

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124963.21 + phase: 0 /pseudo
(739 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
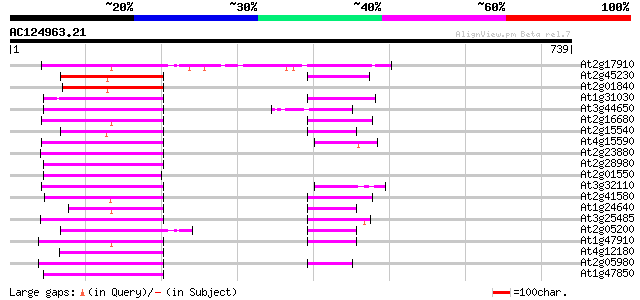
At2g17910 putative non-LTR retroelement reverse transcriptase 124 2e-28
At2g45230 putative non-LTR retroelement reverse transcriptase 102 6e-22
At2g01840 putative non-LTR retroelement reverse transcriptase 97 3e-20
At1g31030 F17F8.5 94 2e-19
At3g44650 putative protein 94 3e-19
At2g16680 putative non-LTR retroelement reverse transcriptase 92 8e-19
At2g15540 putative non-LTR retroelement reverse transcriptase 92 1e-18
At4g15590 reverse transcriptase like protein 91 2e-18
At2g23880 putative non-LTR retroelement reverse transcriptase 91 3e-18
At2g28980 putative non-LTR retroelement reverse transcriptase 90 5e-18
At2g01550 putative non-LTR retroelement reverse transcriptase 90 5e-18
At3g32110 non-LTR reverse transcriptase, putative 89 7e-18
At2g41580 putative non-LTR retroelement reverse transcriptase 89 1e-17
At1g24640 hypothetical protein 88 2e-17
At3g25485 unknown protein 87 3e-17
At2g05200 putative non-LTR retroelement reverse transcriptase 87 3e-17
At1g47910 reverse transcriptase, putative 87 3e-17
At4g12180 putative reverse transcriptase 87 3e-17
At2g05980 putative non-LTR retroelement reverse transcriptase 87 3e-17
At1g47850 hypothetical protein 87 3e-17
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 124 bits (311), Expect = 2e-28
Identities = 130/489 (26%), Positives = 221/489 (44%), Gaps = 54/489 (11%)
Query: 43 LVNFIGMGGLPKESIALSLLLFLRLRA-RRLNDFCLISLVESMYKILAKVLANRLRYVIG 101
+ F G LP++ + L ++ + +R++D ISL +YKI++K+L RL+ +
Sbjct: 449 IFGFFETGVLPQDWNHTHICLIPKITSPQRMSDLRPISLCSVLYKIISKILTQRLKKHLP 508
Query: 102 MVISDSQSTFVKGRQILDGILVANEVVDDAH---RCKKELLLFKVDFEEAYDIVDWSYLD 158
++S +QS FV R I D ILVA+E++ R KE + FK D +AYD V+W +L+
Sbjct: 509 AIVSTTQSAFVPQRLISDNILVAHEMIHSLRTNDRISKEHMAFKTDMSKAYDRVEWPFLE 568
Query: 159 EVMYKMGFPVL*QK*IKECIGTATTSMLVNGSPTDEFPLKRGCKAIRIPLSFFSWLQRPS 218
+M +GF I C+ + + S+L+NG P RG + PLS P+
Sbjct: 569 TMMTALGFNNKWISWIMNCVTSVSYSVLINGQPYGHIIPTRGIRQ-GDPLS-------PA 620
Query: 219 IF*RGLCPKIASFMVIR---WARLTQQSFLTYNLRMIH*L----*LRNCGPTFERCELLS 271
+F LC + ++ + ++T F + + H L L C T + CE L
Sbjct: 621 LF--VLCTEALIHILNKAEQAGKITGIQFQDKKVSVNHLLFADDTLLMCKATKQECEELM 678
Query: 272 VCLKLYPG*K*ISQRVS*LVLMFASLG*SR*HWF*NVRLEIFLLCIWGCLLVGMRVVCLF 331
CL Y +S ++ L + G + +++++ ++ G L G L
Sbjct: 679 QCLSQYGQ---LSGQMINLNKSAITFGKNV-----DIQIKDWIKSRSGISLEGGTGKYLG 730
Query: 332 GNH*LGGSKLDCQGG-KLNIYLLVVIWFC*NLS------------CHLSLFMLF---LSS 375
L GSK D G K + + W+ LS L ++++ L
Sbjct: 731 LPECLSGSKRDLFGFIKEKLQSRLTGWYAKTLSQGGKEVLLKSIALALPVYVMSCFKLPK 790
Query: 376 RLLQVLSPLLNLF*NVFFFCEGSDNHKKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLG 435
L Q L+ ++ + F+ +K+ W+ W + L K+ GG+G + ++ FN ALL
Sbjct: 791 NLCQKLTTVM-----MDFWWNSMQQKRKIHWLSWQRLTLPKDQGGFGFKDLQCFNQALLA 845
Query: 436 KWCWQLLIEKESLWYRVLSARY--NEEGGRLLDGGRSTSAWWRVIATLQREEWFNSCREI 493
K W++L EK SL+ RV +RY N + G R + AW ++ RE R +
Sbjct: 846 KQAWRVLQEKGSLFSRVFQSRYFSNSDFLSATRGSRPSYAWRSIL--FGRELLMQGLRTV 903
