

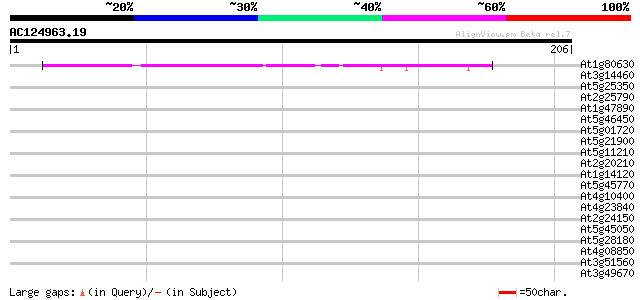
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124963.19 - phase: 0 /pseudo
(206 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g80630 unknown protein 43 1e-04
At3g14460 disease resistance protein, putative 32 0.18
At5g25350 leucine-rich repeats containing protein 32 0.31
At2g25790 putative receptor-like protein kinase 30 0.89
At1g47890 disease resistance protein, putative 29 1.5
At5g46450 disease resistance protein-like 28 2.6
At5g01720 putative F-box protein family, AtFBL3 28 2.6
At5g21900 DNA excision repair protein 28 3.4
At5g11210 putative protein (AtGLR2.5) 28 3.4
At2g20210 hypothetical protein 28 3.4
At1g14120 dioxygenase like protein 28 3.4
At5g45770 predicted GPI-anchored protein 28 4.4
At4g10400 putative protein 28 4.4
At4g23840 leucine rich repeat protein - like 27 5.8
At2g24150 unknown protein 27 5.8
At5g45050 disease resistance protein-like 27 7.5
At5g28180 putative protein 27 9.8
At4g08850 receptor protein kinase like protein 27 9.8
At3g51560 disease resistance-like protein 27 9.8
At3g49670 receptor protein kinase - like protein 27 9.8
>At1g80630 unknown protein
Length = 578
Score = 43.1 bits (100), Expect = 1e-04
Identities = 48/170 (28%), Positives = 78/170 (45%), Gaps = 12/170 (7%)
Query: 13 LVCSNVGSMSRQDISAISRSFQMLEVLDLGHPNLFVDGAHPNTAKALINLSLSTPRMREI 72
L C VG S D+ +I +F +LE LD+ +PN + +I LS + + +I
Sbjct: 112 LKCYGVGGFSDSDLVSIGVNFPLLEKLDISYPN---SSPSRVSDSGVIELSSNLKGLLKI 168
Query: 73 NLSSHSYISDNLFHNLLYNWRHNLEEVFICNCPNITHGIIFNSFSFPDAPPRVKSLVLSD 132
N+S +S+I+D L N L E+ +C I+ I F ++ ++SL ++
Sbjct: 169 NISGNSFITDKSLIALSQNCL-LLREIIFRDCDFISSDCI--KFVLRNS-RNLESLAING 224
Query: 133 TGL--SQPNIPQPY-FLNSLTTIKLFSCNISNQFLSSI--KVLPLHTLLL 177
GL + + + F LT + L +S+ L I LPL LLL
Sbjct: 225 IGLRPRESLLTDAFLFARCLTELDLSDSFLSDDLLCLIASAKLPLKKLLL 274
>At3g14460 disease resistance protein, putative
Length = 1424
Score = 32.3 bits (72), Expect = 0.18
Identities = 32/132 (24%), Positives = 58/132 (43%), Gaps = 23/132 (17%)
Query: 30 SRSFQMLEVLDLGHPNLFVDGAHPNTAKALINLSLST-PRMREINLSSHSYISDNLFHNL 88
+RS+ LE L +G ++ L+N LS P++R +++ H
Sbjct: 1161 TRSYSQLEYLFIG-----------SSCSNLVNFPLSLFPKLRSLSIRDCESFKTFSIHAG 1209
Query: 89 LYNWRHNLEEVFICNCPNITHGIIFNSFSFPDA---PPRVKSLVLSDTGLSQPNIPQPYF 145
L + R LE + I +CPN+ +FP P++ S++LS+ Q + +
Sbjct: 1210 LGDDRIALESLEIRDCPNLE--------TFPQGGLPTPKLSSMLLSNCKKLQALPEKLFG 1261
Query: 146 LNSLTTIKLFSC 157
