

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124215.6 - phase: 0 /pseudo
(679 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
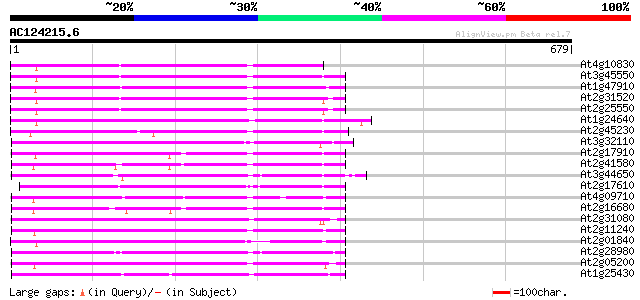
Score E
Sequences producing significant alignments: (bits) Value
At4g10830 putative protein 249 5e-66
At3g45550 putative protein 239 4e-63
At1g47910 reverse transcriptase, putative 234 9e-62
At2g31520 putative non-LTR retroelement reverse transcriptase 233 3e-61
At2g25550 putative non-LTR retroelement reverse transcriptase 232 5e-61
At1g24640 hypothetical protein 223 3e-58
At2g45230 putative non-LTR retroelement reverse transcriptase 220 2e-57
At3g32110 non-LTR reverse transcriptase, putative 220 2e-57
At2g17910 putative non-LTR retroelement reverse transcriptase 218 7e-57
At2g41580 putative non-LTR retroelement reverse transcriptase 211 1e-54
At3g44650 putative protein 210 2e-54
At2g17610 putative non-LTR retroelement reverse transcriptase 210 2e-54
At4g09710 RNA-directed DNA polymerase -like protein 207 1e-53
At2g16680 putative non-LTR retroelement reverse transcriptase 206 3e-53
At2g31080 putative non-LTR retroelement reverse transcriptase 206 3e-53
At2g11240 pseudogene 206 3e-53
At2g01840 putative non-LTR retroelement reverse transcriptase 203 2e-52
At2g28980 putative non-LTR retroelement reverse transcriptase 201 1e-51
At2g05200 putative non-LTR retroelement reverse transcriptase 200 2e-51
At1g25430 hypothetical protein 199 4e-51
>At4g10830 putative protein
Length = 1294
Score = 249 bits (635), Expect = 5e-66
Identities = 146/385 (37%), Positives = 216/385 (55%), Gaps = 11/385 (2%)
Query: 1 DVQSAFIKGRQILDGILVANEVVDDARKRKK---DLLLFKVDFEKAYDSVD*NYLEEVMV 57
D Q+AFI GR + D +++A+E++ + RK+ + K D KAYD V+ N+LE M
Sbjct: 897 DSQAAFIPGRLVNDNVMIAHEMMHSLKTRKRVSQSYMAVKTDVSKAYDRVEWNFLETTMR 956
Query: 58 KMGFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVL 117
GF W KWI V + SVLVNG P RG+RQGDPLSP+LF+L A+ + L
Sbjct: 957 LFGFSETWIKWIMGAVKSVNYSVLVNGIPHGTIQPQRGIRQGDPLSPYLFILCADILNHL 1016
Query: 118 MEALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLK 177
++ V G ++G + + GVTHLQFADD++ C + N +AL+ + +++ SG K
Sbjct: 1017 IKNRVAEGDIRGIRIG-NGVPGVTHLQFADDSLFFCQSNVRNCQALKDVFDVYEYYSGQK 1075
Query: 178 VNFSKSLFV-GVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRIN 236
+N SKS+ G VHG+ +L + YLGLP + + +I R+
Sbjct: 1076 INMSKSMITFGSRVHGTTQNRLKNILGIQSHGGGGKYLGLPEQFGRKKRDMFNYIIERVK 1135
Query: 237 SRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGG 296
R SSW +K+LS G+ ++LKSV S+PVYA+S FK P IVS IE++L NF W
Sbjct: 1136 KRTSSWSAKYLSPAGKEIMLKSVAMSMPVYAMSCFKLPLNIVSEIEALLMNF-----WWE 1190