Query: 494 GGEWERNIL 502
G ++ +
Sbjct: 904 IGNGQKTFV 912
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 102 bits (255), Expect = 6e-22
Identities = 51/139 (36%), Positives = 87/139 (61%), Gaps = 3/139 (2%)
Query: 67 LRARRLNDFCLISLVESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVANE 126
L+A ++ DF ISL +YK++ K++ANRL+ ++ +IS++Q+ FVKGR I D IL+A+E
Sbjct: 496 LKAEKMTDFRPISLCNVIYKVIGKLMANRLKKILPSLISETQAAFVKGRLISDNILIAHE 555
Query: 127 V---VDDAHRCKKELLLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATT 183
+ + ++C +E + K D +AYD V+W +L++ M +GF + I EC+ +
Sbjct: 556 LLHALSSNNKCSEEFIAIKTDISKAYDRVEWPFLEKAMRGLGFADHWIRLIMECVKSVRY 615
Query: 184 SMLVNGSPTDEFPLKRGCK 202
+L+NG+P E RG +
Sbjct: 616 QVLINGTPHGEIIPSRGLR 634
Score = 53.9 bits (128), Expect = 3e-07
Identities = 29/84 (34%), Positives = 43/84 (50%), Gaps = 2/84 (2%)
Query: 393 FFCEGSDNHKKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYRV 452
F+ + + + W W + K VGG G + I FN ALLGK W+++ EK+SL +V
Sbjct: 825 FWWKNKKEGRGLHWKAWCHLSRPKAVGGLGFKEIEAFNIALLGKQLWRMITEKDSLMAKV 884
Query: 453 LSARYNEEGGRLLD--GGRSTSAW 474
+RY + L G R + AW
Sbjct: 885 FKSRYFSKSDPLNAPLGSRPSFAW 908
>At2g01840 putative non-LTR retroelement reverse transcriptase
Length = 1715
Score = 97.1 bits (240), Expect = 3e-20
Identities = 50/136 (36%), Positives = 83/136 (60%), Gaps = 3/136 (2%)
Query: 70 RRLNDFCLISLVESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVANEV-- 127
+ ++D+ ISL + YKI++K+L RL+ +G VISDSQ+ FV G+ I D +LVA+E+
Sbjct: 861 KHMSDYRPISLCTASYKIISKILIKRLKQCLGDVISDSQAAFVPGQNISDNVLVAHELLH 920
Query: 128 -VDDAHRCKKELLLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATTSML 186
+ C+ + K D +AYD V+W++L++VM ++GF K I C+ + + +L
Sbjct: 921 SLKSRRECQSGYVAVKTDISKAYDRVEWNFLEKVMIQLGFAPRWVKWIMTCVTSVSYEVL 980
Query: 187 VNGSPTDEFPLKRGCK 202
+NGSP + RG +
Sbjct: 981 INGSPYGKIFPSRGIR 996
Score = 34.7 bits (78), Expect = 0.20
Identities = 19/58 (32%), Positives = 28/58 (47%), Gaps = 2/58 (3%)
Query: 419 GGYGVRRIREFNSALLGKWCWQLLIEKESLWYRVLSARY--NEEGGRLLDGGRSTSAW 474
G G + + +FN ALL K W++L +SL R+ Y N R GG ++ W
Sbjct: 1195 GDLGFKDLHQFNRALLAKQAWRILTNPQSLLARLYKGLYYPNTTYLRANKGGHASYGW 1252
>At1g31030 F17F8.5
Length = 872
Score = 94.4 bits (233), Expect = 2e-19
Identities = 57/161 (35%), Positives = 91/161 (56%), Gaps = 4/161 (2%)
Query: 45 NFIGMGGLPK--ESIALSLLLFLRLRARRLNDFCLISLVESMYKILAKVLANRLRYVIGM 102
+F G LPK SI L+L+ +L A+ + D+ IS +YK+++K++ANRL+ ++
Sbjct: 145 SFFQKGFLPKGINSIILALIP-KKLAAKEMRDYRPISCCNVLYKVISKIIANRLKLLLPR 203
Query: 103 VISDSQSTFVKGRQILDGILVANEVVDDAHRCK-KELLLFKVDFEEAYDIVDWSYLDEVM 161
I+++QS FVK R +++ +L+A E+V D H+ K+D +A+D V WS+L +
Sbjct: 204 FIAENQSAFVKDRLLIENLLLATELVKDYHKDSISARCAIKIDISKAFDSVQWSFLTNTL 263
Query: 162 YKMGFPVL*QK*IKECIGTATTSMLVNGSPTDEFPLKRGCK 202
M F I CI TA+ S+ VNG F KRG +
Sbjct: 264 VAMNFSPTFIHWINLCITTASFSVQVNGDLVGYFQSKRGLR 304
Score = 42.7 bits (99), Expect = 7e-04
Identities = 25/92 (27%), Positives = 40/92 (43%), Gaps = 2/92 (2%)
Query: 393 FFCEGSDNHKKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYR- 451
F GS+ + W+ VC K GG G+R ++E N K W+++ SLW +