L SL ++ + C
Sbjct: 1262 LTSLLSLFIIKC 1273
>At5g25350 leucine-rich repeats containing protein
Length = 623
Score = 31.6 bits (70), Expect = 0.31
Identities = 35/136 (25%), Positives = 58/136 (41%), Gaps = 9/136 (6%)
Query: 2 AFGNSIPFLTSLVCSNVGSMSRQDISAISRSFQMLEVLDLGHPNLFVDGAHPNTAKALIN 61
A + P L + N+ ++S +S I+RS M+E LDL D A+ +N
Sbjct: 161 AVAHGCPSLRIVSLWNLPAVSDLGLSEIARSCPMIEKLDLSRCPGITDSGLVAIAENCVN 220
Query: 62 LSLSTPRMREINLSSHSYISDNLFHNLLYNWRHNLEEVFICNCPNI-THGIIFNSFSFPD 120
LS ++ + S S + + + NL + I +CP I G+ F
Sbjct: 221 LS-------DLTIDSCSGVGNEGLRAIARRC-VNLRSISIRSCPRIGDQGVAFLLAQAGS 272
Query: 121 APPRVKSLVLSDTGLS 136
+VK +L+ +GLS
Sbjct: 273 YLTKVKLQMLNVSGLS 288
Score = 29.6 bits (65), Expect = 1.2
Identities = 34/140 (24%), Positives = 60/140 (42%), Gaps = 9/140 (6%)
Query: 71 EINLSSHSYISDNLFHNLLYNWRHNLEEVFICNCPNITH----GIIFNSFSFPDAPPRVK 126
++NLS +SDN + LE + + C NIT+ + N +S D +
Sbjct: 489 KVNLSECINVSDNTVSAISVCHGRTLESLNLDGCKNITNASLVAVAKNCYSVNDLD--IS 546
Query: 127 SLVLSDTGLSQPNIPQPYFLNSLTTIKLFSCNISNQFLSSIKVLPLHTLLLGIHLSWFVR 186
+ ++SD G+ + P LN L + + C+ S S + L LLG+++ R
Sbjct: 547 NTLVSDHGI-KALASSPNHLN-LQVLSIGGCS-SITDKSKACIQKLGRTLLGLNIQRCGR 603
Query: 187 LLISAASNPPFGPFRCSLVY 206
+ S +RC ++Y
Sbjct: 604 ISSSTVDTLLENLWRCDILY 623
>At2g25790 putative receptor-like protein kinase
Length = 960
Score = 30.0 bits (66), Expect = 0.89
Identities = 24/106 (22%), Positives = 45/106 (41%), Gaps = 19/106 (17%)
Query: 5 NSIPFLTSLVCSNVGSMSRQDISAISRSFQMLEVLDLGHPNLFVDGAHPNTAKALIN--- 61
N + + +VC+N+ + D+S + S Q+L P L N I
Sbjct: 58 NDVCLWSGVVCNNISRVVSLDLSGKNMSGQILTAATFRLPFLQTINLSNNNLSGPIPHDI 117
Query: 62 LSLSTPRMREINLSSHSY----------------ISDNLFHNLLYN 91
+ S+P +R +NLS++++ +S+N+F +YN
Sbjct: 118 FTTSSPSLRYLNLSNNNFSGSIPRGFLPNLYTLDLSNNMFTGEIYN 163
>At1g47890 disease resistance protein, putative
Length = 1019
Score = 29.3 bits (64), Expect = 1.5
Identities = 37/145 (25%), Positives = 67/145 (45%), Gaps = 32/145 (22%)
Query: 36 LEVLDLGHPNLFVDGAHPNTAKALINLSLSTPRMREINLSSHSYISDNLFHNLLYNWRHN 95
LE LDL +L + + INL L ++ ++LSS + D FH L +
Sbjct: 169 LERLDLSQSSL--------SGQIPINL-LQLTKLVSLDLSSSDFFGDESFHYL------S 213
Query: 96 LEEVFICNCPNITHGIIFNSFSFPDAPPRVKSLVLSDTGLSQPNIPQPYF-LNSLTTIKL 154
+++ F+ P + + ++ L +S +S IP+ + + SL ++ L
Sbjct: 214 IDKSFL---PLLARNL-----------RNLRELDMSYVKISS-EIPEEFSNIRSLRSLNL 258
Query: 155 FSCNISNQFLSSIKVLP-LHTLLLG 178
CN+ +F SSI ++P L ++ LG
Sbjct: 259 NGCNLFGEFPSSILLIPNLQSIDLG 283
