

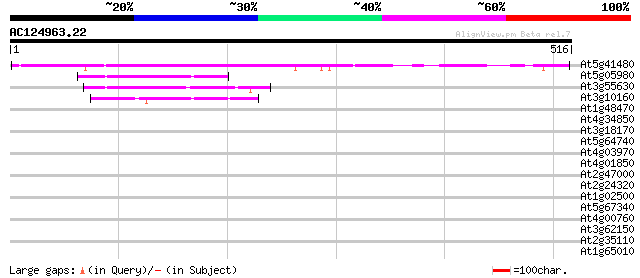
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124963.22 + phase: 0 /pseudo
(516 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g41480 folylpolyglutamate synthase-like protein 348 3e-96
At5g05980 tetrahydrofolylpolyglutamate synthase-like protein 78 1e-14
At3g55630 putative tetrahydrofolylpolyglutamate synthase precursor 75 9e-14
At3g10160 putative folylpolyglutamate synthetase 73 4e-13
At1g48470 glutamine synthetase like protein 30 2.5
At4g34850 chalcone synthase - like protein 30 4.2
At3g18170 hypothetical protein 30 4.2
At5g64740 cellulose synthase 29 5.5
At4g03970 hypothetical protein 29 5.5
At4g01850 S-adenosylmethionine synthase 2 29 7.1
At2g47000 putative ABC transporter 29 7.1
At2g24320 hypothetical protein 29 7.1
At1g02500 s-adenosylmethionine synthetase 29 7.1
At5g67340 unknown proteins 28 9.3
At4g00760 putative protein 28 9.3
At3g62150 P-glycoprotein-like proetin 28 9.3
At2g35110 unknown protein 28 9.3
At1g65010 hypothetical protein 28 9.3
>At5g41480 folylpolyglutamate synthase-like protein
Length = 530
Score = 348 bits (894), Expect = 3e-96
Identities = 235/556 (42%), Positives = 320/556 (57%), Gaps = 101/556 (18%)
Query: 2 RSVRTLCSHREDSEMKDLVDYIDSLKNYEKYGVPTGAGTDSNDGFDLGRMRRLLDRFGNP 61
R++ T+ S ED E++D V +++SLKNYEK GVP GAGTDS+DGFDLGRM+RL+ R NP
Sbjct: 32 RNLSTISS-TEDPELRDFVGFLESLKNYEKSGVPKGAGTDSDDGFDLGRMKRLMLRLRNP 90
Query: 62 HSKFKTE----------------------GYSVGCYSSPHIQTIRERILGRSGDPVSAKL 99
H K+K GYSVGCYSSPHI +I+ERI +G+PVSA
Sbjct: 91 HYKYKVVHVAGTKGKGSTSAFLSNILRAGGYSVGCYSSPHILSIKERI-SCNGEPVSAST 149
Query: 100 LNNLFHRIKQDLDQAIKEENGCISHFEVFTAMAFILFADEKVDIAVIEAGLGGARDATNI 159
LN+LF+ +K L+Q+I+EENG +SHFE+ T +AF LF E VDIAVIEAGLGGARDATN+
Sbjct: 150 LNDLFYSVKPILEQSIQEENGSLSHFEILTGIAFSLFEKENVDIAVIEAGLGGARDATNV 209
Query: 160 ISSSGLAAAVITTVGEEHLAALGGSLESIAMAKAGIIKQGCPLVLGGPFLPHIEYIIREK 219
I SS LAA+VITT+GEEH+AALGGSLESIA AK+GIIK G P+VLGGPFLPHIE I+R K
Sbjct: 210 IESSNLAASVITTIGEEHMAALGGSLESIAEAKSGIIKHGRPVVLGGPFLPHIEGILRSK 269
Query: 220 AVNMDSPVVSASDSGNNLAVKSFSILNG-KPCQICDIEIQTVK---------DLKLAMDP 269
A ++ S V+ AS+ G++ ++K NG CQ CDI IQ K D+ L M
Sbjct: 270 AASVSSSVILASNIGSSSSIKGIINKNGIGLCQSCDIVIQNEKDDQPIVELSDVNLRMLG 329
Query: 270 DPRCACGPVSCMI*SC----KCLVLTN----FKMQQLLLV*HSVFVI*VDGEFLMNL*GV 321