Sbjct: 493 FLWSGSEMSSHKAKISWDIVCKPKAEGGLGLRNLKEANDVSCLKLVWRIISNSNSLWTKW 552
Query: 452 VLSARYNEEGGRLLDGGRSTSAW-WRVIATLQ 482
V ++ L S +W WR I ++
Sbjct: 553 VAEYLIRKKSIWSLKQSTSMGSWIWRKILKIR 584
>At3g44650 putative protein
Length = 762
Score = 94.0 bits (232), Expect = 3e-19
Identities = 55/160 (34%), Positives = 90/160 (55%), Gaps = 2/160 (1%)
Query: 45 NFIGMGGLPKESIALSLLLF-LRLRARRLNDFCLISLVESMYKILAKVLANRLRYVIGMV 103
+F +G LPK + L L +L ++ + D+ IS MYK+++K+LANRL+ ++
Sbjct: 51 SFFALGFLPKGVNSTILALIPKKLESKEMKDYRPISCCNVMYKVISKILANRLKLLLPQF 110
Query: 104 ISDSQSTFVKGRQILDGILVANEVVDDAHRCK-KELLLFKVDFEEAYDIVDWSYLDEVMY 162
I+ +QS+FVK R +++ +L+A ++V D H+ E K+D +A D V WS+L +
Sbjct: 111 IAGNQSSFVKDRLLIENVLLATDLVKDYHKDSISERCAIKIDISKASDSVQWSFLINTLT 170
Query: 163 KMGFPVL*QK*IKECIGTATTSMLVNGSPTDEFPLKRGCK 202
M FP + I+ CI T + S+ VNG F RG +
Sbjct: 171 AMHFPEMFIHWIRLCITTPSFSVQVNGELAGFFQSSRGLR 210
Score = 46.6 bits (109), Expect = 5e-05
Identities = 29/106 (27%), Positives = 49/106 (45%), Gaps = 10/106 (9%)
Query: 346 GKLNIYLLVVIWFC*NLSCHLSLFMLFLSSRLLQVLSPLLNLF*NVFFFCEGSDNHKKVT 405
G+ N+ + +IW SC+ L L +Q + L + F G++ + K
Sbjct: 362 GRFNL-ISSIIWS----SCNFWLSAFQLPRACIQEIEKLCSSF-----LWSGTNLNSKKA 411
Query: 406 WVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYR 451
+ WN VC K GG G+R ++E N K W+++ +SLW +
Sbjct: 412 KISWNQVCKPKSEGGLGLRSLKEANDVCCLKLVWRIISHGDSLWVK 457
>At2g16680 putative non-LTR retroelement reverse transcriptase
Length = 1319
Score = 92.4 bits (228), Expect = 8e-19
Identities = 55/164 (33%), Positives = 90/164 (54%), Gaps = 4/164 (2%)
Query: 43 LVNFIGMGGLPKESIALSLLLFLRL-RARRLNDFCLISLVESMYKILAKVLANRLRYVIG 101
++ F G LP+E L L + +R++D ISL +YKI++K+L+ +L+ +
Sbjct: 423 ILGFFETGVLPQEWNHTHLYLIPKFTNPQRMSDIRPISLCSVLYKIISKILSFKLKKHLP 482
Query: 102 MVISDSQSTFVKGRQILDGILVANEVVDDAH---RCKKELLLFKVDFEEAYDIVDWSYLD 158
++S SQS F R I D IL+A+E+V + KE ++FK D +AYD V+WS+L
Sbjct: 483 SIVSPSQSAFFAERLISDNILIAHEIVHSLRTNDKISKEFMVFKTDMSKAYDRVEWSFLQ 542
Query: 159 EVMYKMGFPVL*QK*IKECIGTATTSMLVNGSPTDEFPLKRGCK 202
E++ +GF I C+ + T S+L+NG +RG +
Sbjct: 543 EILVALGFNDKWISWIMGCVTSVTYSVLINGQHFGHITPERGIR 586
Score = 46.6 bits (109), Expect = 5e-05
Identities = 23/88 (26%), Positives = 44/88 (49%), Gaps = 2/88 (2%)
Query: 393 FFCEGSDNHKKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYRV 452
F+ + K+ W+ + L K +GG+G + ++ FN ALL K W+L + +S+ ++
Sbjct: 777 FWWNSMEFSNKIHWIGGKKLTLPKSLGGFGFKDLQCFNQALLAKQAWRLFSDSKSIVSQI 836
Query: 453 LSARY--NEEGGRLLDGGRSTSAWWRVI 478
+RY N + G R + W ++
Sbjct: 837 FKSRYFMNTDFLNARQGTRPSYTWRSIL 864
>At2g15540 putative non-LTR retroelement reverse transcriptase
Length = 1225
Score = 91.7 bits (226), Expect = 1e-18
Identities = 53/138 (38%), Positives = 80/138 (57%), Gaps = 3/138 (2%)
Query: 68 RARRLNDFCLISLVESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVANE- 126
R R++ +F ISL YK+++KVL++RL+ ++ +IS++QS FV R I D IL+A E
Sbjct: 444 RPRKMAEFRPISLCNVSYKVISKVLSSRLKRLLPELISETQSAFVAERLITDNILIAQEN 503
Query: 127 --VVDDAHRCKKELLLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATTS 184
+ CKK+ + K D +AYD V+WS+L +M KMGF I CI + +