>At5g46450 disease resistance protein-like
Length = 1123
Score = 28.5 bits (62), Expect = 2.6
Identities = 43/187 (22%), Positives = 73/187 (38%), Gaps = 47/187 (25%)
Query: 10 LTSLVCSNVGSMSR----QDISAISRSFQMLEVL------DLGHPNLFVDGAHPNTAKAL 59
L SL C N+ S+ DIS + E +L NL+ G + ++ L
Sbjct: 697 LESLYCLNLNGCSKLRSFPDISTTISELYLSETAIEEFPTELHLENLYYLGLYDMKSEKL 756
Query: 60 IN--------LSLSTPRMREINLSSHSYISD--NLFHNLLYNWRHNLEEVFICNCPNITH 109
+++ +P + ++ LS + + + F NL HNLE + I C N+
Sbjct: 757 WKRVQPLTPLMTMLSPSLTKLFLSDIPSLVELPSSFQNL-----HNLEHLNIARCTNLET 811
Query: 110 ---GIIFNSF------------SFPDAPPRVKSLVLSDTGLSQPNIPQPYFLNS---LTT 151
G+ SFPD + SLVL TG+ + P+++ L+
Sbjct: 812 LPTGVNLELLEQLDFSGCSRLRSFPDISTNIFSLVLDGTGIEE----VPWWIEDFYRLSF 867
Query: 152 IKLFSCN 158
+ + CN
Sbjct: 868 LSMIGCN 874
>At5g01720 putative F-box protein family, AtFBL3
Length = 665
Score = 28.5 bits (62), Expect = 2.6
Identities = 30/121 (24%), Positives = 47/121 (38%), Gaps = 18/121 (14%)
Query: 5 NSIPFLTSLVCSNVGSMSRQDISAISRSFQMLEVLDLGHPNLFVDGAHP----------- 53
NS P L SL + +SR+ I + ++LE LDL + +G
Sbjct: 378 NSCPLLVSLKMESCSLVSREAFWLIGQKCRLLEELDLTDNEIDDEGLKSISSCLSLSSLK 437
Query: 54 ------NTAKALINLSLSTPRMREINLSSHSYISDNLFHNLLYNWRHNLEEVFICNCPNI 107
T K L + + +RE++L I+D + H LE + I C +I
Sbjct: 438 LGICLNITDKGLSYIGMGCSNLRELDLYRSVGITDVGISTIAQGCIH-LETINISYCQDI 496
Query: 108 T 108
T
Sbjct: 497 T 497
>At5g21900 DNA excision repair protein
Length = 544
Score = 28.1 bits (61), Expect = 3.4
Identities = 32/118 (27%), Positives = 52/118 (43%), Gaps = 9/118 (7%)
Query: 46 LFVDGAHPNTAKALINLSLSTPRMREINLSSHSYISDNLFHNLLYNWRHNLEEVFICNCP 105
L + GA T AL+ +S S+P ++ INL+ S ++ L + L + I C
Sbjct: 260 LSLQGAFCLTDNALLLISKSSPLLQYINLTECSLLTYRALRILADKFGSTLRGLSIGGCQ 319
Query: 106 NITHGIIFNSFSFPDAPPRVKSLVLSDTGLSQPN--IPQPYFL---NSLTTIKLFSCN 158
I F+S + K LS GL N + + +F+ + LT + L +CN
Sbjct: 320 GIKKHKGFSSSLYKFE----KLNYLSVAGLVSVNDGVVRSFFMFRSSILTDLSLANCN 373
>At5g11210 putative protein (AtGLR2.5)
Length = 829
Score = 28.1 bits (61), Expect = 3.4
Identities = 30/107 (28%), Positives = 45/107 (42%), Gaps = 10/107 (9%)
Query: 1 MAFGNSIPFLTSLV------CSNVGSMSRQDISAISRSFQMLEVLDLGHPNLFVDGAHPN 54
++F + P L SL ++ S Q ISAI SF+ EV+ + N F +G PN
Sbjct: 55 ISFSATSPLLDSLRSPYFIRATHDDSSQVQAISAIIESFRWREVVPIYVDNEFGEGILPN 114
Query: 55 TAKALINLSLSTPRMREINLSSHSYISDNLFHNLLYNWRHNLEEVFI 101
A +++ I+L + SD+ LY VFI
Sbjct: 115 LVDAFQEINVRIRYRSAISL----HYSDDQIKKELYKLMTMPTRVFI 157
