

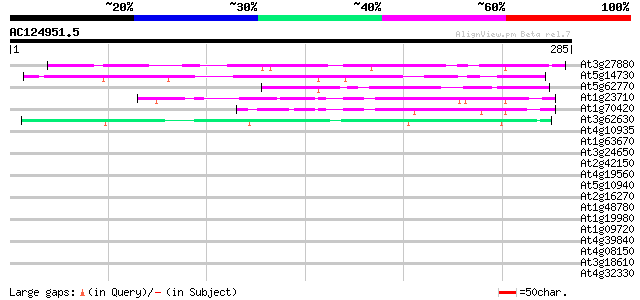
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124951.5 + phase: 0
(285 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g27880 unknown protein 113 1e-25
At5g14730 unknown protein 74 1e-13
At5g62770 putative protein 69 3e-12
At1g23710 unknown protein 67 8e-12
At1g70420 unknown protein 65 5e-11
At3g62630 unknown protein 47 1e-05
At4g10935 unknown protein 40 0.001
At1g63670 unknown protein 40 0.001
At3g24650 abscisic acid-insensitive protein 3 34 0.10
At2g42150 unknown protein 33 0.23
At4g19560 putative protein 32 0.30
At5g10940 unknown protein 32 0.51
At2g16270 unknown protein 32 0.51
At1g48780 hypothetical protein 32 0.51
At1g19980 unknown protein 32 0.51
At1g09720 hypothetical protein 31 0.67
At4g39840 unknown protein 31 0.87
At4g08150 KNAT1 homeobox-like protein 31 0.87
At3g18610 unknown protein 31 0.87
At4g32330 unknown protein 30 1.1
>At3g27880 unknown protein
Length = 242
Score = 113 bits (283), Expect = 1e-25
Identities = 93/280 (33%), Positives = 136/280 (48%), Gaps = 68/280 (24%)
Query: 20 PSFSVYSSNNINDVAQQVTNENDNSHSQNDDFEF-VAFRSHRRNAADVSPIFNRDERRNS 78
PSFS YS + + ++A++V + + + +FEF A S + V P+FN
Sbjct: 12 PSFSTYSGDRLVEIAERVCYKEKS----DAEFEFSTAATSSSGDGGLVFPVFN------- 60
Query: 79 DAMDISISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLFIGNEKEK------WTSS- 131
KNL+ GD P E + LK LF+ ++ ++SS
Sbjct: 61 ---------KNLISGDVSP-------------EKVIPLKDLFLRERNDQQPPQQTYSSSS 98
Query: 132 ---EVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNT--SSSR 186
E +D+ DSIP+E YC WTP S+ SP C+KS STGSSS + S+ R
Sbjct: 99 DEEEEDDEFDSIPSEIYCPWTPARSTADMSPSGG-------CRKSKSTGSSSTSTWSTKR 151
Query: 187 WKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGNVVGKKIPATEKKTPATVSAMEV 246
W+ L+RSKSDGK+SL + P +++ +S KK+ + TVSA E
Sbjct: 152 WRLRDFLKRSKSDGKQSLKFLNPTNRDDDDESSKSK-----KKVSVS-----VTVSAHEK 201
Query: 247 FYGR----KKETRVKSYLPYKKELIGFSVGFNANVGRGFP 282
FY R K+E + KSYLPYK++L+G + G+ FP
Sbjct: 202 FYLRNKAIKEEDKRKSYLPYKQDLVGLFSNIH-RYGKTFP 240
>At5g14730 unknown protein
Length = 246
Score = 73.6 bits (179), Expect = 1e-13
Identities = 78/279 (27%), Positives = 124/279 (43%), Gaps = 57/279 (20%)
Query: 8 EETSSTMDSFLCPSFSVYSSNNINDVAQQVTNENDNSHS-----QND-DFEFVAFRSHRR 61
++ SST+ PSFS Y+S ++ + A +V E+++ +S +ND +FEF
Sbjct: 5 QDLSSTVS--FSPSFSFYASGDLVETAVRVIRESESYYSVKVDGENDKEFEFETLPLGDE 62
Query: 62 NAADVSPIFNRDERRNSD--AMDISISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKL 119
+ S + +R + D D ++ KNL G +S+
Sbjct: 63 SLFHFSTMTTTFKRSSDDDDVADDRVTSKNLFYGG-------------------WSVDPP 103
Query: 120 FIGNEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSL---IASPKTSPMNSTIK--CKKSN 174
