score = 131 bits (330), Expect = 8e-31
Identities = 99/363 (27%), Positives = 169/363 (46%), Gaps = 45/363 (12%)
Query: 119 SIVFTVFLLAFAAISVDSNHQTSTKAAMVRM-----HPPI-ASRILLPDGRYMAYQDQGV 172
S +F ++ I V +Q+ K ++ PPI A RI L DGRY+AY++ G+
Sbjct: 18 SFLFPSVVIVIVGIIVALTYQSKLKPPQPKLCGSSSGPPITAPRIKLQDGRYLAYKEHGL 77
Query: 173 PPGRARFSLVAPHSFLSSRLAGIPGVKAS--LLEDYGVRLVTYDLPGFGESDPHPSRNFN 230
P +A +V H R + S L+E+ GV +V++D PG+ ESDPHPSR
Sbjct: 78 PREKANRKIVFIHGSDCCRHDAVFATLLSPDLVEELGVYMVSFDRPGYCESDPHPSRTPR 137
Query: 231 SSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWA--SLKYIPERIAGAAMLAPMVSPYESH 288
S D+ L D +++ KF+V+ S G AW +LKYIP R+AG ++AP+V+ Y +
Sbjct: 138 SLVSDIEELDDQLSLGSKFYVIGKSMGGQAAWGCLNLKYIPHRLAGVTLVAPVVNYYWRN 197
Query: 289 MTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSFLPEKHERIDKQFSLSLGKKDE 348
+ + + R ++ +A+ P L+ ++ + + P ++ LS +D
Sbjct: 198 LPLNVSTEGFNFQQKRDQWAVRVAHYAPWLIYWWNTQKWFPGS-SIANRDSLLSQSDRDI 256
Query: 349 I----LVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRWDFNIEELHVHKKCQTGG 404
I +P + E Q+ + ES+ + + ++ W+F+ +L
Sbjct: 257 ISKRGYTRKPHWAEVRQQGIHESINR--------DMIVGFGNWEFDPLDL---------- 298
Query: 405 LLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIERVLPEAVIHKLPNEGHF 464
+ G +H+WQG +D +VP + Y+ LP H++P GHF
Sbjct: 299 ------------DNPFLNNEGFVHLWQGDEDMLVPVKLQRYLAHQLPWVHYHEVPRSGHF 346
Query: 465 SYF 467
+F
Sbjct: 347 FHF 349
>At1g74300 unknown protein
Length = 346
Score = 130 bits (327), Expect = 2e-30
Identities = 93/326 (28%), Positives = 154/326 (46%), Gaps = 47/326 (14%)
Query: 151 PPI-ASRILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPGVKAS--LLEDYG 207
PPI A RI L DGR++AY++ G+P +A+ +V H S R + S L+++ G
Sbjct: 38 PPITAPRIKLRDGRHLAYKEYGLPREKAKHKIVFIHGSDSCRHDAVFATLLSPDLVQERG 97
Query: 208 VRLVTYDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKY 267
V +V++D PG+GESDP P R S A+D+ L D +++ KF+V+ S G AW LKY
Sbjct: 98 VYMVSFDKPGYGESDPDPIRTPKSLALDIEELADQLSLGSKFYVIGKSMGGQAAWGCLKY 157
Query: 268 IPERIAGAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSF 327
P R+AG ++AP+V+ Y ++ + + R ++ +A+ P L+ ++ +++
Sbjct: 158 TPHRLAGVTLVAPVVNYYWRNLPLNISTEGFNLQQKRDQWAVRVAHYAPWLIYWWNTQNW 217
Query: 328 LPEKH---------ERIDKQFSLSLGKKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIE 378
P + DK L LG +P E Q+ + ES+ +
Sbjct: 218 FPGSSVVNRDGGVLSQPDKDIILKLGSS-----RKPHLAEVRQQGIHESINR-------- 264
Query: 379 EALLQVSRWDFNIEELHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMV 438
+ ++ W+F+ EL E G +H+WQG +D +V
Sbjct: 265 DMIVGFGNWEFDPLEL----------------------ENPFLNREGSVHLWQGDEDMLV 302
Query: 439 PPSMTEYIERVLPEAVIHKLPNEGHF 464
P ++ YI LP H++ GHF
Sbjct: 303 PVTLQRYIADKLPWLHYHEVAGGGHF 328
>At2g36290 unknown protein
Length = 364
Score = 126 bits (316), Expect = 3e-29
Identities = 88/314 (28%), Positives = 148/314 (47%), Gaps = 28/314 (8%)