>At2g20210 hypothetical protein
Length = 271
Score = 28.1 bits (61), Expect = 3.4
Identities = 16/60 (26%), Positives = 35/60 (57%), Gaps = 3/60 (5%)
Query: 53 PNTAKALINLSLSTPR--MREINLSSHSYISDNLFHN-LLYNWRHNLEEVFICNCPNITH 109
P A ++ SL T + ++ ++++ +S IS H+ LL+ ++HN E +F+ +++H
Sbjct: 205 PPEALLMLCDSLRTAKGDLKRLDMTGNSCISHEADHSSLLHEFQHNGEPIFVLPSSSVSH 264
>At1g14120 dioxygenase like protein
Length = 312
Score = 28.1 bits (61), Expect = 3.4
Identities = 16/45 (35%), Positives = 23/45 (50%), Gaps = 5/45 (11%)
Query: 40 DLGHPNLFVDGAHPNTAKALINLSLSTPRMREINLSSHSYISDNL 84
DL P+ FVD HP K +S +R+I LS H + ++L
Sbjct: 258 DLEAPDEFVDAEHPRLYK-----PISDGGLRKIRLSKHLHAGESL 297
>At5g45770 predicted GPI-anchored protein
Length = 425
Score = 27.7 bits (60), Expect = 4.4
Identities = 34/128 (26%), Positives = 57/128 (43%), Gaps = 18/128 (14%)
Query: 52 HPNTAKALINLSLSTPRMREINLSSHSYISDNLFHNLLYNWRHNLEEVFICNCPNITHGI 111
+P KAL +L++STP N +++ S ++ + +RH + + NC +
Sbjct: 29 NPKQLKALQSLNISTPTNDPCNNNNNQSSSSSITCDDASPYRH-ITSISFTNCSSTL--- 84
Query: 112 IFNSFSFPDAPPRVKSLV-LSDTGLSQPNIPQPYF----LNSLTTIKLFSCN----ISNQ 162
S P KSL+ LS T P++ PY L+S + + F N +S
Sbjct: 85 ---SLPSKTLKPLSKSLISLSFTNC--PSLSPPYHLPISLHSFSAVSSFLQNNRTKLSGL 139
Query: 163 FLSSIKVL 170
FL+ +K L
Sbjct: 140 FLARLKNL 147
>At4g10400 putative protein
Length = 326
Score = 27.7 bits (60), Expect = 4.4
Identities = 47/186 (25%), Positives = 74/186 (39%), Gaps = 34/186 (18%)
Query: 7 IPFLTSLVCSNVGSMSRQDISAISRSFQMLE--VLDLGHPNLFVDGAHPNTAK------A 58
+P L +L V + + + + +LE V++L H H N K +
Sbjct: 150 LPSLKTLELKGVRYFKQGSLQRLLCNCPVLEDLVVNLSH--------HDNMGKLTVIVPS 201
Query: 59 LINLSLSTPRMREINLSSHSYISDNLFHNLLYNWRHNLEEVFICNCPNITHGIIFNSFSF 118
L LSLSTP RE + + S +S L N I N P + I + F
Sbjct: 202 LQRLSLSTPSSREFVIDTPSLLSFQLVDR-----NDNSHTFLIENMPKLREAYI--NVPF 254
Query: 119 PDAPPRVKSLVLSDTGL------SQPNIPQPYFLNSLTTIKLFSCNISNQFLSSIKVLP- 171
D +KSL+ S T + S+ + + N L + L++ SN + +K P
Sbjct: 255 AD----IKSLIGSITSVKRLAISSEVGYGEGFIFNHLEELTLWNKYSSNLLVWFLKNSPN 310
Query: 172 LHTLLL 177
L L+L
Sbjct: 311 LRELML 316
>At4g23840 leucine rich repeat protein - like
Length = 192
Score = 27.3 bits (59), Expect = 5.8
Identities = 15/48 (31%), Positives = 28/48 (58%)
Query: 125 VKSLVLSDTGLSQPNIPQPYFLNSLTTIKLFSCNISNQFLSSIKVLPL 172
+K L +S TG+++ I L L+ + L +++Q L S++VLP+
Sbjct: 138 LKKLWISQTGVTEVGISLLASLKKLSLLDLGGLPVTDQNLISLQVLPV 185
>At2g24150 unknown protein
Length = 344
Score = 27.3 bits (59), Expect = 5.8