+ + E +SS+ +D + P++ YCFW+P S K S IK ++S+
Sbjct: 104 SLTSPSESLSSSDSDDSENISPSKFYCFWSPIRSPARDDSLKSKGSSRRCRIKEFLRRSH 163
Query: 175 STGSSSNTSSS-RWKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGNVVGKKIPAT 233
S G+ S S++ R F LLRRS SDG E +++ GNG++V K
Sbjct: 164 SDGAVSTVSATKRCSFKDLLRRSHSDG----------GGEGSSISTSGNGSLVVK----G 209
Query: 234 EKKTPATVSAMEVFYGRKKETRVKSYLPYKKELIGFSVG 272
KT A Y + R KSYLPY+++LIG G
Sbjct: 210 RNKTAA--------YKPDVDKRRKSYLPYRQDLIGVFAG 240
>At5g62770 putative protein
Length = 268
Score = 68.9 bits (167), Expect = 3e-12
Identities = 51/152 (33%), Positives = 78/152 (50%), Gaps = 27/152 (17%)
Query: 129 TSSEVEDDLDSIPAESYCFWTPKSSSL----IASPKTSPMNSTIKCKKSNSTGSSSNTSS 184
+SSE E+DL +P E+YC W PK S+ + +SP +S I KS+S G S
Sbjct: 129 SSSEAEEDLTGVPPETYCVWKPKQSNSGDDDLQRLSSSPSHSKI---KSHSAG-----FS 180
Query: 185 SRWKFLSLLR-RSKSDGKESLNMVTPAKKENLKLNSGGNGNVVGKKIPATEKKTPATVSA 243
RWK +LL RS S+G + L P KK + ++ E++ P+ V
Sbjct: 181 KRWKLRNLLYVRSSSEGNDKLVFPAPVKKNDETVSD-----------QREEEEPPSKVDG 229
Query: 244 MEVFYGR-KKETRVKSYLPYKKELIGFSVGFN 274
E GR ++ET+ ++Y+PY+K++IG N
Sbjct: 230 EE--EGREREETKRQTYVPYRKDMIGILKNVN 259
>At1g23710 unknown protein
Length = 295
Score = 67.4 bits (163), Expect = 8e-12
Identities = 68/237 (28%), Positives = 102/237 (42%), Gaps = 67/237 (28%)
Query: 66 VSPIFNRD---ERRNSDAMDISISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLFIG 122
V P+FNRD E N D + ++S+ + + +P+L +KLF+
Sbjct: 91 VFPLFNRDLLFEYENEDDKNDNVSVTD----ENRPRL-----------------RKLFVE 129
Query: 123 NEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNT 182
+ E E + P YC WT + + ASP+T C+KSNSTG S
Sbjct: 130 DRNGNGDGEETEGS-EKEPLGPYCSWTGGTVAE-ASPET--------CRKSNSTGFSK-- 177
Query: 183 SSSRWKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGNVVG----KKI-------- 230
W+F L+ RS SDG+++ + + + +S + + KK+
Sbjct: 178 ---LWRFRDLVLRSNSDGRDAFVFLNNSNDKTRTRSSSSSSSTAAEENDKKVITEKKKGK 234
Query: 231 ------PATEKKTPATVSAMEVFYGR----KKETRVKSYLPYKKELIGFSVGFNANV 277
T+KKT T SA E Y R K+E + +SYLPYK+ VGF NV
Sbjct: 235 EKTSTSSETKKKTTTTKSAHEKLYMRNRAMKEEVKHRSYLPYKQ------VGFFTNV 285
>At1g70420 unknown protein
Length = 272
Score = 64.7 bits (156), Expect = 5e-11
Identities = 59/175 (33%), Positives = 84/175 (47%), Gaps = 39/175 (22%)
Query: 116 LKKLFIGNEKEKWTSSEVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNS 175
LKKLF+ E T+ E E++ D++ YC WT ++ ASP+T C+KSNS
Sbjct: 114 LKKLFV----ESTTTEEEEEESDTVGP--YCSWTNRTVEQ-ASPET--------CRKSNS 158
Query: 176 TGSSSNTSSSRWKFLSLLRRSKSDGKESL-------NMVTPAKKENLKLNSGGNGNVVGK 228
TG S W+F L+ RS SDGK++ + + +L+ + GK
