

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124215.9 - phase: 0
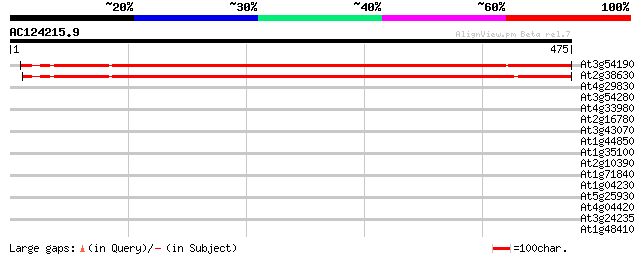
(475 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g54190 unknown protein 730 0.0
At2g38630 unknown protein 708 0.0
At4g29830 unknown protein 34 0.20
At3g54280 TATA box binding protein (TBP) associated factor (TAF)... 33 0.45
At4g33980 putative protein 31 1.3
At2g16780 putative WD-40 repeat protein, MSI2 31 1.3
At3g43070 putative protein 30 2.2
At1g44850 hypothetical protein 30 2.2
At1g35100 hypothetical protein 30 2.2
At2g10390 pseudogene 30 3.8
At1g71840 unknown protein (At1g71840) 30 3.8
At1g04230 Unknown protein 29 5.0
At5g25930 receptor-like protein kinase - like 28 8.4
At4g04420 putative transposon protein 28 8.4
At3g24235 leucine-rich repeat protein, putative, 3' partial 28 8.4
At1g48410 Argonaute protein (AGO1) 28 8.4
>At3g54190 unknown protein
Length = 467
Score = 730 bits (1884), Expect = 0.0
Identities = 368/466 (78%), Positives = 412/466 (87%), Gaps = 12/466 (2%)
Query: 10 SCRRIVATKGESSNNGSKPNIKINNKNIIKKLQTREISPKPHRSFVAATSPHRFQNMRLT 69
S RRIVA K S+P+ +N+ +KKLQ REIS + R+F +T+ RF+NMRL
Sbjct: 14 SGRRIVAKKR------SRPDGFVNS---VKKLQRREISSRKDRAFSISTAQERFRNMRLV 64
Query: 70 HQFDTHDPKHHSSPSPFLPFLMKRTKVVEIVAAKNIVFALAHSGLCAAFSRETNERICFL 129
Q+DTHDPK H + LPFLMKRTKV+EIVAA++IVFALAHSG+CAAFSRE+N+RICFL
Sbjct: 65 EQYDTHDPKGHCLVA--LPFLMKRTKVIEIVAARDIVFALAHSGVCAAFSRESNKRICFL 122
Query: 130 NICPDEVIRSLFYNKNNDSLITVSVYASENFSSLKCRSTRIEYIKRAKPDAGFPLFQSES 189
N+ PDEVIRSLFYNKNNDSLITVSVYAS+NFSSLKCRSTRIEYI R +PDAGF LF+SES
Sbjct: 123 NVSPDEVIRSLFYNKNNDSLITVSVYASDNFSSLKCRSTRIEYILRGQPDAGFALFESES 182
Query: 190 LKWPGFVEFDDVNAKVLTYSAQDSIYKVFDLKNYTLLYSISDRNVQEIKISPGIMLLIFN 249
LKWPGFVEFDDVN KVLTYSAQDS+YKVFDLKNYT+LYSISD+NVQEIKISPGIMLLIF
Sbjct: 183 LKWPGFVEFDDVNGKVLTYSAQDSVYKVFDLKNYTMLYSISDKNVQEIKISPGIMLLIFK 242
Query: 250 RASGHIPLKIISIEDGTVLKAFNHLLHRNKKVDFIEQFNEKLLVKQENENLQILDVRSSE 309
RA+ H+PLKI+SIEDGTVLK+FNHLLHRNKKVDFIEQFNEKLLVKQENENLQILDVR++E
Sbjct: 243 RAASHVPLKILSIEDGTVLKSFNHLLHRNKKVDFIEQFNEKLLVKQENENLQILDVRNAE 302
Query: 310 LMEVSRTEFMTPSAFIFLYENQLFLTFRNRTVSVWNFRGELVTSFEDHLLWHPDCNTNNI 369
LMEVSR EFMTPSAFIFLYENQLFLTFRNR VSVWNFRGELVTSFEDHLLWHPDCNTNNI
Sbjct: 303 LMEVSRAEFMTPSAFIFLYENQLFLTFRNRNVSVWNFRGELVTSFEDHLLWHPDCNTNNI 362
Query: 370 YITSDQDLIISYCKAESEDQWMEANAGSINVSNILTGKCVAKINAANCISKVDDCSSTCS 429