Identities = 20/52 (38%), Positives = 25/52 (47%), Gaps = 11/52 (21%)
Query: 155 FSC--NISNQFLSSIKVLPLHTLLLGIHLSWFVRLLISAASNPPFGPFRCSL 204
FSC N +LSSI +L L + + LL A S P F PFR +L
Sbjct: 203 FSCHPNFRLLYLSSISILGLLAI---------ITLLSPALSTPRFRPFRANL 245
>At5g45050 disease resistance protein-like
Length = 1372
Score = 26.9 bits (58), Expect = 7.5
Identities = 23/65 (35%), Positives = 36/65 (55%), Gaps = 8/65 (12%)
Query: 117 SFPDAPPRVKSLVLSDTGLSQ--PNIPQPYF---LNSLTTIKLFSCNISNQFLSSIKVLP 171
SFP+ PP +++L L TG+ + +I +P + LN L I S +SN S +K P
Sbjct: 653 SFPEIPPNIETLNLQGTGIIELPLSIVKPNYRELLNLLAEIPGLS-GVSNLEQSDLK--P 709
Query: 172 LHTLL 176
L +L+
Sbjct: 710 LTSLM 714
>At5g28180 putative protein
Length = 352
Score = 26.6 bits (57), Expect = 9.8
Identities = 14/35 (40%), Positives = 19/35 (54%), Gaps = 7/35 (20%)
Query: 74 LSSHSYISDNLFHNLLYNWRHNLEEVFICNCPNIT 108
LS H+ S N+F N E++F CN PN+T
Sbjct: 156 LSQHNDPSSNMFVR-------NKEDLFWCNAPNMT 183
>At4g08850 receptor protein kinase like protein
Length = 1045
Score = 26.6 bits (57), Expect = 9.8
Identities = 17/39 (43%), Positives = 23/39 (58%), Gaps = 2/39 (5%)
Query: 25 DISAISRSFQMLEVLDLGHPNLFVDGAHPNTAKALINLS 63
+IS+ RS Q LE LDL H NL G P + K ++ L+
Sbjct: 613 EISSQFRSLQNLERLDLSHNNL--SGQIPPSFKDMLALT 649
>At3g51560 disease resistance-like protein
Length = 1253
Score = 26.6 bits (57), Expect = 9.8
Identities = 9/19 (47%), Positives = 13/19 (68%)
Query: 117 SFPDAPPRVKSLVLSDTGL 135
SFP+ PP ++ L L TG+
Sbjct: 653 SFPEVPPNIEELYLKQTGI 671
>At3g49670 receptor protein kinase - like protein
Length = 1002
Score = 26.6 bits (57), Expect = 9.8
Identities = 17/59 (28%), Positives = 27/59 (44%)
Query: 114 NSFSFPDAPPRVKSLVLSDTGLSQPNIPQPYFLNSLTTIKLFSCNISNQFLSSIKVLPL 172
+SF+ + P + S LS T S + L +T++ L N+S S + LPL
Sbjct: 36 SSFTIDEHSPLLTSWNLSTTFCSWTGVTCDVSLRHVTSLDLSGLNLSGTLSSDVAHLPL 94
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.139 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,776,447
Number of Sequences: 26719
Number of extensions: 192864
Number of successful extensions: 629
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 617
Number of HSP's gapped (non-prelim): 26
length of query: 206
length of database: 11,318,596
effective HSP length: 95
effective length of query: 111
effective length of database: 8,780,291
effective search space: 974612301
effective search space used: 974612301
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC124963.19