Sbjct: 159 TGFSK-----LWRFRDLVLRSNSDGKDAFVFLSNGSSSTSSTSSTAARLSGVVKSSEKGK 213
Query: 229 KIPATEKKTP--ATVSAMEVFYGR----KKETRVKSYLPYKKELIGFSVGFNANV 277
+ T+KK T SA E Y R ++E + +SYLPYK VGF NV
Sbjct: 214 EKTKTDKKKDKMRTKSAHEKLYMRNRAMREEGKRRSYLPYK------HVGFFTNV 262
>At3g62630 unknown protein
Length = 380
Score = 47.0 bits (110), Expect = 1e-05
Identities = 65/317 (20%), Positives = 117/317 (36%), Gaps = 68/317 (21%)
Query: 7 NEETSSTMDSFLCPSFSVYSSNNINDVAQQVTNENDNSHSQ---NDDFEFVAFRSHRRNA 63
+ E +D+ C F S+ ++ + + + S + N + + SH +
Sbjct: 65 SSENPKKLDTSSCGDFEFDFSSRLSSSSGPLGGVSMTSAEELFSNGQIKPMKLSSHLQRP 124
Query: 64 ADVSPIFNRDERRNSDAMDISISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLF--- 120
+SP+ + + D D++ K NG +R D S+ +
Sbjct: 125 QILSPLLDLENEEEDD--------------DDETKPNGEMKRGRDLKLRSRSVHRKARSL 170
Query: 121 --IGNEKEKWTSSEVEDDLDSIPAE-SYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTG 177
+ N +W E E++ + E C + + S +T+P C S+S
Sbjct: 171 SPLRNAAYQWNQEEGEEEEVAGEREVKECIRKLQEDENVPSAETTP-----SCSASSSRS 225
Query: 178 SSSNTSSSRWKFLS-LLRRSKSDGK----------------------------------E 202
SS +S +W FL LL RSKS+G+ E
Sbjct: 226 SSYGRNSKKWIFLKDLLHRSKSEGRGNGKEKFWSNISFSPSNFKDKKLKSSQLEEKPIQE 285
Query: 203 SLNMVTPAKKENLKLNSGGNGNVVGKKIPATEKKTPATVSAMEVFY----GRKKETRVKS 258
+++ +KK+ K V GK K+ SA E+ Y + +E + ++
Sbjct: 286 TVDAAVESKKQKQKQPPAKKAPVNGKPTNGIAKRRGLQPSAHELHYTTNRAQAEEMKKRT 345
Query: 259 YLPYKKELIGFSVGFNA 275
YLPY+ L G +GF++
Sbjct: 346 YLPYRHGLFG-CLGFSS 361
>At4g10935 unknown protein
Length = 984
Score = 40.4 bits (93), Expect = 0.001
Identities = 50/216 (23%), Positives = 89/216 (41%), Gaps = 34/216 (15%)
Query: 26 SSNNINDVAQQVTNENDNSH----SQNDDFEFVAFRSHRRNAA--DVSPIFNRDERRNSD 79
S N V ++ + D+SH + ++ E VA +S + D+S + D +
Sbjct: 63 SDTNPLPVQAKMDGDPDSSHLKILCREENTEQVAVKSELNQSLPHDISSRLSSDSDQPFF 122
Query: 80 AMDIS-----ISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLFI---GNEKEKWTSS 131
A DI + K+ L D+K L+ NSD A ++ SLK+ G++ T
Sbjct: 123 AADIENRADPVVEKDCLSIDDKKNLSTAAISNSDVASVI-SLKRKHSDCSGDDGNSETKP 181
Query: 132 EVEDDLDSIPAESYCFWTPKSSSLIASPKTSPMNSTI--------------KCKKSNSTG 177
E+ + L+ + E + + + SP N+T +SN T
Sbjct: 182 EIYESLNELKLEE-----EEELTTVHHESRSPSNNTTVDIFSIVKGTGRRKNLMRSNPTD 236
Query: 178 SSSNTSSSRWKFLSLLRRSKSDGKESLNMVTPAKKE 213
SS ++ + ++R+ D KES+ +V +KE
Sbjct: 237 KSSEAENAAGLRVKKIKRTPEDEKESMVLVEKLRKE 272
>At1g63670 unknown protein
Length = 689
Score = 40.4 bits (93), Expect = 0.001
Identities = 56/228 (24%), Positives = 97/228 (41%), Gaps = 47/228 (20%)
Query: 37 VTNENDNSHSQND--------DFEFVAFRSHRRNAADV------SPIFNRDERRNSDAMD 82
V + D++H++N + E AFR+ + + V SP+F R +R SD