+ + + C C +T+ ++ L+ S F+ + E L+ L G
Sbjct: 330 HHQLQNAVTATCVSLCLRDQGCGRVTDEAIRIGLENTRLLGRSQFLTPKEAETLL-LPGA 388
Query: 322 G*SVHTSLEGVSS*NLKKPRPWDFLEQQYCLI*SVAHTKESAKALMNTIKMASPKARLAF 381
+ + S+ LK+ DF E++ + + MAS K ++F
Sbjct: 389 TVLLDGAHTKESARALKEMIKKDFPEKRLVFV----------------VAMASDKDHVSF 432
Query: 382 VVAMASDKDHAGFAREILSDAYVKTVILTEAAIAGAVTRTAPASLLRDSWIKASEELGTD 441
A+E+LS + VILTEA I G R+ +S+L++SWIKA++ELG
Sbjct: 433 -------------AKELLSGLKPEAVILTEADIGGGKIRSTESSVLKESWIKAADELG-- 477
Query: 442 ICHDGMTEYRELFKEQPVSSESNLTDGKTILATESSLKDCLRMANEIL--NRRRDEKGVI 499
S ++ KT+L + L++A +IL + + G++
Sbjct: 478 ------------------SRSMEASENKTVLGS-------LKLAYKILSDDTTSSDSGMV 512
Query: 500 VITGSLHIVSSVLASL 515
++TGSLHIVSSVLASL
Sbjct: 513 IVTGSLHIVSSVLASL 528
>At5g05980 tetrahydrofolylpolyglutamate synthase-like protein
Length = 571
Score = 78.2 bits (191), Expect = 1e-14
Identities = 48/139 (34%), Positives = 72/139 (51%), Gaps = 3/139 (2%)
Query: 63 SKFKTEGYSVGCYSSPHIQTIRERILGRSGDPVSAKLLNNLFHRIKQDLDQAIKEENGCI 122
S + G+ G ++SPH+ +RER G +S + F L + EE
Sbjct: 132 SIIRNYGFRTGLFTSPHLIDVRERFR-LDGVDISEEKFLGYFWWCYNRLKERTNEEIPMP 190
Query: 123 SHFEVFTAMAFILFADEKVDIAVIEAGLGGARDATNIISSSGLAAAVITTVGEEHLAALG 182
++F +AF +FA E+VD A++E GLGG DATN + + I+++G +H+ LG
Sbjct: 191 TYFRFLALLAFKIFAAEEVDAAILEVGLGGKFDATNAVQKPVVCG--ISSLGYDHMEILG 248
Query: 183 GSLESIAMAKAGIIKQGCP 201
+L IA KAGI K G P
Sbjct: 249 DTLGKIAGEKAGIFKLGVP 267
>At3g55630 putative tetrahydrofolylpolyglutamate synthase precursor
Length = 491
Score = 75.1 bits (183), Expect = 9e-14
Identities = 55/175 (31%), Positives = 85/175 (48%), Gaps = 9/175 (5%)
Query: 69 GYSVGCYSSPHIQTIRERILGRSGDPVSAKLLNNLFHRIKQDLDQAIKEENGCISHFEVF 128
G G ++SPH+ +RER +G +S + N F L + E ++F
Sbjct: 84 GLRTGLFTSPHLIDVRERFR-LNGIEISQEKFVNYFWCCFHKLKEKTSNEVPMPTYFCFL 142
Query: 129 TAMAFILFADEKVDIAVIEAGLGGARDATNIISSSGLAAAVITTVGEEHLAALGGSLESI 188
+AF +F E+VD+ ++E GLGG DATNI I+++G +H+ LG +L I
Sbjct: 143 ALLAFKIFTTEQVDVVILEVGLGGRFDATNIQKP---VVCGISSLGYDHMEILGYTLAEI 199
Query: 189 AMAKAGIIKQGCP-LVLGGPFLPHIEYIIREKA--VNMDSPVVSASDSGNNLAVK 240
A KAGI K G P + P ++ EKA + ++ VV DS L ++
Sbjct: 200 AAEKAGIFKSGVPAFTVAQP--DEAMRVLNEKASKLEVNLQVVEPLDSSQRLGLQ 252
>At3g10160 putative folylpolyglutamate synthetase
Length = 442
Score = 72.8 bits (177), Expect = 4e-13