Query: 297 GSADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVR 356
+A R+I W+ W + S++ GGL R + +FN ALL K WR++ +SL+ R++ R
Sbjct: 1191 KNAKKREIPWIAWKRLQYSKKEGGLGFRDLAKFNDALLAKQVWRMINNPNSLFARIMKAR 1250
Query: 357 NRVEGGHL-CSGGRNESMWWRNIVA 380
E L R +S W +++A
Sbjct: 1251 YFREDSILDAKRQRYQSYGWTSMLA 1275
>At3g45550 putative protein
Length = 851
Score = 239 bits (610), Expect = 4e-63
Identities = 149/411 (36%), Positives = 223/411 (54%), Gaps = 13/411 (3%)
Query: 1 DVQSAFIKGRQILDGILVANEVVDDARKRK---KDLLLFKVDFEKAYDSVD*NYLEEVMV 57
D Q+AFI GR I D +++A+E++ + RK K + K D KAYD V+ ++LE M
Sbjct: 156 DSQAAFIPGRIINDNVMIAHEIMHSLKVRKRVSKTYMAVKTDVSKAYDRVEWDFLETTMR 215
Query: 58 KMGFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVL 117
GF W WI V + SVL+NGSP S RG+RQGDPLSP+LF+L + L
Sbjct: 216 LFGFCDKWIGWIMAAVKSVHYSVLINGSPHGYISPTRGIRQGDPLSPYLFILCGDILSHL 275
Query: 118 MEALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLK 177
++ + G ++G + +THLQFADD++ C + N +AL+ + +++ SG K
Sbjct: 276 IKVKASSGDIRGVRIG-NGAPAITHLQFADDSLFFCQANVRNCQALKDVFDVYEYYSGQK 334
Query: 178 VNFSKSLFV-GVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRIN 236
+N KSL G V+GS T+LN YLGLP + + +I+R+
Sbjct: 335 INVQKSLITFGSRVYGSTQTRLKTLLNIPNQGGGGKYLGLPEQFGRKKKEMFNYIIDRVK 394
Query: 237 SRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGG 296
R +SW +K LS G+ +LLKSV ++PVYA+S FK P GIVS IES+L NF W
Sbjct: 395 ERTASWSAKFLSPAGKEILLKSVALAMPVYAMSCFKLPQGIVSEIESLLMNF-----WWE 449
Query: 297 GSADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVR 356
+++ R I WV W + S++ GGL R + +FN ALL K WR++ +SL+ RV+ R
Sbjct: 450 KASNKRGIPWVAWKRLQYSKKEGGLGFRDLAKFNDALLAKQAWRIIQYPNSLFARVMKAR 509
Query: 357 NRVEGGHLCSGGRN-ESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTD 406
+ + + R+ +S W ++ L + +GDG ++ D
Sbjct: 510 YFKDNSIIDAKTRSQQSYGWSSL--LSGIALLRKGTRYVIGDGKTIRLGID 558
>At1g47910 reverse transcriptase, putative
Length = 1142
Score = 234 bits (598), Expect = 9e-62
Identities = 144/411 (35%), Positives = 214/411 (52%), Gaps = 13/411 (3%)
Query: 1 DVQSAFIKGRQILDGILVANEVVDDARKR---KKDLLLFKVDFEKAYDSVD*NYLEEVMV 57
+ QSAF+ GR I D IL+A E+ R K + K D KAYD V+ N++E ++
Sbjct: 319 ETQSAFVDGRLISDNILIAQEMFHGLRTNSSCKDKFMAIKTDMSKAYDQVEWNFIEALLR 378
Query: 58 KMGFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVL 117
KMGF W WI C+ T VL+NG P RGLRQGDPLSP+LF+L E
Sbjct: 379 KMGFCEKWISWIMWCITTVQYKVLINGQPKGLIIPERGLRQGDPLSPYLFILCTEVLIAN 438
Query: 118 MEALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLK 177
+ NL G KV + V+HL FADD++ C + + IL ++ +SG +
Sbjct: 439 IRKAERQNLITGIKVAT-PSPAVSHLLFADDSLFFCKANKEQCGIILEILKQYESVSGQQ 497
Query: 178 VNFSK-SLFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRIN 236