YITSDQDLIISYCKA++EDQW+E NAGSIN+SNILTGKC+AKI ++ K DD SS+ +
Sbjct: 363 YITSDQDLIISYCKADTEDQWIEGNAGSINISNILTGKCLAKITPSSGPPK-DDESSSSN 421
Query: 430 CKHTDSSQLRSSVAEALEDITALFYDEDRNEIYTGNRHGFVHVWSN 475
C +S Q R++VAEALEDITALFYDE+RNEIYTGNRHG VHVWSN
Sbjct: 422 CMGKNSKQRRNAVAEALEDITALFYDEERNEIYTGNRHGLVHVWSN 467
>At2g38630 unknown protein
Length = 467
Score = 708 bits (1828), Expect = 0.0
Identities = 355/464 (76%), Positives = 408/464 (87%), Gaps = 14/464 (3%)
Query: 12 RRIVATKGESSNNGSKPNIKINNKNIIKKLQTREISPKPHRSFVAATSPHRFQNMRLTHQ 71
RR++A K S+P+ +N+ +KKLQ REIS + R+F +T+ RF+NMRL Q
Sbjct: 18 RRVIAKKR------SRPDGFVNS---VKKLQRREISSRMDRAFSISTAQERFRNMRLVEQ 68
Query: 72 FDTHDPKHHSSPSPFLPFLMKRTKVVEIVAAKNIVFALAHSGLCAAFSRETNERICFLNI 131
+DTHDPK + S LP L+KR+KV+EIVAA++IVFAL SG+CA+FSRETN+++CFLN+
Sbjct: 69 YDTHDPKGYCLVS--LPNLLKRSKVIEIVAARDIVFALTLSGVCASFSRETNKKVCFLNV 126
Query: 132 CPDEVIRSLFYNKNNDSLITVSVYASENFSSLKCRSTRIEYIKRAKPDAGFPLFQSESLK 191
PDEVIRSLFYNKNNDSLITVSVYAS+N+SSLKCRSTRIEYI R + DAGFPLF+SESLK
Sbjct: 127 SPDEVIRSLFYNKNNDSLITVSVYASDNYSSLKCRSTRIEYILRGQADAGFPLFESESLK 186
Query: 192 WPGFVEFDDVNAKVLTYSAQDSIYKVFDLKNYTLLYSISDRNVQEIKISPGIMLLIFNRA 251
WPGFVEFDDVN KVLTYSAQDS+YKVFDLKNY LLYSISD+NVQEIKISPGIMLLIF RA
Sbjct: 187 WPGFVEFDDVNGKVLTYSAQDSVYKVFDLKNYALLYSISDKNVQEIKISPGIMLLIFKRA 246
Query: 252 SGHIPLKIISIEDGTVLKAFNHLLHRNKKVDFIEQFNEKLLVKQENENLQILDVRSSELM 311
+ H+PLKI+SIEDGT+LK+F+HLLHRNKKVDFIEQFNEKLLVKQENENLQILDVR++EL+
Sbjct: 247 ASHVPLKILSIEDGTLLKSFHHLLHRNKKVDFIEQFNEKLLVKQENENLQILDVRNAELI 306
Query: 312 EVSRTEFMTPSAFIFLYENQLFLTFRNRTVSVWNFRGELVTSFEDHLLWHPDCNTNNIYI 371
EVSRT+FMTPSAFIFLYENQLFLTFRNR VSVWNFRGELVTSFEDHLLWHPDCNTNNIYI
Sbjct: 307 EVSRTDFMTPSAFIFLYENQLFLTFRNRNVSVWNFRGELVTSFEDHLLWHPDCNTNNIYI 366
Query: 372 TSDQDLIISYCKAESEDQWMEANAGSINVSNILTGKCVAKINAANCISKVDDCSSTCSCK 431
TSDQDLIISYCKA++EDQW+E NAGSIN+SNILTGKC+AKI A N K +DCSS+
Sbjct: 367 TSDQDLIISYCKADTEDQWIEGNAGSINISNILTGKCLAKIKANNGPPKEEDCSSS---D 423
Query: 432 HTDSSQLRSSVAEALEDITALFYDEDRNEIYTGNRHGFVHVWSN 475
+SS+ RS+VAEALEDITALFYDE+RNEIYTGNRHG +HVWSN
Sbjct: 424 LGNSSRRRSAVAEALEDITALFYDEERNEIYTGNRHGLLHVWSN 467
>At4g29830 unknown protein
Length = 321
Score = 33.9 bits (76), Expect = 0.20
Identities = 39/145 (26%), Positives = 61/145 (41%), Gaps = 13/145 (8%)
Query: 118 FSRETNERICFLNICPDEVIRSLFYNKNNDSLITVSVYASENFSSLKCRSTR----IEYI 173