Identities = 52/157 (33%), Positives = 81/157 (51%), Gaps = 8/157 (5%)
Query: 75 YSSPHIQTIRERILGRSGDPVSAKLLNNLFHRIKQDLDQAIKEENGCISH--FEVFTAMA 132
++SPH+ +RER D K L + K ++A+ +G F+ T +A
Sbjct: 2 FTSPHLIDVRERFRIDGLDISEEKFLQYFWECWKLLKEKAV---DGLTMPPLFQFLTVLA 58
Query: 133 FILFADEKVDIAVIEAGLGGARDATNIISSSGLAAAVITTVGEEHLAALGGSLESIAMAK 192
F +F EKVD+AVIE GLGG D+TN+I + I ++G +H+ LG +L IA K
Sbjct: 59 FKIFVCEKVDVAVIEVGLGGKLDSTNVIQKPVVCG--IASLGMDHMDILGNTLADIAFHK 116
Query: 193 AGIIKQGCPLVLGGPFLPHIEYIIREKAVNMDSPVVS 229
AGI K P P L ++++ A N++ +V+
Sbjct: 117 AGIFKPQIP-AFTVPQLSEAMDVLQKTANNLEVTIVA 152
>At1g48470 glutamine synthetase like protein
Length = 353
Score = 30.4 bits (67), Expect = 2.5
Identities = 38/158 (24%), Positives = 65/158 (41%), Gaps = 28/158 (17%)
Query: 336 NLKKPRPWDFLEQQYCLI*SVAHTKESAKALMNTIKMASPKARLAFVVAMASDKDHAGFA 395
N+K PW +EQ+Y L+ K+ K + P + + A+ +DK F
Sbjct: 118 NVKAEEPWFGIEQEYTLL------KKDVKWPLGWPLGGFPGPQGPYYCAVGADK---AFG 168
Query: 396 REILSDAYVKTVILTEAAIAGAVTRTAPA----SLLRDSWIKASEELG---------TDI 442
R+I+ DA+ K + + +I GA P + I A ++L T+I
Sbjct: 169 RDIV-DAHYKACLYSGLSIGGANGEVMPGQWEFQISPTVGIGAGDQLWVARYILERITEI 227
Query: 443 CHDGMTEYRELFKEQPVSSESNLTDGKTILATESSLKD 480
C ++ F +P+ + N T +T+S KD
Sbjct: 228 CGVIVS-----FDPKPIQGDWNGAAAHTNFSTKSMRKD 260
>At4g34850 chalcone synthase - like protein
Length = 392
Score = 29.6 bits (65), Expect = 4.2
Identities = 23/102 (22%), Positives = 43/102 (41%), Gaps = 19/102 (18%)
Query: 365 ALMNTIKMASPKARLAFVVAMASDKDHAGFAREILSDAYVKTVILTEAAIAGAVTRTAPA 424
A++ + K ++P A ++A+ H +E L D Y KT + + +TR
Sbjct: 7 AVLGSEKKSNPGK--ATILALGKAFPHQLVMQEYLVDGYFKTTKCDDPELKQKLTRLCKT 64
Query: 425 SLLRDSWIKASEEL-----------------GTDICHDGMTE 449
+ ++ ++ SEE+ DIC+D +TE
Sbjct: 65 TTVKTRYVVMSEEILKKYPELAIEGGSTVTQRLDICNDAVTE 106
>At3g18170 hypothetical protein
Length = 519
Score = 29.6 bits (65), Expect = 4.2
Identities = 17/50 (34%), Positives = 28/50 (56%), Gaps = 3/50 (6%)
Query: 446 GMTEYRELFKE---QPVSSESNLTDGKTILATESSLKDCLRMANEILNRR 492
G+T +RE FKE P +SE +++D ++ L SL++ +I RR
Sbjct: 304 GLTRHREYFKELTIDPSNSEYSMSDFRSFLRDTYSLRNDAVATRQIRRRR 353
>At5g64740 cellulose synthase
Length = 1084
Score = 29.3 bits (64), Expect = 5.5
Identities = 15/44 (34%), Positives = 24/44 (54%), Gaps = 6/44 (13%)