+NFSK S+ G V S A+ +L + YLGLP S+ + + +R+
Sbjct: 498 INFSKSSIQFGHKVEDSIKADIKLILGIHNLGGMGSYLGLPESLGGSKTKVFSFVRDRLQ 557
Query: 237 SRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGG 296
SR++ W +K LS GG+ V++KSV ++LP Y +S F+ P I S + S + F W
Sbjct: 558 SRINGWSAKFLSKGGKEVMIKSVAATLPRYVMSCFRLPKAITSKLTSAVAKF-----WWS 612
Query: 297 GSADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVR 356
+ D R + W+ W+ +C S+ GGL R + +FN ALL K WR++T DSL+ +V R
Sbjct: 613 SNGDSRGMHWMAWDKLCSSKSDGGLGFRNVDDFNSALLAKQLWRLITAPDSLFAKVFKGR 672
Query: 357 NRVEGGHLCS-GGRNESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTD 406
+ L S + S WR++++ S + + +G G+S+ W D
Sbjct: 673 YFRKSNPLDSIKSYSPSYGWRSMISARS--LVYKGLIKRVGSGASISVWND 721
>At2g31520 putative non-LTR retroelement reverse transcriptase
Length = 1524
Score = 233 bits (593), Expect = 3e-61
Identities = 147/414 (35%), Positives = 225/414 (53%), Gaps = 19/414 (4%)
Query: 1 DVQSAFIKGRQILDGILVANEVVDDARKRK---KDLLLFKVDFEKAYDSVD*NYLEEVMV 57
D Q+AFI GR I D +++A+EV+ + RK K + K D KAYD V+ ++LE M
Sbjct: 691 DSQAAFIPGRIINDNVMIAHEVMHSLKVRKRVSKTYMAVKTDVSKAYDRVEWDFLETTMR 750
Query: 58 KMGFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVL 117
GF W WI V + SVL+NGSP + RG+RQGDPLSP+LF+L + L
Sbjct: 751 LFGFCNKWIGWIMAAVKSVHYSVLINGSPHGYITPTRGIRQGDPLSPYLFILCGDILSHL 810
Query: 118 MEALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLK 177
+ + G ++G + +THLQFADD++ C + N +AL+ + +++ SG K
Sbjct: 811 INGRASSGDLRGVRIG-NGAPAITHLQFADDSLFFCQANVRNCQALKDVFDVYEYYSGQK 869
Query: 178 VNFSKSLFV-GVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRIN 236
+N KS+ G V+GS ++ +L YLGLP + ++ +I+R+
Sbjct: 870 INVQKSMITFGSRVYGSTQSKLKQILEIPNQGGGGKYLGLPEQFGRKKKEMFEYIIDRVK 929
Query: 237 SRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGG 296
R S+W ++ LS G+ ++LKSV ++PVYA+S FK P GIVS IES+L NF W
Sbjct: 930 KRTSTWSARFLSPAGKEIMLKSVALAMPVYAMSCFKLPKGIVSEIESLLMNF-----WWE 984
Query: 297 GSADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVR 356
+++ R I WV W + S++ GGL R + +FN ALL K WR++ +SL+ RV+ R
Sbjct: 985 KASNQRGIPWVAWKRLQYSKKEGGLGFRDLAKFNDALLAKQAWRLIQYPNSLFARVMKAR 1044
Query: 357 NRVEGGHLCSGGR-NESMWWRNI---VALHSEGWFQNHVSRSLGDGSSVLFWTD 406
+ L + R +S W ++ +AL +G +GDG ++ D
Sbjct: 1045 YFKDVSILDAKVRKQQSYGWASLLDGIALLKKG-----TRHLIGDGQNIRIGLD 1093
>At2g25550 putative non-LTR retroelement reverse transcriptase
Length = 1750
Score = 232 bits (592), Expect = 5e-61
Identities = 147/414 (35%), Positives = 224/414 (53%), Gaps = 19/414 (4%)
Query: 1 DVQSAFIKGRQILDGILVANEVVDDARKRK---KDLLLFKVDFEKAYDSVD*NYLEEVMV 57
D Q+AFI GR I D +++A+EV+ + RK K + K D KAYD V+ ++LE M