F +TN I L P EV F K T+ A + +S+K T I +
Sbjct: 87 FDVDTNATIAVLEAPPSEVWGMQFEPKG-----TILAVAGGSSASVKLWDTASWRLISTL 141
Query: 174 KRAKPDAGFPLFQSESLKWPGFVEFDDVNAKVLTYSAQDSIYKVFDLKNYTLLYSISDRN 233
+PDA P ++ S K+ V + N K L + D VFD+ LL+ + N
Sbjct: 142 SIPRPDAPKPSDKTSSKKFVLSVAWSP-NGKRLACGSMDGTICVFDVDRSKLLHQLEGHN 200
Query: 234 --VQEIKISPGIMLLIFNRA-SGHI 255
V+ + SP ++F+ + GH+
Sbjct: 201 MPVRSLVFSPVDPRVLFSGSDDGHV 225
>At3g54280 TATA box binding protein (TBP) associated factor (TAF)
-like protein
Length = 2049
Score = 32.7 bits (73), Expect = 0.45
Identities = 19/68 (27%), Positives = 32/68 (46%), Gaps = 1/68 (1%)
Query: 261 SIEDGTVLKAFNHLLHRNKKVDFIEQFNEKLLVKQENENLQILD-VRSSELMEVSRTEFM 319
S++D T NH+ N + E ++ KQ+ ENL++LD V+ + + EF+
Sbjct: 438 SMDDSTSHSEINHVAEVNNHFEDKSFIEEPVIPKQQEENLEVLDLVKQARHSWIKNFEFL 497
Query: 320 TPSAFIFL 327
FL
Sbjct: 498 QDCTIRFL 505
>At4g33980 putative protein
Length = 247
Score = 31.2 bits (69), Expect = 1.3
Identities = 19/70 (27%), Positives = 35/70 (49%), Gaps = 10/70 (14%)
Query: 363 DCNTNNIYITSDQDLIISYCKAESEDQWMEANAGSINVSNILTGKCVAKINAANCISKVD 422
DC N + + D+D +S AES+ + +SN L A+ + ++ SK+D
Sbjct: 4 DCTVNIVSLEKDRD--VSEASAESQSE--------STLSNSLDSGVTAETSRSDADSKLD 53
Query: 423 DCSSTCSCKH 432
+C++ + KH
Sbjct: 54 ECTAWTNEKH 63
>At2g16780 putative WD-40 repeat protein, MSI2
Length = 415
Score = 31.2 bits (69), Expect = 1.3
Identities = 36/133 (27%), Positives = 59/133 (44%), Gaps = 20/133 (15%)
Query: 195 FVEFDDVNAKVLTYSAQDSIYKVFDLKNYTL-LYSIS-----------DRNVQEIKISPG 242
++ F+ N VL ++ DS +FDL+ L+ +S D N + + S G
Sbjct: 265 YLSFNPFNEWVLATASSDSTVALFDLRKLNAPLHVMSSHEGEVFQVEWDPNHETVLASSG 324
Query: 243 ----IMLLIFNRASGHIPLKI-ISIEDGTVLKAFNHLLHRNKKVDFIEQFNEKLLVKQ-- 295
+M+ NR G L+I + EDG F+H H+ K DF NE ++
Sbjct: 325 EDRRLMVWDLNRV-GEEQLEIELDAEDGPPELLFSHGGHKAKISDFAWNKNEPWVIASVA 383
Query: 296 ENENLQILDVRSS 308
E+ +LQ+ + S
Sbjct: 384 EDNSLQVWQMAES 396
>At3g43070 putative protein
Length = 426
Score = 30.4 bits (67), Expect = 2.2
Identities = 21/68 (30%), Positives = 31/68 (44%), Gaps = 7/68 (10%)
Query: 83 PSPFLPFLMKRTKVVEIVAAKNIVFALAHSGLCAAFSRETNERI------CFLNICPDEV 136
P P L ++ I A KN + + H G AAFS ET R+ F+N+ +
Sbjct: 222 PEPILALIVSHVSEEGIEALKNWIKS-GHDGKSAAFSVETLSRVRLDKSPDFINMSSPDS 280
Query: 137 IRSLFYNK 144
+ FY+K
Sbjct: 281 VHFSFYSK 288
>At1g44850 hypothetical protein