Query: 156 RILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRL--AGIPGVKASLLEDYGVRLVTY 213
RI L DGR +AY++ GVP A ++ H S R A + + E GV +V++
Sbjct: 57 RIKLRDGRQLAYKEHGVPRDEATHKIIVVHGSDSCRHDNAFAALLSPDIKEGLGVYMVSF 116
Query: 214 DLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIPERIA 273
D PG+ ESDP P+R S A+D+ L D +++ KF+V+ +S G WA LKYIP R+A
Sbjct: 117 DRPGYAESDPDPNRTPKSLALDIEELADQLSLGSKFYVIGYSMGGQATWACLKYIPHRLA 176
Query: 274 GAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSFLPEKHE 333
G ++AP+V+ + + + + + ++ +A+ P L ++ +S+ P
Sbjct: 177 GVTLVAPVVNYWWKNFPSEISTEAFNQQGRNDQWAVRVAHYAPWLTHWWNSQSWFPGSSV 236
Query: 334 RIDKQFSLSLGKKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRWDFNIEE 393
LS K EI+ A + + + QG + + ++ W+F+ E
Sbjct: 237 VARNLGMLSKADK-EIMFKLGAARSQHEAQIRQ---QGTHETLHRDMIVGFGTWEFDPME 292
Query: 394 LHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIERVLPEA 453
L++++ E G +H+WQG DD +VP ++ YI + LP
Sbjct: 293 ---------------LENLFPNNE-------GSVHLWQGDDDVLVPVTLQRYIAKKLPWI 330
Query: 454 VIHKLPNEGHFSYF 467
H++P GH F
Sbjct: 331 HYHEIPGAGHLFPF 344
>At3g03240 hypothetical protein
Length = 326
Score = 121 bits (303), Expect = 1e-27
Identities = 82/312 (26%), Positives = 150/312 (47%), Gaps = 38/312 (12%)
Query: 153 IASRILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPGVKASLLEDYGVRLVT 212
I+ RI L DGRY+AY++ G P +A+ ++ H F SS+L +++++ + +
Sbjct: 34 ISPRIKLNDGRYLAYKELGFPKDKAKNKIIILHGFGSSKL--------EMIDEFEIYFLL 85
Query: 213 YDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIPERI 272
+D G+GESDPHPSR + D+ L D + + KF VL S G + LKYIP R+
Sbjct: 86 FDRAGYGESDPHPSRTLKTDTYDIEELADKLQIGPKFHVLGMSLGAYPVYGCLKYIPHRL 145
Query: 273 AGAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFY-RKSFLPEK 331
+GA ++ P+++ + S + + ++K + ++ +A+ FP LL ++ +K F P
Sbjct: 146 SGATLVVPILNFWWSCLPLNLSISAFKKLPIQNQWTLGVAHYFPWLLYWWMTQKWFSPFS 205
Query: 332 HERIDKQFSLSLGKKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRWDFNI 391
+ ++ ++D L D+ Y + E ++RQG + + W+F+
Sbjct: 206 QNPRE-----TMTERDIELADKHTKHAYIK---ESALRQGEYVSMQRDIIAGYENWEFDP 257
Query: 392 EELHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIERVLP 451
E L + + G +HIW L+D+ + + Y+ LP
Sbjct: 258 TE---------------LSNPFSDDN------KGSVHIWCALEDKQISHEVLLYLCDKLP 296
Query: 452 EAVIHKLPNEGH 463
+H++P+ GH
Sbjct: 297 WIKLHEVPDAGH 308
>At3g03230 hypothetical protein
Length = 333
Score = 115 bits (288), Expect = 6e-26
Identities = 75/312 (24%), Positives = 152/312 (48%), Gaps = 30/312 (9%)
Query: 153 IASRILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPGVKASLLEDYGVRLVT 212
I+ RI L DGR++AY++ G P +A+ ++ H +S+ + + +++++ + +
Sbjct: 34 ISPRIKLNDGRHLAYKELGFPKDKAKNKIIIVHGNGNSKDVDLY-ITQEMIDEFKIYFLF 92
Query: 213 YDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIPERI 272