Sbjct: 287 VGSNKDSNHAENKPEGSSRGREIEAAAFRTSDVSTSTVGHRSPESPVFRRKKRVESDVFK 346
Query: 83 ISIS----LKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLFIGNEKEKWTSSEVEDDLD 138
+SI + ++ ++ +LN +S E+ Y+L L K K ++E
Sbjct: 347 LSIENDVLPRRFMVERQQERLN-----SSPVYEVPYALSSL---QTKLKERRQKLEK--- 395
Query: 139 SIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKS 198
ES+ W+ + + P +P NS ++ ++N TGSS S LR
Sbjct: 396 --KRESFKLWSLDKNFEVFDP--NPYNSNLRFLENNCTGSSEYKS---------LRTHVE 442
Query: 199 DG-KESLNMVTPAKKENLKLNSGGNGNVVGKKIPATEKKTPATVSAME 245
DG E + + + NL L + N+ P E++ P+ VS +E
Sbjct: 443 DGFVEDRYLESSLAETNLSLER-QDCNI---HDPKQEQEQPSPVSVLE 486
>At3g24650 abscisic acid-insensitive protein 3
Length = 720
Score = 33.9 bits (76), Expect = 0.10
Identities = 22/80 (27%), Positives = 37/80 (45%), Gaps = 6/80 (7%)
Query: 152 SSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKESLNMVTPAK 211
SSS +S +P+N+ + S+S SSS +S++ W L +SDG++
Sbjct: 83 SSSSSSSTSPAPVNAIVSSASSSSAASSSTSSAASWAIL------RSDGEDPTPNQNQYA 136
Query: 212 KENLKLNSGGNGNVVGKKIP 231
N +SG + +IP
Sbjct: 137 SGNCDDSSGALQSTASMEIP 156
>At2g42150 unknown protein
Length = 631
Score = 32.7 bits (73), Expect = 0.23
Identities = 40/207 (19%), Positives = 82/207 (39%), Gaps = 18/207 (8%)
Query: 20 PSFSVYSSNNINDVAQQVTNENDNSHSQNDDFEFVAFRSHRRN-AADVSPIFNRDERRNS 78
P ++++ D ++ ++ D S DD + + + R + + NR+++
Sbjct: 432 PDKKAKKTDHVVDYDEKPVSDKDGEASGKDDDDSLIVKIMTRGRTSSTGKVANRNDKNRD 491
Query: 79 DAMDISISLKNLLIGDEKPKLNGGGRRNSDAAEILYSLKKLFIGNEKEKWTSSEVEDDLD 138
++++ S + DE+ K GG + AA L +K +G+ + S D
Sbjct: 492 SSLNVDDSKDKVKKTDEEKK---GGSKKKRAASFLRRMK---VGSSDDTLKRSSAAD--S 543
Query: 139 SIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKK--------SNSTGSSSNTSSSRWKFL 190
S + K++S A K +P+ + K +N S + S +
Sbjct: 544 STTGKGGGAEQRKNNSNKADNKKTPIPKIRQTNKKASPVKRSNNGRNSEREAAPSSSSYP 603
Query: 191 SLLRRSKSDG-KESLNMVTPAKKENLK 216
L +RS+ G KE + +P K+ +
Sbjct: 604 ILAKRSRDAGEKEEASSYSPRLKKRAR 630
>At4g19560 putative protein
Length = 474
Score = 32.3 bits (72), Expect = 0.30
Identities = 23/69 (33%), Positives = 38/69 (54%), Gaps = 1/69 (1%)
Query: 154 SLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKESLNMVTPAKKE 213
+++ P + M ST KC S+ G SS + S+ S+ S+S+G +N + A+K
Sbjct: 295 AVVHQPISRDMASTEKCPSSDIEGGSSQVNLSQSDDHSVHDGSRSEGIGEVNSESEAQK- 353
Query: 214 NLKLNSGGN 222
NL+ +S GN
Sbjct: 354 NLQDHSVGN 362
>At5g10940 unknown protein
Length = 757
Score = 31.6 bits (70), Expect = 0.51
Identities = 27/108 (25%), Positives = 50/108 (46%), Gaps = 13/108 (12%)
Query: 5 QQNEETSSTMDSFLCPSFSVYSSNNINDVAQQVTNENDNSHSQNDDFEFVAFRSHRRNAA 64
++NEET + L S +Y S +D + +D S S+ +D ++