Sbjct: 917 DSQAAFIPGRIINDNVMIAHEVMHSLKVRKRVSKTYMAVKTDVSKAYDRVEWDFLETTMR 976
Query: 58 KMGFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVL 117
GF W WI V + SVL+NGSP + RG+RQGDPLSP+LF+L + L
Sbjct: 977 LFGFCNKWIGWIMAAVKSVHYSVLINGSPHGYITPTRGIRQGDPLSPYLFILCGDILSHL 1036
Query: 118 MEALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLK 177
+ + G ++G + +THLQFADD++ C + N +AL+ + +++ SG K
Sbjct: 1037 INGRASSGDLRGVRIG-NGAPAITHLQFADDSLFFCQANVRNCQALKDVFDVYEYYSGQK 1095
Query: 178 VNFSKSLFV-GVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRIN 236
+N KS+ G V+GS + +L YLGLP + ++ +I+R+
Sbjct: 1096 INVQKSMITFGSRVYGSTQSRLKQILEIPNQGGGGKYLGLPEQFGRKKKEMFEYIIDRVK 1155
Query: 237 SRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGG 296
R S+W ++ LS G+ ++LKSV ++PVYA+S FK P GIVS IES+L NF W
Sbjct: 1156 KRTSTWSARFLSPAGKEIMLKSVALAMPVYAMSCFKLPKGIVSEIESLLMNF-----WWE 1210
Query: 297 GSADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVR 356
+++ R I WV W + S++ GGL R + +FN ALL K WR++ +SL+ RV+ R
Sbjct: 1211 KASNQRGIPWVAWKRLQYSKKEGGLGFRDLAKFNDALLAKQAWRLIQYPNSLFARVMKAR 1270
Query: 357 NRVEGGHLCSGGR-NESMWWRNI---VALHSEGWFQNHVSRSLGDGSSVLFWTD 406
+ L + R +S W ++ +AL +G +GDG ++ D
Sbjct: 1271 YFKDVSILDAKVRKQQSYGWASLLDGIALLKKG-----TRHLIGDGQNIRIGLD 1319
>At1g24640 hypothetical protein
Length = 1270
Score = 223 bits (568), Expect = 3e-58
Identities = 156/451 (34%), Positives = 226/451 (49%), Gaps = 22/451 (4%)
Query: 1 DVQSAFIKGRQILDGILVANEVVDDARKRKK---DLLLFKVDFEKAYDSVD*NYLEEVMV 57
D QSAF+ R I D ILVA+E+V + + + + K D KAYD V+ +YL +++
Sbjct: 498 DTQSAFVSERLITDNILVAHELVHSLKVHPRISSEFMAVKSDMSKAYDRVEWSYLRSLLL 557
Query: 58 KMGFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVL 117
+GF W WI CV + T SVL+N P L RGLRQGDPLSPFLF+L EG L
Sbjct: 558 SLGFHLKWVNWIMVCVSSVTYSVLINDCPFGLIILQRGLRQGDPLSPFLFVLCTEGLTHL 617
Query: 118 MEALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLK 177
+ G + + + V HL FADD++ LC S L+ IL ++ +G
Sbjct: 618 LNKAQWEGALEGIQFSENGPM-VHHLLFADDSLFLCKASREQSLVLQKILKVYGNATGQT 676
Query: 178 VNFSK-SLFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRIN 236
+N +K S+ G V T L YLGLP + S++ L +R+
Sbjct: 677 INLNKSSITFGEKVDEQLKGTIRTCLGIFTEGGAGTYLGLPECFSGSKVDMLHYLKDRLK 736
Query: 237 SRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGG 296
+L W ++ LS GG+ VLLKSV ++PV+A+S FK P ++ES + +F++
Sbjct: 737 EKLDVWFTRCLSQGGKEVLLKSVALAMPVFAMSCFKLPITTCENLESAMASFWW------ 790
Query: 297 GSADH-RKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAV 355
S DH RKI W W +C ++ GGL R I+ FN ALL K WR+L D L R+L
Sbjct: 791 DSCDHSRKIHWQSWERLCLPKDSGGLGFRDIQSFNQALLAKQAWRLLHFPDCLLSRLLKS 850
Query: 356 RNRVEGGHLCSG-GRNESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTDV*VGELSL 414