Sbjct: 504 FHALRTNPACKKKYMAIKTDMSKAYDRVEWSFLRALMLKMGFAQKWVDWIIFCISSVSYK 563
Query: 185 MLVNGSPTDEFPLKRGCK 202
+L+NGSP RG +
Sbjct: 564 ILLNGSPKGFIKPSRGIR 581
Score = 47.8 bits (112), Expect = 2e-05
Identities = 23/65 (35%), Positives = 33/65 (50%)
Query: 393 FFCEGSDNHKKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYRV 452
F+ + + + W+ W+ +C GG G R + EFN LL K W+L+ SL RV
Sbjct: 705 FWWKTREESNGIHWIAWDKLCTPFSDGGLGFRTLEEFNLVLLAKQLWRLIRFPNSLLSRV 764
Query: 453 LSARY 457
L RY
Sbjct: 765 LRGRY 769
>At4g15590 reverse transcriptase like protein
Length = 929
Score = 91.3 bits (225), Expect = 2e-18
Identities = 55/163 (33%), Positives = 87/163 (52%), Gaps = 3/163 (1%)
Query: 43 LVNFIGMGGLPKESIALSLLLFLRL-RARRLNDFCLISLVESMYKILAKVLANRLRYVIG 101
++ F G LPK + + L+L ++ + R+ F +SL ++KI+ K++ RL+ VI
Sbjct: 287 VMEFFESGVLPKSTNDVLLVLLAKVAKPERITQFRPVSLCNVLFKIITKMMVIRLKNVIS 346
Query: 102 MVISDSQSTFVKGRQILDGILVANEVVDDAHRCK--KELLLFKVDFEEAYDIVDWSYLDE 159
+I +Q++F+ GR D I+V E V R K K +L K+D E+AYD + W +L E
Sbjct: 347 KLIGPAQASFIPGRLSFDNIVVVQEAVHSMRRKKGRKGWMLLKLDLEKAYDRIRWDFLAE 406
Query: 160 VMYKMGFPVL*QK*IKECIGTATTSMLVNGSPTDEFPLKRGCK 202
+ G K I EC+ S+L NG TD F +RG +
Sbjct: 407 TLEAAGLSEGWIKRIMECVAGPEMSLLWNGEKTDSFTPERGLR 449
Score = 52.4 bits (124), Expect = 9e-07
Identities = 32/86 (37%), Positives = 43/86 (49%), Gaps = 3/86 (3%)
Query: 402 KKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYRVLSARY---N 458
+K + W VC K GG G+R ++ N ALL K W+LL +K SLW RVL +Y +
Sbjct: 632 RKQHLLSWKKVCRPKAAGGLGLRASKDMNRALLAKVGWRLLNDKVSLWARVLRRKYKVTD 691
Query: 459 EEGGRLLDGGRSTSAWWRVIATLQRE 484
L + S+ WR I RE
Sbjct: 692 VHDSSWLVPKATWSSTWRSIGVGLRE 717
>At2g23880 putative non-LTR retroelement reverse transcriptase
Length = 1216
Score = 90.5 bits (223), Expect = 3e-18
Identities = 55/164 (33%), Positives = 88/164 (53%), Gaps = 2/164 (1%)
Query: 41 VLLVNFIGMGGLPKESIALSLLLFLRLR-ARRLNDFCLISLVESMYKILAKVLANRLRYV 99
+ + +F G LPK + L L + + AR + D+ IS +YK ++K+LANRL+ +
Sbjct: 215 IAIQSFFAKGFLPKGVNSTILALIPKKKEAREIKDYRPISCCNVLYKAISKILANRLKRI 274
Query: 100 IGMVISDSQSTFVKGRQILDGILVANEVVDDAHRCK-KELLLFKVDFEEAYDIVDWSYLD 158
+ I +QS FVK R +++ +L+A E+V D H+ K+D +A+D + WS+L
Sbjct: 275 LPKFIVGNQSAFVKDRLLIENVLLATELVKDYHKDSISTRCAMKIDISKAFDSLQWSFLT 334
Query: 159 EVMYKMGFPVL*QK*IKECIGTATTSMLVNGSPTDEFPLKRGCK 202
V+ M FP I C+ TA+ S+ VNG F RG +
Sbjct: 335 HVLAAMNFPGEFIHWISLCMSTASFSIQVNGELAGYFRSARGLR 378
Score = 41.2 bits (95), Expect = 0.002
Identities = 19/55 (34%), Positives = 31/55 (55%)
Query: 397 GSDNHKKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYR 451
G + + K V W+ +C K+ GG G++ +RE N K W+LL ++SLW +
Sbjct: 571 GPELNPKKAKVSWDEICKPKKEGGLGLQSLREANKVSSLKLIWRLLSCQDSLWVK 625
>At2g28980 putative non-LTR retroelement reverse transcriptase
Length = 1529
Score = 89.7 bits (221), Expect = 5e-18
Identities = 56/160 (35%), Positives = 85/160 (53%), Gaps = 2/160 (1%)
Query: 45 NFIGMGGLPKESIALSLLLFLRL-RARRLNDFCLISLVESMYKILAKVLANRLRYVIGMV 103
+F G LPK A L L + A + D+ IS +YK+++K+LANRL+ ++
Sbjct: 795 SFFVKGFLPKGLNATILALIPKKDEAIEMKDYRPISCCNVLYKVISKILANRLKLLLPSF 854
Query: 104 ISDSQSTFVKGRQILDGILVANEVVDDAHR-CKKELLLFKVDFEEAYDIVDWSYLDEVMY 162