+D G+GESDP+P+R + D+ L D + V KF V+ S G + LKYIP R+
Sbjct: 93 FDRAGYGESDPNPTRTLKTDTYDIEELADKLQVGPKFHVIGMSLGAYPVYGCLKYIPNRL 152
Query: 273 AGAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSFLPEKH 332
+GA+++ P+V+ + S + ++ + +K + +A+ P LL ++ + + P
Sbjct: 153 SGASLVVPLVNFWWSRVPQNLLNAAMKKLPIGFQLTLRVAHYSPWLLYWWMTQKWFPNSR 212
Query: 333 ERIDKQFSLSLGKKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRWDFNIE 392
D ++ ++D L ++ Y + E ++RQG ++ + W+F+
Sbjct: 213 NPKD-----TMTERDLELAEKHTKHSYIK---ESALRQGGYVTTQQDIIAGYGNWEFDPT 264
Query: 393 ELHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIERVLPE 452
E LK+ + + G +H+W L+D+ + + YI LP
Sbjct: 265 E---------------LKNPFSDSN------KGSVHMWCALEDKQISRDVLLYICDKLPW 303
Query: 453 AVIHKLPNEGHF 464
+H++P+ GH+
Sbjct: 304 IKLHEVPDGGHY 315
>At3g48410 unknown protein
Length = 385
Score = 115 bits (287), Expect = 8e-26
Identities = 81/322 (25%), Positives = 150/322 (46%), Gaps = 34/322 (10%)
Query: 151 PPIAS-RILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLS----SRLAGIPGVKASLLED 205
P + S RI L DGR++AY + G+P A+F ++ H F S S A + +L+E+
Sbjct: 66 PTVTSPRIKLRDGRHLAYTEFGIPRDEAKFKIINIHGFDSCMRDSHFANF--LSPALVEE 123
Query: 206 YGVRLVTYDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASL 265
+ +V++D PG+GESDP+ + + S A+D+ L D + + +F++ +S G WA L
Sbjct: 124 LRIYIVSFDRPGYGESDPNLNGSPRSIALDIEELADGLGLGPQFYLFGYSMGGEITWACL 183
Query: 266 KYIPERIAGAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRK 325
YIP R+AGAA++AP ++ + ++ D + + P ++ +A+ P L ++ +
Sbjct: 184 NYIPHRLAGAALVAPAINYWWRNLPGDLTREAFSLMHPADQWSLRVAHYAPWLTYWWNTQ 243
Query: 326 SFLPEKHERIDKQFSLSLGKKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVS 385
+ P + S ++D ++ + F + + + QG + + S
Sbjct: 244 KWFPISNVIAGNPIIFS--RQDMEILSKLGFVNPNRAYIRQ---QGEYVSLHRDLNVAFS 298
Query: 386 RWDFNIEELHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEY 445
W+F+ +L + G +H+W G +D+ VP + Y
Sbjct: 299 SWEFDPLDL----------------------QDPFPNNNGSVHVWNGDEDKFVPVKLQRY 336
Query: 446 IERVLPEAVIHKLPNEGHFSYF 467
+ LP H++ GHF F
Sbjct: 337 VASKLPWIRYHEISGSGHFVPF 358
>At1g08310 unknown protein
Length = 315
Score = 111 bits (277), Expect = 1e-24
Identities = 80/323 (24%), Positives = 144/323 (43%), Gaps = 33/323 (10%)
Query: 151 PPIASRILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPGVKASLLEDYGVRL 210
P ++R+ L DGR++AY+++GVP +A++ ++ H F SS+ K L+E+ V L
Sbjct: 5 PASSNRVKLRDGRFLAYKERGVPKEKAKYKIILVHGFGSSKDMNFSASK-ELIEELEVYL 63
Query: 211 VTYDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKYIPE 270
+ YD G+G SD + R+ S D+ L D + + KF+++ S G W L++IP
Sbjct: 64 LFYDRSGYGASDSNTKRSLESEVEDIAELADQLELGPKFYLIGISMGSYPTWGCLRHIPH 123
Query: 271 RIAGAAMLAPMVSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSFLPE 330
R++G A +AP+V+ + K +K+ + + K+ ++ P LL ++ +