R L + + S WR+I L + + +GDG+S+ W D + +
Sbjct: 851 RYFDATDFLDAALSQRPSFGWRSI--LFGRELLSKGLQKRVGDGASLFVWIDPWIDDNGF 908
Query: 415 RD--RFSRLYDL-----SVLKGGVGRYDESV 438
R R + +YD+ ++L G +DE V
Sbjct: 909 RAPWRKNLIYDVTLKVKALLNPRTGFWDEEV 939
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 220 bits (561), Expect = 2e-57
Identities = 140/418 (33%), Positives = 219/418 (51%), Gaps = 19/418 (4%)
Query: 1 DVQSAFIKGRQILDGILVANEVV---DDARKRKKDLLLFKVDFEKAYDSVD*NYLEEVMV 57
+ Q+AF+KGR I D IL+A+E++ K ++ + K D KAYD V+ +LE+ M
Sbjct: 535 ETQAAFVKGRLISDNILIAHELLHALSSNNKCSEEFIAIKTDISKAYDRVEWPFLEKAMR 594
Query: 58 KMGFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVL 117
+GF W + I +CV + VL+NG+P E RGLRQGDPLSP+LF++ E +
Sbjct: 595 GLGFADHWIRLIMECVKSVRYQVLINGTPHGEIIPSRGLRQGDPLSPYLFVICTEMLVKM 654
Query: 118 MEALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQE---LS 174
+++ N G KV + ++HL FADD++ C N AL I+ + +E S
Sbjct: 655 LQSAEQKNQITGLKV-ARGAPPISHLLFADDSMFYCK---VNDEALGQIIRIIEEYSLAS 710
Query: 175 GLKVNFSK-SLFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLIN 233
G +VN+ K S++ G ++ L +YLGLP S++ L +
Sbjct: 711 GQRVNYLKSSIYFGKHISEERRCLVKRKLGIEREGGEGVYLGLPESFQGSKVATLSYLKD 770
Query: 234 RINSRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGG 293
R+ ++ W+S LS GG+ +LLK+V +LP Y +S FK P I IES++ F
Sbjct: 771 RLGKKVLGWQSNFLSPGGKEILLKAVAMALPTYTMSCFKIPKTICQQIESVMAEF----- 825
Query: 294 WGGGSADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVL 353
W + R + W W + R + VGGL + I+ FN+ALLGK WR++TE+DSL +V
Sbjct: 826 WWKNKKEGRGLHWKAWCHLSRPKAVGGLGFKEIEAFNIALLGKQLWRMITEKDSLMAKVF 885
Query: 354 AVRNRVEGGHLCSG-GRNESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTDV*VG 410
R + L + G S W++I ++ + + +G+G ++ WTD +G
Sbjct: 886 KSRYFSKSDPLNAPLGSRPSFAWKSI--YEAQVLIKQGIRAVIGNGETINVWTDPWIG 941
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 220 bits (560), Expect = 2e-57
Identities = 147/421 (34%), Positives = 214/421 (49%), Gaps = 15/421 (3%)
Query: 3 QSAFIKGRQILDGILVANEVVDDARKRK--KDLLLFKVDFEKAYDSVD*NYLEEVMVKMG 60
Q++FI GR D I+V EVV R++K K +L K+D EKAYD + + LE+ + G
Sbjct: 1077 QTSFIPGRLSTDNIVVVQEVVHSMRRKKGVKGWMLLKLDLEKAYDRIRWDLLEDTLKAAG 1136
Query: 61 FPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEA 120
P W +WI KCV + +L NG T F RGLRQGDPLSP+LF+L E L+E+
Sbjct: 1137 LPGTWVQWIMKCVEGPSMRLLWNGEKTDAFKPLRGLRQGDPLSPYLFVLCIERLCHLIES 1196
Query: 121 LVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVNF 180
+ + K+ S ++H+ FADD I+ + S IR LR +L F SG KV+
Sbjct: 1197 SIAAKKWKPIKI-SQSGPRLSHICFADDLILFAEASIDQIRVLRGVLEKFCGASGQKVSL 1255