Length = 266
Score = 30.4 bits (67), Expect = 2.2
Identities = 21/68 (30%), Positives = 31/68 (44%), Gaps = 7/68 (10%)
Query: 83 PSPFLPFLMKRTKVVEIVAAKNIVFALAHSGLCAAFSRETNERI------CFLNICPDEV 136
P P L ++ I A KN + + H G AAFS ET R+ F+N+ +
Sbjct: 62 PEPILALIVSHVSEEGIEALKNWIKS-GHEGKSAAFSVETLSRVRLDKSPDFINMSSPDS 120
Query: 137 IRSLFYNK 144
+ FY+K
Sbjct: 121 VHFSFYSK 128
>At1g35100 hypothetical protein
Length = 231
Score = 30.4 bits (67), Expect = 2.2
Identities = 21/68 (30%), Positives = 31/68 (44%), Gaps = 7/68 (10%)
Query: 83 PSPFLPFLMKRTKVVEIVAAKNIVFALAHSGLCAAFSRETNERI------CFLNICPDEV 136
P P L ++ I A KN + + H G AAFS ET R+ F+N+ +
Sbjct: 27 PEPILALIVSHVSEEGIEALKNWIKS-GHEGKSAAFSVETLSRVRLDKSPDFINMSSPDS 85
Query: 137 IRSLFYNK 144
+ FY+K
Sbjct: 86 VHFSFYSK 93
>At2g10390 pseudogene
Length = 194
Score = 29.6 bits (65), Expect = 3.8
Identities = 21/68 (30%), Positives = 31/68 (44%), Gaps = 7/68 (10%)
Query: 83 PSPFLPFLMKRTKVVEIVAAKNIVFALAHSGLCAAFSRETNERI------CFLNICPDEV 136
P P L ++ I A KN + + H G AAFS ET R+ F+N+ +
Sbjct: 27 PEPILALIVSHVSEEGIEALKNWIKS-GHEGKSAAFSVETLSRVRLDKSPDFINMSLPDS 85
Query: 137 IRSLFYNK 144
+ FY+K
Sbjct: 86 VHFSFYSK 93
>At1g71840 unknown protein (At1g71840)
Length = 407
Score = 29.6 bits (65), Expect = 3.8
Identities = 23/95 (24%), Positives = 41/95 (42%), Gaps = 16/95 (16%)
Query: 396 GSINVSNILTGKCVAKIN----AANCISKVDDCSSTCSCKHTDSSQLR-----------S 440
GS+++ NI+TGK V+ +N + C+ K S+T T +
Sbjct: 264 GSVHIVNIVTGKVVSSLNSHTDSVECV-KFSPSSATIPLAATGGMDKKLIIWDLQHSTPR 322
Query: 441 SVAEALEDITALFYDEDRNEIYTGNRHGFVHVWSN 475
+ E E +T+L + + TG +G V +W +
Sbjct: 323 FICEHEEGVTSLTWIGTSKYLATGCANGTVSIWDS 357
>At1g04230 Unknown protein
Length = 278
Score = 29.3 bits (64), Expect = 5.0
Identities = 22/82 (26%), Positives = 33/82 (39%), Gaps = 3/82 (3%)
Query: 2 DERRRGVWSCRRIVATKGESSNNGSKPNIKINNKNIIKKLQTREISPKPHRSFVAATSPH 61
DER + +S R ++ + S+ + I T S + +S AATS H
Sbjct: 193 DERNQKPYSTRVLMPPPRSRFASTSRQYSSVKRNEIPSSSNT---SHRRSQSSHAATSSH 249
Query: 62 RFQNMRLTHQFDTHDPKHHSSP 83
Q+ L+ D H PK P
Sbjct: 250 TSQSSNLSSNSDAHKPKRKRRP 271
>At5g25930 receptor-like protein kinase - like
Length = 1005
Score = 28.5 bits (62), Expect = 8.4
Identities = 26/85 (30%), Positives = 38/85 (44%), Gaps = 8/85 (9%)
Query: 196 VEFDDVNAKVLTYSAQDSIYKVFDLKNYTLLYSISDRNVQEIKISPGIMLLIF-----NR 250