Query: 232 DSGNNLAVKSFSILNGKPCQICDIEIQTVKDLKLAMDPDPRCAC 275
++ N ++S L+G+ CQIC EI+ L +D +P AC
Sbjct: 21 NADENARIRSVQELSGQTCQICRDEIE------LTVDGEPFVAC 58
>At4g03970 hypothetical protein
Length = 817
Score = 29.3 bits (64), Expect = 5.5
Identities = 19/67 (28%), Positives = 32/67 (47%), Gaps = 1/67 (1%)
Query: 405 KTVILTEAAIAGAVTRTAPASLLRDSWIKASEELGTDICHDGMTEYRELFKEQPVSSESN 464
K V L AA++ A+++T P D +KA I HD ++ + + P +E
Sbjct: 485 KKVTLESAAVSQALSQTPPEVNFDDDTMKAMVIYSHPI-HDTNSKPNKQLESSPTPTEDQ 543
Query: 465 LTDGKTI 471
+TD + I
Sbjct: 544 ITDSQDI 550
>At4g01850 S-adenosylmethionine synthase 2
Length = 393
Score = 28.9 bits (63), Expect = 7.1
Identities = 14/35 (40%), Positives = 18/35 (51%)
Query: 241 SFSILNGKPCQICDIEIQTVKDLKLAMDPDPRCAC 275
S S+ G P ++CD V D L DPD + AC
Sbjct: 8 SESVNEGHPDKLCDQISDAVLDACLEQDPDSKVAC 42
>At2g47000 putative ABC transporter
Length = 1286
Score = 28.9 bits (63), Expect = 7.1
Identities = 24/71 (33%), Positives = 33/71 (45%), Gaps = 11/71 (15%)
Query: 144 AVIEAGLGGARDATNIISS----------SGLAAAVITTVGEEHLAALGGSLESIAMAKA 193
A I G GG + I+SS SGL T VGE + GG + +A+A+A
Sbjct: 1136 ANIAYGKGGDASESEIVSSAELSNAHGFISGLQQGYDTMVGERGIQLSGGQKQRVAIARA 1195
Query: 194 GIIKQGCPLVL 204
I+K L+L
Sbjct: 1196 -IVKDPKVLLL 1205
>At2g24320 hypothetical protein
Length = 316
Score = 28.9 bits (63), Expect = 7.1
Identities = 23/94 (24%), Positives = 40/94 (42%), Gaps = 11/94 (11%)
Query: 7 LCSHREDSEMKDLVDYIDSLK----------NYEKYGVPTGAGTDSNDGFDLGRMRRLL- 55
L SH +++ +VD L+ +Y YG TG T+ N +D+ + L
Sbjct: 71 LYSHGNAADLGQMVDLFIELRAHLRVNIMSYDYSGYGASTGKPTELNTYYDIEAVYNCLR 130
Query: 56 DRFGNPHSKFKTEGYSVGCYSSPHIQTIRERILG 89
+G + G SVG + H+ + +R+ G
Sbjct: 131 TEYGIMQEEMILYGQSVGSGPTLHLASRVKRLRG 164
>At1g02500 s-adenosylmethionine synthetase
Length = 393
Score = 28.9 bits (63), Expect = 7.1
Identities = 14/35 (40%), Positives = 18/35 (51%)
Query: 241 SFSILNGKPCQICDIEIQTVKDLKLAMDPDPRCAC 275
S S+ G P ++CD V D L DPD + AC
Sbjct: 8 SESVNEGHPDKLCDQISDAVLDACLEQDPDSKVAC 42
>At5g67340 unknown proteins
Length = 707
Score = 28.5 bits (62), Expect = 9.3
Identities = 16/58 (27%), Positives = 27/58 (45%), Gaps = 6/58 (10%)
Query: 56 DRFGNPHSKFKTEGYSVGCYSSPHIQTIRERILGRSGDPVSAKLLNNLFHRIKQDLDQ 113
D NP K+ T G +G P ++ + + D ++LLNN F + Q +D+
Sbjct: 28 DLSSNPAHKYYTRGEDIGKLIKPVLENLID------SDAAPSELLNNGFEELAQYVDE 79