Sbjct: 495 EKNEETGAGTTRVLSLSDILYRSEANSD------SSHDMSRSEREDSDY-----DEELEL 543
Query: 65 DVSPIFNRDERRNSDAMDI--SISLKNLLIGDEKPKLNGGGRRNSDAA 110
D+ + DE R++D+ + S++L+ +GD+KP N +S A
Sbjct: 544 DIQTSLSDDEGRDTDSNSMRGSLNLRIHRVGDDKPMENTVDNASSGTA 591
>At2g16270 unknown protein
Length = 759
Score = 31.6 bits (70), Expect = 0.51
Identities = 26/108 (24%), Positives = 48/108 (44%), Gaps = 11/108 (10%)
Query: 125 KEKWTSSEVEDDLD-----SIPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTGSS 179
KEK S E+++D S +S C P+S K + +S+ K ++ + S+
Sbjct: 653 KEKLFSLNFEEEVDDKMSNSFQKKSSCHKEPQSKG---GKKNNNNSSSSKLRRESMASSA 709
Query: 180 SNTSSSRWKFLSLLRRSK---SDGKESLNMVTPAKKENLKLNSGGNGN 224
S S + + S K G+E M+TP ++ + N+ +G+
Sbjct: 710 SEYSIGSFSYGSFTTYEKIPIKSGREEEEMITPVRRSSRIKNNKPSGS 757
>At1g48780 hypothetical protein
Length = 251
Score = 31.6 bits (70), Expect = 0.51
Identities = 26/84 (30%), Positives = 40/84 (46%), Gaps = 1/84 (1%)
Query: 144 SYCFWTPK-SSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKE 202
S FW+ K SSSL K S + S + +SNSTGS +N+ + + ++ R S
Sbjct: 135 SKSFWSFKRSSSLNCDIKKSLICSFPRLTRSNSTGSVTNSKRAMLRDVNNHRPSSRSSCC 194
Query: 203 SLNMVTPAKKENLKLNSGGNGNVV 226
+ P K K GG+ +V+
Sbjct: 195 NAYQFRPQKHTGKKGEGGGSFSVI 218
>At1g19980 unknown protein
Length = 342
Score = 31.6 bits (70), Expect = 0.51
Identities = 32/116 (27%), Positives = 49/116 (41%), Gaps = 8/116 (6%)
Query: 151 KSSSLIA-----SPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKESLN 205
K SSLI+ + TI K+ +N S+ + + +S L R K+S
Sbjct: 203 KISSLISYQEQLQSSNQEKDETISRLKAKMAEMETN-STKKDEEISKLTRDLESAKKSRG 261
Query: 206 MVTPAKKENLKLNSGGNGNVVGKKIPATEKKTPATVSAMEVFYGRKKETRVKSYLP 261
TP KL NGN VG I + K+ A+ + +V K++ R K+ P
Sbjct: 262 F-TPVLTRCTKLEKRSNGNTVGSHISTKKDKSAASTTNEKVSKSSKRK-RAKNMTP 315
>At1g09720 hypothetical protein
Length = 947
Score = 31.2 bits (69), Expect = 0.67
Identities = 37/162 (22%), Positives = 68/162 (41%), Gaps = 14/162 (8%)
Query: 32 DVAQQVTNENDNSHSQNDDFEFVAFRSHRRNAADVSPIFNRDERRNSDAMDISISLKNLL 91
++A Q+ + ++ DF F+ + P+ ER ++++ IS S +
Sbjct: 597 ELALQLRELKNAVSCEDVDFHFLHQKPELPGQGFPHPV----ERNRAESVSISHSSNSSF 652
Query: 92 IGDEKPKLNGGGRRNSDAAEILYSLKKLFIGNEKEK---WTSSEVEDDLDSIPAESYCFW 148
P+ R + E + +K I + K +V D+D++ E+ FW
Sbjct: 653 SMPPLPQRGDLKRASEQEKEDGFKVKFAGISDSLRKKIPTVEEKVRGDIDAVLEENIEFW 712
Query: 149 TPKSSSL--IASPKTSPMN-----STIKCKKSNSTGSSSNTS 183
S+S+ I TS + S I+ K+ + GSSSNT+
Sbjct: 713 LRFSTSVHQIQKYHTSVQDLKAELSKIESKQQGNAGSSSNTA 754
>At4g39840 unknown protein
Length = 451
Score = 30.8 bits (68), Expect = 0.87
Identities = 32/138 (23%), Positives = 58/138 (41%), Gaps = 21/138 (15%)
Query: 138 DSIPAESYCFWTPKSSSLIASPKTSPMNSTIKCK-----------KSNSTGSSSNTSSSR 186