Query: 181 SKS-LFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSRL 239
KS ++ NV + A YLG+PI + +I R +SRL
Sbjct: 1256 EKSKIYFSKNVLRDLGKRISEESGMKATKDLGKYLGVPILQKRINKETFGEVIKRFSSRL 1315
Query: 240 SSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGGGSA 299
+ WK + LS GRL L K+VL+S+ V+ +S K P + ++ + + F W G +
Sbjct: 1316 AGWKGRMLSFAGRLTLTKAVLTSILVHTMSTIKLPQSTLDGLDKV-SRAFLW----GSTL 1370
Query: 300 DHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVRNRV 359
+ +K V W VC + GGL +R N AL+ K WRVL + SLW +V+ + +V
Sbjct: 1371 EKKKQHLVAWTRVCLPRREGGLGIRSATAMNKALIAKVGWRVLNDGSSLWAQVVRSKYKV 1430
Query: 360 EGGHLCSGGRNESMW---WRNI-VALHSEGWFQNHVSRSLGDGSSVLFWTDV*VGELSLR 415
H + +S W WR++ V L W + H +GDG + FWTD + E +
Sbjct: 1431 GDVHDRNWTVAKSNWSSTWRSVGVGLRDVIWREQH--WVIGDGRQIRFWTDRWLSETPIA 1488
Query: 416 D 416
D
Sbjct: 1489 D 1489
>At2g17910 putative non-LTR retroelement reverse transcriptase
Length = 1344
Score = 218 bits (556), Expect = 7e-57
Identities = 145/414 (35%), Positives = 209/414 (50%), Gaps = 23/414 (5%)
Query: 3 QSAFIKGRQILDGILVANEVVDDARKR---KKDLLLFKVDFEKAYDSVD*NYLEEVMVKM 59
QSAF+ R I D ILVA+E++ R K+ + FK D KAYD V+ +LE +M +
Sbjct: 515 QSAFVPQRLISDNILVAHEMIHSLRTNDRISKEHMAFKTDMSKAYDRVEWPFLETMMTAL 574
Query: 60 GFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEG-FHVLM 118
GF W WI CV + + SVL+NG P RG+RQGDPLSP LF+L E H+L
Sbjct: 575 GFNNKWISWIMNCVTSVSYSVLINGQPYGHIIPTRGIRQGDPLSPALFVLCTEALIHILN 634
Query: 119 EALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKV 178
+A + G + + V V HL FADDT+++C + L L + +LSG +
Sbjct: 635 KAEQAGKI-TGIQF-QDKKVSVNHLLFADDTLLMCKATKQECEELMQCLSQYGQLSGQMI 692
Query: 179 NFSKS-LFVGVNVH---GSWLAEAATV-LNCNAGSIPFLYLGLPIGGNASRMVFWKPLIN 233
N +KS + G NV W+ + + L G YLGLP + S+ + +
Sbjct: 693 NLNKSAITFGKNVDIQIKDWIKSRSGISLEGGTGK----YLGLPECLSGSKRDLFGFIKE 748
Query: 234 RINSRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGG 293
++ SRL+ W +K LS GG+ VLLKS+ +LPVY +S FK P + + +++ +F
Sbjct: 749 KLQSRLTGWYAKTLSQGGKEVLLKSIALALPVYVMSCFKLPKNLCQKLTTVMMDF----- 803
Query: 294 WGGGSADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVL 353
W RKI W+ W + ++ GG + ++ FN ALL K WRVL E+ SL+ RV
Sbjct: 804 WWNSMQQKRKIHWLSWQRLTLPKDQGGFGFKDLQCFNQALLAKQAWRVLQEKGSLFSRVF 863
Query: 354 AVRNRVEGGHL-CSGGRNESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTD 406
R L + G S WR+I L + +G+G WTD
Sbjct: 864 QSRYFSNSDFLSATRGSRPSYAWRSI--LFGRELLMQGLRTVIGNGQKTFVWTD 915
>At2g41580 putative non-LTR retroelement reverse transcriptase
Length = 1094
Score = 211 bits (536), Expect = 1e-54
Identities = 136/417 (32%), Positives = 218/417 (51%), Gaps = 29/417 (6%)
Query: 3 QSAFIKGRQILDGILVANEVVDDAR---KRKKDLLLFKVDFEKAYDSVD*NYLEEVMVKM 59