>At4g00760 putative protein
Length = 448
Score = 28.5 bits (62), Expect = 9.3
Identities = 16/49 (32%), Positives = 25/49 (50%), Gaps = 4/49 (8%)
Query: 9 SHREDSEMKDLVDYIDSL----KNYEKYGVPTGAGTDSNDGFDLGRMRR 53
++ + E +DL Y D L K E+ TG T+ N+G DLG ++
Sbjct: 245 NNNSEVETEDLDKYKDELGQGNKRKERADTDTGEHTEKNNGSDLGDQKK 293
>At3g62150 P-glycoprotein-like proetin
Length = 1292
Score = 28.5 bits (62), Expect = 9.3
Identities = 24/71 (33%), Positives = 33/71 (45%), Gaps = 11/71 (15%)
Query: 144 AVIEAGLGGARDATNIISS----------SGLAAAVITTVGEEHLAALGGSLESIAMAKA 193
A I G GG T I+S+ SGL T VGE + GG + +A+A+A
Sbjct: 1142 ANIAYGKGGDATETEIVSAAELSNAHGFISGLQQGYDTMVGERGVQLSGGQKQRVAIARA 1201
Query: 194 GIIKQGCPLVL 204
I+K L+L
Sbjct: 1202 -IVKDPKVLLL 1211
>At2g35110 unknown protein
Length = 1339
Score = 28.5 bits (62), Expect = 9.3
Identities = 25/81 (30%), Positives = 39/81 (47%), Gaps = 6/81 (7%)
Query: 144 AVIEAGLGGARDATNIISSSGLAAAVITTVGEEHLAALGGSLESIAMAKAGIIKQGCPLV 203
++IE+ +GG NI+ S G A+ + + E AA L + + A +K P V
Sbjct: 652 SLIESIMGGLEGLINILDSEGGFGALESQLLPEQAAAY---LNNASRISAPSVKS--PRV 706
Query: 204 LGGPFLP-HIEYIIREKAVNM 223
+GG LP H Y K++ M
Sbjct: 707 VGGFTLPGHESYPENNKSIKM 727
>At1g65010 hypothetical protein
Length = 1318
Score = 28.5 bits (62), Expect = 9.3
Identities = 23/97 (23%), Positives = 45/97 (45%), Gaps = 11/97 (11%)
Query: 106 RIKQDLDQAIKEENGCISHFEVFTAMAFILFADEKVDIAVIEAG----LGGARDATNIIS 161
R+K +L +N +SH E T +A I EK +I E G L G+++ I
Sbjct: 168 RVKHELSMTADAKNKALSHAEEATKIAEI--HAEKAEILASELGRLKALLGSKEEKEAIE 225
Query: 162 SSGLAAAVITTVGEEHLAALGGSLESIAMAKAGIIKQ 198
+ + + + + + L G LE +++ ++ + +Q
Sbjct: 226 GNEIVSKL-----KSEIELLRGELEKVSILESSLKEQ 257
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.327 0.140 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,551,389
Number of Sequences: 26719
Number of extensions: 434190
Number of successful extensions: 1394
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 1373
Number of HSP's gapped (non-prelim): 24
length of query: 516
length of database: 11,318,596
effective HSP length: 104
effective length of query: 412
effective length of database: 8,539,820
effective search space: 3518405840
effective search space used: 3518405840
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC124963.22