I +QS FVK R +++ +L+A E+V D H+ K+D +A+D V W +L +
Sbjct: 855 ILQNQSAFVKERLLMENVLLATELVKDYHKESVTPRCAMKIDISKAFDSVQWQFLLNTLE 914
Query: 163 KMGFPVL*QK*IKECIGTATTSMLVNGSPTDEFPLKRGCK 202
+ FP + IK CI TAT S+ VNG F RG +
Sbjct: 915 ALNFPETFRHWIKLCISTATFSVQVNGELAGFFGSSRGLR 954
Score = 38.9 bits (89), Expect = 0.010
Identities = 18/57 (31%), Positives = 28/57 (48%)
Query: 393 FFCEGSDNHKKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLW 449
F G + K + W+ +C K+ GG G++ + E N K W+LL + SLW
Sbjct: 1143 FLWSGPVLNPKKAKIAWSSICQPKKEGGLGIKSLAEANKVSCLKLIWRLLSTQPSLW 1199
>At2g01550 putative non-LTR retroelement reverse transcriptase
Length = 1449
Score = 89.7 bits (221), Expect = 5e-18
Identities = 52/157 (33%), Positives = 88/157 (55%), Gaps = 2/157 (1%)
Query: 45 NFIGMGGLPKE-SIALSLLLFLRLRARRLNDFCLISLVESMYKILAKVLANRLRYVIGMV 103
+F G LPK + + L+ +L A+ + D+ IS +YK+++K++ANRL++V+
Sbjct: 913 SFFEKGFLPKGVNTTILALIPKKLEAKEMKDYRPISCCNVIYKVISKIIANRLKHVLPNF 972
Query: 104 ISDSQSTFVKGRQILDGILVANEVVDDAHR-CKKELLLFKVDFEEAYDIVDWSYLDEVMY 162
I+ +QS FVK R +++ +L+A E+V D H+ K+D +A+D V WS+L V+
Sbjct: 973 IAGNQSAFVKDRLLIENLLLATELVKDYHKDTISGRCAIKIDISKAFDSVQWSFLKNVLS 1032
Query: 163 KMGFPVL*QK*IKECIGTATTSMLVNGSPTDEFPLKR 199
+ FP + C+ TA+ S+ VNG F R
Sbjct: 1033 ALDFPPEFVHWVMLCVTTASFSVQVNGELAGYFQSSR 1069
Score = 35.0 bits (79), Expect = 0.15
Identities = 25/93 (26%), Positives = 43/93 (45%), Gaps = 10/93 (10%)
Query: 346 GKLNIYLLVVIWFC*NLSCHLSLFMLFLSSRLLQVLSPLLNLF*NVFFFCEGSDNHKKVT 405
G+LN+ + V+W C+ L L + ++ + L + F G D +
Sbjct: 1088 GRLNL-ISSVLWSI----CNFWLAAFRLPRKCIREIDKLCSTF-----LWSGPDLNSHKA 1137
Query: 406 WVDWNFVCLSKEVGGYGVRRIREFNSALLGKWC 438
V W+ VC K+ GG G+R I+E N + ++C
Sbjct: 1138 KVAWDDVCKPKKEGGLGLRSIKEANDDMAKQFC 1170
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 89.4 bits (220), Expect = 7e-18
Identities = 52/163 (31%), Positives = 88/163 (53%), Gaps = 3/163 (1%)
Query: 43 LVNFIGMGGLPKESIALSLLLFLR-LRARRLNDFCLISLVESMYKILAKVLANRLRYVIG 101
+++F G P+E+ + ++L + L+ ++ F ISL ++K + KV+ RL+ VI
Sbjct: 1011 VMDFFSSGSFPQETNDVLVVLIAKVLKPEKITQFRPISLCNVLFKTITKVMVGRLKGVIN 1070
Query: 102 MVISDSQSTFVKGRQILDGILVANEVVDDAHRCK--KELLLFKVDFEEAYDIVDWSYLDE 159
+I +Q++F+ GR D I+V EVV R K K +L K+D E+AYD + W L++
Sbjct: 1071 KLIGPAQTSFIPGRLSTDNIVVVQEVVHSMRRKKGVKGWMLLKLDLEKAYDRIRWDLLED 1130
Query: 160 VMYKMGFPVL*QK*IKECIGTATTSMLVNGSPTDEFPLKRGCK 202
+ G P + I +C+ + +L NG TD F RG +
Sbjct: 1131 TLKAAGLPGTWVQWIMKCVEGPSMRLLWNGEKTDAFKPLRGLR 1173
Score = 48.5 bits (114), Expect = 1e-05
Identities = 28/93 (30%), Positives = 43/93 (46%), Gaps = 12/93 (12%)
Query: 402 KKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYRVLSARYNEEG 461
KK V W VCL + GG G+R N AL+ K W++L + SLW +V+ ++Y
Sbjct: 1373 KKQHLVAWTRVCLPRREGGLGIRSATAMNKALIAKVGWRVLNDGSSLWAQVVRSKYKV-- 1430
Query: 462 GRLLDGGRSTSAWWRVIATLQREEWFNSCREIG 494
G W T+ + W ++ R +G
Sbjct: 1431 -----GDVHDRNW-----TVAKSNWSSTWRSVG 1453
>At2g41580 putative non-LTR retroelement reverse transcriptase
Length = 1094
Score = 88.6 bits (218), Expect = 1e-17
Identities = 50/161 (31%), Positives = 89/161 (55%), Gaps = 4/161 (2%)