+E D++ LT D + F LKN T Y ++ EI S L+F N
Sbjct: 237 LEHVDLSVNNLTGRIPDVL---FGLKNLTEFYLFANGLTGEIPKSISATNLVFLDLSANN 293
Query: 251 ASGHIPLKIISIEDGTVLKAFNHLL 275
+G IP+ I ++ VL FN+ L
Sbjct: 294 LTGSIPVSIGNLTKLQVLNLFNNKL 318
>At4g04420 putative transposon protein
Length = 1008
Score = 28.5 bits (62), Expect = 8.4
Identities = 29/125 (23%), Positives = 53/125 (42%), Gaps = 7/125 (5%)
Query: 196 VEFDDVNAKVLTYSAQDSIYKVFDLKNYTLLYSISDRNVQEIKISPGIMLLIFNRASGHI 255
++F++V + Y + + K K L S +QE+K + +M F+RA +
Sbjct: 197 IDFEEVVGMLQAYELEITSGKGGYSKGLALAASAKKNEIQELKDTMSMMAKDFSRAMRRV 256
Query: 256 PLKIISIEDGTVLKAFNHLLHRNKKVDFIE--QFNEKLLVKQENENLQILDVRSSELMEV 313
K GT + R+ K D I+ + +K E +L+ D++ SE +
Sbjct: 257 EKKGFGRNQGT-----DRYRDRSSKRDEIQCHECQGYGHIKAECPSLKRKDLKCSECNGL 311
Query: 314 SRTEF 318
T+F
Sbjct: 312 GHTKF 316
>At3g24235 leucine-rich repeat protein, putative, 3' partial
Length = 576
Score = 28.5 bits (62), Expect = 8.4
Identities = 36/140 (25%), Positives = 63/140 (44%), Gaps = 15/140 (10%)
Query: 193 PGFVEFDDVNAKVLTYSAQDSIYK--VFDLKNYTLLYSISDRNV----QEIKISPGIMLL 246
PG + D+ A L+ ++ +F L+N T L IS+ QEI ++ L
Sbjct: 412 PGLADCTDLQALDLSRNSLTGTIPSGLFMLRNLTKLLLISNSLSGFIPQEIGNCSSLVRL 471
Query: 247 I--FNRASGHIPLKIISIEDGTVLKAFNHLLHRNKKVDFIEQFNEKLLVKQENENLQ--- 301
FNR +G IP I S++ L ++ LH K D I +E ++ N +L+
Sbjct: 472 RLGFNRITGEIPSGIGSLKKINFLDFSSNRLH-GKVPDEIGSCSELQMIDLSNNSLEGSL 530
Query: 302 ---ILDVRSSELMEVSRTEF 318
+ + ++++VS +F
Sbjct: 531 PNPVSSLSGLQVLDVSANQF 550
>At1g48410 Argonaute protein (AGO1)
Length = 870
Score = 28.5 bits (62), Expect = 8.4
Identities = 13/32 (40%), Positives = 19/32 (58%)
Query: 244 MLLIFNRASGHIPLKIISIEDGTVLKAFNHLL 275
+L+ F R++GH PL+II DG F +L
Sbjct: 648 LLIAFRRSTGHKPLRIIFYRDGVSEGQFYQVL 679
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.134 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,696,774
Number of Sequences: 26719
Number of extensions: 448575
Number of successful extensions: 1296
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 1287
Number of HSP's gapped (non-prelim): 16
length of query: 475
length of database: 11,318,596
effective HSP length: 103
effective length of query: 372
effective length of database: 8,566,539
effective search space: 3186752508
effective search space used: 3186752508
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC124215.9