DS+ + S T K+ + + P +S ++ + K K NST SSSNT+ +
Sbjct: 79 DSVSSSSSSSGTKKNQTKLLKPISSSSSTKNQTKLAKTTTMGTSHKLNSTKSSSNTTKTS 138
Query: 187 WKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGNVVGKKIPATEKKTP-----ATV 241
+ L + G +S N + KK S + N K P+++ +P +
Sbjct: 139 SELKKL-----NSGTKSTNSTSSIKKSADLSKSSSSKNKTTIKPPSSKLSSPPSEKKSQP 193
Query: 242 SAMEVFYGRKKETRVKSY 259
S+ V ++ E +K +
Sbjct: 194 SSKPVTKSKQSEKEIKPF 211
>At4g08150 KNAT1 homeobox-like protein
Length = 398
Score = 30.8 bits (68), Expect = 0.87
Identities = 17/48 (35%), Positives = 25/48 (51%), Gaps = 3/48 (6%)
Query: 1 MQNQQQNEETSSTMDSFLCPSFSVYSSNNINDVAQQVTNENDNSHSQN 48
M+ Q + T+ SFL +S SS+N ND N N+N++S N
Sbjct: 1 MEEYQHDNSTTPQRVSFL---YSPISSSNKNDNTSDTNNNNNNNNSSN 45
>At3g18610 unknown protein
Length = 636
Score = 30.8 bits (68), Expect = 0.87
Identities = 31/124 (25%), Positives = 43/124 (34%), Gaps = 16/124 (12%)
Query: 124 EKEKWTSSEVEDDLDS------IPAESYCFWTPKSSSLIASPKTSPMNSTIKCKKSNSTG 177
EK K SS +DD S + + K S + +S T+ KK +
Sbjct: 144 EKAKVESSSSDDDSSSDEETVPVKKQPAVLEKAKIESSSSDDDSSSDEETVPMKKQTAVL 203
Query: 178 SSSNTSSSRWKFLSLLRRSKSDGKESLNMVTPAKKENLKLNSGGNGNVVGKKIPATEKKT 237
+ SS S DG S TPAKKE + + + + KK
Sbjct: 204 EKAKAESS----------SSDDGSSSDEEPTPAKKEPIVVKKDSSDESSSDEETPVVKKK 253
Query: 238 PATV 241
P TV
Sbjct: 254 PTTV 257
>At4g32330 unknown protein
Length = 436
Score = 30.4 bits (67), Expect = 1.1
Identities = 35/135 (25%), Positives = 54/135 (39%), Gaps = 8/135 (5%)
Query: 150 PKSSSLIASPKTSPMNSTIKCKKSNSTGSSSNTSSSRWKFLSLLRRSKSDGKESLNMV-- 207
P + L P T P + + KK+ S S T + R LSL R+ D + ++
Sbjct: 273 PPKTELKKIPPTRPKSPKLGRKKTASGADSEETQTPRLGRLSLDERASKDNPTAKGIMPT 332
Query: 208 -----TPAKKENLKLNSGGNGNVVGKKIPATEKKTPATVSAMEVFYGRKKETRVKSYLPY 262
P +K +L S GK PA PA V + + ET ++ P
Sbjct: 333 VDLKKQPVRKSLPRLPSQKTVLPDGKPAPAKAAIIPAKVRPEKKKLEKDAETVNQTSHPT 392
Query: 263 KKELIGFSVGFNANV 277
++E +V NA+V
Sbjct: 393 EEE-AQVTVSSNADV 406
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.309 0.126 0.351
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,727,780
Number of Sequences: 26719
Number of extensions: 306517
Number of successful extensions: 1108
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 46
Number of HSP's that attempted gapping in prelim test: 1063
Number of HSP's gapped (non-prelim): 76
length of query: 285
length of database: 11,318,596
effective HSP length: 98
effective length of query: 187
effective length of database: 8,700,134
effective search space: 1626925058
effective search space used: 1626925058
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC124951.5