Query: 46 FIGMGGLPKESIALSLLLFLRL-RARRLNDFCLISLVESMYKILAKVLANRLRYVIGMVI 104
F G LP++ L L ++ + R+ D ISL MYKI++K+L+ RL+ + +++
Sbjct: 203 FFQTGILPEDWNHTHLCLIPKITKPARMADIRPISLCSVMYKIISKILSARLKKYLPVIV 262
Query: 105 SDSQSTFVKGRQILDGILVANEVVDDA---HRCKKELLLFKVDFEEAYDIVDWSYLDEVM 161
S +QS FV R + D I++A+E+V + + K+ ++FK D +AYD V+W +L ++
Sbjct: 263 SPTQSAFVAERLVSDNIILAHEIVHNLRTNEKISKDFMVFKTDMSKAYDRVEWPFLKGIL 322
Query: 162 YKMGFPVL*QK*IKECIGTATTSMLVNGSPTDEFPLKRGCK 202
+GF + C+ + + S+L+NG P RG +
Sbjct: 323 LALGFNSTWINWMMACVSSVSYSVLINGQPFGHITPHRGLR 363
Score = 46.2 bits (108), Expect = 7e-05
Identities = 26/88 (29%), Positives = 46/88 (51%), Gaps = 2/88 (2%)
Query: 393 FFCEGSDNHKKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYRV 452
F+ + KK+ W+ + L K +GG+G + ++ FN ALL K +L + +SL ++
Sbjct: 554 FWWNSVQDKKKIHWIGAQKLMLPKFLGGFGFKDLQCFNQALLAKQASRLHTDSDSLLSQI 613
Query: 453 LSARY--NEEGGRLLDGGRSTSAWWRVI 478
L +RY N + G R + AW ++
Sbjct: 614 LKSRYYMNSDFLSATKGTRPSYAWQSIL 641
>At1g24640 hypothetical protein
Length = 1270
Score = 87.8 bits (216), Expect = 2e-17
Identities = 49/128 (38%), Positives = 74/128 (57%), Gaps = 3/128 (2%)
Query: 78 ISLVESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVANEVVDDAH---RC 134
ISL +YKI++K++A RL+ + ++SD+QS FV R I D ILVA+E+V R
Sbjct: 470 ISLCSVLYKIISKIMAKRLQPWLPEIVSDTQSAFVSERLITDNILVAHELVHSLKVHPRI 529
Query: 135 KKELLLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATTSMLVNGSPTDE 194
E + K D +AYD V+WSYL ++ +GF + I C+ + T S+L+N P
Sbjct: 530 SSEFMAVKSDMSKAYDRVEWSYLRSLLLSLGFHLKWVNWIMVCVSSVTYSVLINDCPFGL 589
Query: 195 FPLKRGCK 202
L+RG +
Sbjct: 590 IILQRGLR 597
Score = 59.7 bits (143), Expect = 6e-09
Identities = 27/65 (41%), Positives = 39/65 (59%)
Query: 393 FFCEGSDNHKKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYRV 452
F+ + D+ +K+ W W +CL K+ GG G R I+ FN ALL K W+LL + L R+
Sbjct: 788 FWWDSCDHSRKIHWQSWERLCLPKDSGGLGFRDIQSFNQALLAKQAWRLLHFPDCLLSRL 847
Query: 453 LSARY 457
L +RY
Sbjct: 848 LKSRY 852
>At3g25485 unknown protein
Length = 979
Score = 87.4 bits (215), Expect = 3e-17
Identities = 52/164 (31%), Positives = 88/164 (52%), Gaps = 2/164 (1%)
Query: 41 VLLVNFIGMGGLPKESIALSLLLF-LRLRARRLNDFCLISLVESMYKILAKVLANRLRYV 99
V + +F G LPK + L L + + + D+ IS +YK+++K++A RL+ +
Sbjct: 410 VAVKSFFEKGFLPKGVNSTILALIPKKFETKEMKDYRPISCCNVIYKVISKLIAKRLKEI 469
Query: 100 IGMVISDSQSTFVKGRQILDGILVANEVVDDAHR-CKKELLLFKVDFEEAYDIVDWSYLD 158
+ I+ +QS FVK R ++ +L+A E+V D H+ + K+D +A+D V WS+L+
Sbjct: 470 LPQFIAGNQSAFVKDRLLIQNLLLATEIVKDYHKESVSDRCAIKIDISKAFDSVQWSFLE 529
Query: 159 EVMYKMGFPVL*QK*IKECIGTATTSMLVNGSPTDEFPLKRGCK 202
V++ + F I CI TA+ S+ VNG F RG +
Sbjct: 530 NVLHSLNFSQEFIHWIMLCITTASFSVQVNGELVGFFNSSRGLR 573
Score = 44.7 bits (104), Expect = 2e-04
Identities = 27/86 (31%), Positives = 42/86 (48%), Gaps = 4/86 (4%)
Query: 393 FFCEGSDNHKKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYRV 452
F G + + K V+W +C K GG G+R ++E N K W+ + +SLW ++
Sbjct: 762 FLWSGPELNSKRAKVNWEAICKPKREGGLGLRSLKEANDVSCLKLIWRWVSRGDSLWRKL 821
Query: 453 LSARYNEEGGRLLD---GGRSTSAWW 475
L R + +D GGR TS W+
Sbjct: 822 LKYRDTAKQFCKVDIRNGGR-TSFWF 846