Sbjct: 124 RLSGVAFVAPVVNYRWPSLPKKLIKKDYRTGI--IKWGLRISKYAPGLLHWWIIQKLFAS 181
Query: 331 KHERIDKQFSLSLGKKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRWDFN 390
++ E+L + F + L E L+ ++ ++ +WDF
Sbjct: 182 TSSVLESNPVYFNSHDIEVLKRKTGFPMLTKEKLRERNVFDTLR---DDFMVCFGQWDFE 238
Query: 391 IEELHVHKKCQTGGLLLWLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIERVL 450
+L + K IHIW G +D++VP + I +
Sbjct: 239 PADLSISTK-------------------------SYIHIWHGKEDKVVPFQLQRCILQKQ 273
Query: 451 PEAVIHKLPNEGHF--SYFFFCD 471
P H++P GH Y CD
Sbjct: 274 PLINYHEIPQGGHLIVHYDGICD 296
>At3g44520 putative protein
Length = 122
Score = 97.4 bits (241), Expect = 2e-20
Identities = 48/123 (39%), Positives = 73/123 (59%), Gaps = 1/123 (0%)
Query: 149 MHPPIASRILLPDGRYMAYQDQGVPPGRARFSLVAPHSFLSSRLAGIPGVKASLLEDYGV 208
M + RI L DGRY+AY+++GVP A+F++V H F SS+ V L+ + G+
Sbjct: 1 MEEHLQGRIKLHDGRYLAYKERGVPKDDAKFTIVLVHGFGSSKDMNF-NVSQELVNEIGI 59
Query: 209 RLVTYDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWASLKYI 268
V YD G+GESDP+P R+ S A D+ L D + + +F+++ S G W+ LK+I
Sbjct: 60 YFVLYDRAGYGESDPNPKRSLKSEAYDVQELADGLEIGSRFYLIGISMGSYTVWSCLKHI 119
Query: 269 PER 271
P+R
Sbjct: 120 PQR 122
>At3g51000 epoxide hydrolase-like protein
Length = 323
Score = 40.0 bits (92), Expect = 0.003
Identities = 25/80 (31%), Positives = 37/80 (46%), Gaps = 4/80 (5%)
Query: 203 LEDYGVRLVTYDLPGFGESD---PHPSRNFNSSAMDMLHLVDAVNVTDKFWVLCHSSGCI 259
L +G +V DL G+G+SD H S + D++ L+D T F V H G I
Sbjct: 50 LSSHGYHVVAPDLRGYGDSDSLPSHESYTVSHLVADVIGLLDHYGTTQAF-VAGHDWGAI 108
Query: 260 HAWASLKYIPERIAGAAMLA 279
W + P+R+ G L+
Sbjct: 109 IGWCLCLFRPDRVKGFISLS 128
>At4g15955 unknown protein
Length = 178
Score = 39.7 bits (91), Expect = 0.004
Identities = 25/87 (28%), Positives = 44/87 (49%), Gaps = 4/87 (4%)
Query: 203 LEDYGVRLVTYDLPGFGESDPHPSRNFNSSAM---DMLHLVDAV-NVTDKFWVLCHSSGC 258
L G R + DL G+G++D S + +S D++ L+DAV +K +V+ H G
Sbjct: 57 LSSLGYRTIAPDLRGYGDTDAPESVDAYTSLHVVGDLIGLIDAVVGDREKVFVVGHDWGA 116
Query: 259 IHAWASLKYIPERIAGAAMLAPMVSPY 285
I AW + P+R+ ++ + P+
Sbjct: 117 IIAWHLCLFRPDRVKALVNMSVVFDPW 143
>At4g02340 unknown protein
Length = 324
Score = 38.5 bits (88), Expect = 0.009
Identities = 24/81 (29%), Positives = 42/81 (51%), Gaps = 4/81 (4%)
Query: 207 GVRLVTYDLPGFGESDPHPSRNFNS---SAMDMLHLVDAVNVTDKFWVLCHSSGCIHAWA 263
G R + DL G+G+SD PSR + D++ L+D++ V D+ +++ H G I AW
Sbjct: 51 GYRAIAPDLRGYGDSDAPPSRESYTILHIVGDLVGLLDSLGV-DRVFLVGHDWGAIVAWW 109
Query: 264 SLKYIPERIAGAAMLAPMVSP 284
P+R+ + + +P
Sbjct: 110 LCMIRPDRVNALVNTSVVFNP 130
>At3g44510 putative protein
Length = 198
Score = 38.1 bits (87), Expect = 0.012
Identities = 18/53 (33%), Positives = 28/53 (51%), Gaps = 2/53 (3%)
Query: 427 IHIWQGLDDRMVPPSMTEYIERVLPEAVIHKLPNEGHF--SYFFFCDECHKQI 477
+HIWQG +D+++P + + R LP H++P GH Y CD K +