>At2g05200 putative non-LTR retroelement reverse transcriptase
Length = 1229
Score = 87.4 bits (215), Expect = 3e-17
Identities = 58/178 (32%), Positives = 96/178 (53%), Gaps = 13/178 (7%)
Query: 67 LRARRLNDFCLISLVESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVANE 126
L R++ D+ I+L YKI+AK++ R++ ++ +IS++QS FV GR I D +L+ +E
Sbjct: 391 LGPRKVADYRPIALCNIFYKIVAKIMTKRMQLILPKLISENQSAFVPGRVISDNVLITHE 450
Query: 127 VVD--DAHRCKKEL-LLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATT 183
V+ KK + K D +AYD V+W +L +V+ + GF + + EC+ + +
Sbjct: 451 VLHFLRTSSAKKHCSMAVKTDMSKAYDRVEWDFLKKVLQRFGFHSIWIDWVLECVTSVSY 510
Query: 184 SMLVNGSPTDEFPLKRGCKAIRIPLSFFSWLQRPSIF*RGLCPKIASFMVIRWARLTQ 241
S L+NG+P + RG + PLS P +F LC ++ S + R RL Q
Sbjct: 511 SFLINGTPQGKVVPTRGLRQ-GDPLS-------PCLF--ILCTEVLSGLCTRAQRLRQ 558
Score = 48.1 bits (113), Expect = 2e-05
Identities = 24/65 (36%), Positives = 35/65 (52%)
Query: 393 FFCEGSDNHKKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYRV 452
F+ + + +K +WV W+ + K GG G R I N +LL K W+LL ESL R+
Sbjct: 720 FWWDTKPDVRKTSWVAWSKLTNPKNAGGLGFRDIERCNDSLLAKLGWRLLNSPESLLSRI 779
Query: 453 LSARY 457
L +Y
Sbjct: 780 LLGKY 784
>At1g47910 reverse transcriptase, putative
Length = 1142
Score = 87.4 bits (215), Expect = 3e-17
Identities = 55/168 (32%), Positives = 88/168 (51%), Gaps = 4/168 (2%)
Query: 39 LCVLLVNFIGMGGLPKESIALSLLLFLRL-RARRLNDFCLISLVESMYKILAKVLANRLR 97
L L+ +F+ G K ++ L + R R+ + ISL YK+++K+L RL+
Sbjct: 251 LLSLVNSFLQEGVFDKRLNTTNICLIPKTERPTRMTELRPISLCNVGYKVISKILCQRLK 310
Query: 98 YVIGMVISDSQSTFVKGRQILDGILVANEVVDDAH---RCKKELLLFKVDFEEAYDIVDW 154
V+ +IS++QS FV GR I D IL+A E+ CK + + K D +AYD V+W
Sbjct: 311 TVLPNLISETQSAFVDGRLISDNILIAQEMFHGLRTNSSCKDKFMAIKTDMSKAYDQVEW 370
Query: 155 SYLDEVMYKMGFPVL*QK*IKECIGTATTSMLVNGSPTDEFPLKRGCK 202
++++ ++ KMGF I CI T +L+NG P +RG +
Sbjct: 371 NFIEALLRKMGFCEKWISWIMWCITTVQYKVLINGQPKGLIIPERGLR 418
Score = 55.8 bits (133), Expect = 8e-08
Identities = 24/65 (36%), Positives = 39/65 (59%)
Query: 393 FFCEGSDNHKKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYRV 452
F+ + + + + W+ W+ +C SK GG G R + +FNSALL K W+L+ +SL+ +V
Sbjct: 609 FWWSSNGDSRGMHWMAWDKLCSSKSDGGLGFRNVDDFNSALLAKQLWRLITAPDSLFAKV 668
Query: 453 LSARY 457
RY
Sbjct: 669 FKGRY 673
>At4g12180 putative reverse transcriptase
Length = 662
Score = 87.0 bits (214), Expect = 3e-17
Identities = 46/138 (33%), Positives = 79/138 (56%), Gaps = 1/138 (0%)
Query: 66 RLRARRLNDFCLISLVESMYKILAKVLANRLRYVIGMVISDSQSTFVKGRQILDGILVAN 125
++ A+ + D+ IS +YK+++K++ANRL+ V+ I+ +QS F+K R +++ +L+A
Sbjct: 38 KMEAKEIKDYRPISCCNVLYKVISKIIANRLKRVLPQFIAGNQSAFIKDRLLIENLLLAT 97
Query: 126 EVVDDAHR-CKKELLLFKVDFEEAYDIVDWSYLDEVMYKMGFPVL*QK*IKECIGTATTS 184
E+V D H+ E K+D +A+D V WS+L V+ + FP I C+ TA+ S
Sbjct: 98 ELVKDYHKDSVSERCAIKIDISKAFDSVQWSFLRNVLLTLDFPQEFVHWIMLCVTTASFS 157
Query: 185 MLVNGSPTDEFPLKRGCK 202
+ VN F RG +
Sbjct: 158 VQVNRELAGYFNSLRGLR 175
Score = 38.5 bits (88), Expect = 0.014
Identities = 29/104 (27%), Positives = 49/104 (46%), Gaps = 10/104 (9%)
Query: 346 GKLNIYLLVVIWFC*NLSCHLSLFMLFLSSRLLQVLSPLLNLF*NVFFFCEGSDNHKKVT 405
G+LN+ + V+W C+ L L ++ + L + F ++ E S N K+
Sbjct: 327 GRLNL-VSSVLWSI----CNFWLSAFRLPRECVREIDKLCSAF--LWSGPELSTNKAKIA 379
Query: 406 WVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLW 449
W VC K GG G++ I+E N K W+++ + +SLW
Sbjct: 380 W---ETVCRPKREGGLGLQSIKEANDVCCLKLIWRIVSQGDSLW 420
>At2g05980 putative non-LTR retroelement reverse transcriptase
Length = 1352
Score = 87.0 bits (214), Expect = 3e-17
Identities = 52/166 (31%), Positives = 93/166 (55%), Gaps = 2/166 (1%)
Query: 39 LCVLLVNFIGMGGLPKE-SIALSLLLFLRLRARRLNDFCLISLVESMYKILAKVLANRLR 97
L + + +F G LPK + + L+ + + D+ IS +YKI++K++ANRL+
Sbjct: 639 LVIAIQSFFLKGFLPKGINTTILALISKKHEVSGMKDYRPISCCNVLYKIVSKLMANRLK 698
Query: 98 YVIGMVISDSQSTFVKGRQILDGILVANEVVDDAHR-CKKELLLFKVDFEEAYDIVDWSY 156
++ I+ +QS F+K R +++ +L+A+E+V D H+ K+D +A+D V W +
Sbjct: 699 EILPASIAPNQSAFIKDRLMMENLLLASELVKDYHKESISSRSALKIDISKAFDFVQWPF 758
Query: 157 LDEVMYKMGFPVL*QK*IKECIGTATTSMLVNGSPTDEFPLKRGCK 202
L V+ + P + I+ CIGTA+ S+ VNG + F +RG +
Sbjct: 759 LINVLKAIHLPEMFIHWIELCIGTASFSVQVNGELSGFFRSERGLR 804
Score = 46.6 bits (109), Expect = 5e-05
Identities = 21/59 (35%), Positives = 33/59 (55%)
Query: 393 FFCEGSDNHKKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYR 451
F G D + K + W+ VC KE GG G++ ++E N L K W++L ++SLW +
Sbjct: 993 FLWSGPDLNTKKAKIAWSEVCKLKEEGGLGLKPLKEANEVSLLKLIWRILSARDSLWVK 1051
>At1g47850 hypothetical protein
Length = 799
Score = 87.0 bits (214), Expect = 3e-17
Identities = 55/160 (34%), Positives = 85/160 (52%), Gaps = 2/160 (1%)
Query: 45 NFIGMGGLPKESIALSLLLFLRL-RARRLNDFCLISLVESMYKILAKVLANRLRYVIGMV 103
+F G LPK A L L + A + D+ IS +YK+++K++ANRL+ ++
Sbjct: 71 SFFIKGFLPKGLNATILALIPKKDEATLMRDYRPISCCNVIYKVISKIIANRLKVMLPTF 130
Query: 104 ISDSQSTFVKGRQILDGILVANEVVDDAHRCK-KELLLFKVDFEEAYDIVDWSYLDEVMY 162
I +QS FV+ R +++ +L+A E+V D H+ K+D +A+D V W +L +
Sbjct: 131 ILQNQSAFVRERLLIENVLLATELVKDYHKDSISPRCAMKIDISKAFDSVQWQFLLNTLE 190
Query: 163 KMGFPVL*QK*IKECIGTATTSMLVNGSPTDEFPLKRGCK 202
+ FP IK CI TAT S+ VNG F KRG +
Sbjct: 191 ALNFPENFCHWIKLCISTATFSVQVNGELAGFFGSKRGLR 230
Score = 39.7 bits (91), Expect = 0.006
Identities = 21/83 (25%), Positives = 35/83 (41%)
Query: 393 FFCEGSDNHKKVTWVDWNFVCLSKEVGGYGVRRIREFNSALLGKWCWQLLIEKESLWYRV 452
F G + + K + W +C K+ GG G++ + E N K W+L+ + SLW
Sbjct: 419 FLWSGPELNPKKAKITWTSLCKLKQEGGLGIKSLLEANKVSCLKLIWRLVSRQSSLWVNW 478
Query: 453 LSARYNEEGGRLLDGGRSTSAWW 475
+ +G RS+ W
Sbjct: 479 VWTYIIRKGSFWSANDRSSLGSW 501
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.350 0.157 0.564
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,064,427
Number of Sequences: 26719
Number of extensions: 608063
Number of successful extensions: 2400
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 65
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 2175
Number of HSP's gapped (non-prelim): 153
length of query: 739
length of database: 11,318,596
effective HSP length: 107
effective length of query: 632
effective length of database: 8,459,663
effective search space: 5346507016
effective search space used: 5346507016
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC124963.21