Sbjct: 127 VHIWQGYEDKVMPFQLQRCLCRKLPWIRYHEVPKGGHLIVHYDGICDAILKSL 179
>At5g41900 unknown protein
Length = 471
Score = 37.0 bits (84), Expect = 0.026
Identities = 68/307 (22%), Positives = 121/307 (39%), Gaps = 63/307 (20%)
Query: 176 RARFSLVAPHSFLSSRLAG----IPGVKASLLEDYGVRLVTYDLPGFGESDPHPSRNFNS 231
+AR ++V H F+SS P S +Y R + DL G+G S P P+ + +
Sbjct: 188 KARDNVVFIHGFVSSSAFWTETLFPNFSDSAKSNY--RFIAVDLLGYGRS-PKPNDSLYT 244
Query: 232 SAMDMLHLVDAVNVTD----KFWVLCHSSGCIHAWASLKYIPERIAGAAMLAPM------ 281
+ L +++ ++ F ++ HS GCI A A P I +LAP
Sbjct: 245 -LREHLEMIEKSVISKFKLKTFHIVAHSLGCILALALAVKHPGAIKSLTLLAPPYYKVPK 303
Query: 282 -VSPYESHMTKDEMKRTWEKWLPRRKYMYSLAYRFPKLLSFFYRKSFLPEKHERIDKQFS 340
V P + M + K W P ++ S +LS++ E + +
Sbjct: 304 GVQPAQYVMREVARKEVW----PPMQFGAS-------VLSWY----------EHLGRTIG 342
Query: 341 LSLGKKDEILVDEPAFEEYWQRDLEESVRQGNLKPFIEEALLQVSRWDFNIEELHVHKKC 400
L L K +++ E+ R L + + L IE L F+ +H
Sbjct: 343 LVLIKNHQLI-------EFVTRLLTLNRMRTYL---IEGFLCHTHNGSFHT----LHNII 388
Query: 401 QTGGLLL--WLKSMYGQAECELAGYLGRIHIWQGLDDRMVPPSMTEYIERVLPEAVIHKL 458
G L +L + +C++A I+ G D ++P + ++ +P A +H +
Sbjct: 389 FGSGAKLDSYLDHVRDHVDCDVA-------IFHGGKDELIPVECSYSVKSKVPRATVHVI 441
Query: 459 PNEGHFS 465
P++ H +
Sbjct: 442 PDKDHIT 448
>At3g05600 putative epoxide hydrolase
Length = 331
Score = 36.2 bits (82), Expect = 0.044
Identities = 29/93 (31%), Positives = 44/93 (47%), Gaps = 7/93 (7%)
Query: 203 LEDYGVRLVTYDLPGFGESD-PHPSRNFN--SSAMDMLHLVDAV-NVTDKFWVLCHSSGC 258
L G R V DL G+G+SD P + + D++ L+D+V +K +++ H G
Sbjct: 49 LSSLGYRAVAPDLRGYGDSDSPESFSEYTCLNVVGDLVALLDSVAGNQEKVFLVGHDWGA 108
Query: 259 IHAWASLKYIPERIAGAAMLAPMVSPYESHMTK 291
I W + PE+I G L+ PY S K
Sbjct: 109 IIGWFLCLFRPEKINGFVCLS---VPYRSRNPK 138
>At5g17780 putative protein
Length = 419
Score = 33.9 bits (76), Expect = 0.22
Identities = 32/105 (30%), Positives = 44/105 (41%), Gaps = 17/105 (16%)
Query: 204 EDYGVRLVTYDLPGFGESDPHPSRNFNSSAMDMLHLVDAVNVT-------DKFWVLCHSS 256
+DY RL+ DL GFGES P P S + VD + + D F V+ HS
Sbjct: 176 DDY--RLLAIDLLGFGES-PKP----RDSLYTLKDHVDTIERSVIKPYQLDSFHVVAHSM 228
Query: 257 GCIHAWASLKYIPERIAGAAMLAPMVSPYE---SHMTKDEMKRTW 298
GC+ A A + ++AP P S + + KR W
Sbjct: 229 GCLIALALAAKHSNIVKSVTLVAPPYFPSSVEGSVLNRIARKRLW 273
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.136 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,000,154
Number of Sequences: 26719
Number of extensions: 525993
Number of successful extensions: 1407
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 1359
Number of HSP's gapped (non-prelim): 44
length of query: 512
length of database: 11,318,596
effective HSP length: 104
effective length of query: 408
effective length of database: 8,539,820
effective search space: 3484246560
effective search space used: 3484246560
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC124963.7