QSAF+ R + D I++A+E+V + R K KD ++FK D KAYD V+ +L+ +++ +
Sbjct: 266 QSAFVAERLVSDNIILAHEIVHNLRTNEKISKDFMVFKTDMSKAYDRVEWPFLKGILLAL 325
Query: 60 GFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEG-FHVLM 118
GF + W W+ CV + + SVL+NG P + HRGLRQGDPLSPFLF+L E H+L
Sbjct: 326 GFNSTWINWMMACVSSVSYSVLINGQPFGHITPHRGLRQGDPLSPFLFVLCTEALIHILN 385
Query: 119 EALVVNNL----FNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELS 174
+A + + FNG V HL FADDT+++C S + L + +S
Sbjct: 386 QAEKIGKISGIQFNG------TGPSVNHLLFADDTLLICKASQLECAEIMHCLSQYGHIS 439
Query: 175 GLKVNFSKS-LFVGVNVH---GSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKP 230
G +N KS + G V+ W+ + + G+ YLGLP S+ V +
Sbjct: 440 GQMINSEKSAITFGAKVNEETKQWIMNRSGI-QTEGGT--GKYLGLPECFQGSKQVLFGF 496
Query: 231 LINRINSRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFF 290
+ ++ SRLS W +K LS GG+ +LLKS+ + PVYA++ F+ + + + S++ +F
Sbjct: 497 IKEKLQSRLSGWYAKTLSQGGKDILLKSIAMAFPVYAMTCFRLSKTLCTKLTSVMMDF-- 554
Query: 291 WGGWGGGSADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWF 350
W D +KI W+ + + +GG + ++ FN ALL K R+ T+ DSL
Sbjct: 555 ---WWNSVQDKKKIHWIGAQKLMLPKFLGGFGFKDLQCFNQALLAKQASRLHTDSDSLLS 611
Query: 351 RVLAVRNRVEGGHL-CSGGRNESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTD 406
++L R + L + G S W++I L+ + + + +G+G + W D
Sbjct: 612 QILKSRYYMNSDFLSATKGTRPSYAWQSI--LYGRELLVSGLKKIIGNGENTYVWMD 666
>At3g44650 putative protein
Length = 762
Score = 210 bits (535), Expect = 2e-54
Identities = 139/435 (31%), Positives = 219/435 (49%), Gaps = 25/435 (5%)
Query: 3 QSAFIKGRQILDGILVANEVVDDARKRK-KDLLLFKVDFEKAYDSVD*NYLEEVMVKMGF 61
QS+F+K R +++ +L+A ++V D K + K+D KA DSV ++L + M F
Sbjct: 115 QSSFVKDRLLIENVLLATDLVKDYHKDSISERCAIKIDISKASDSVQWSFLINTLTAMHF 174
Query: 62 PTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEAL 121
P ++ WI+ C+ T + SV VNG F RGLRQG LSP+LF++ + L++ +
Sbjct: 175 PEMFIHWIRLCITTPSFSVQVNGELAGFFQSSRGLRQGCALSPYLFVICMDVLSKLLDKV 234
Query: 122 VVNNLFNGYKVGSH---EMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKV 178
V ++G H + +G+THL FADD +IL D +I + + LF + SGLK+
Sbjct: 235 VGIG-----RIGYHPHCKRMGLTHLSFADDLMILTDGQCRSIEGIIEVFDLFSKWSGLKI 289
Query: 179 NFSKSLFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSR 238
+ KS + + A+ T G +P YLGLP+ V + PLI +I R
Sbjct: 290 SMEKSTIFSAGLSSTSRAQLHTHFPFEVGELPIRYLGLPLVTKRLSSVDYAPLIEQIRKR 349
Query: 239 LSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGGGS 298
+ SW S+ LS GR L+ S++ S + LS F+ P + IE + ++F W G +
Sbjct: 350 IGSWSSRFLSFAGRFNLISSIIWSSCNFWLSAFQLPRACIQEIEKLCSSFL----WSGTN 405
Query: 299 ADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFR-VLAVRN 357
+ +K + WN VC+ + GGL +R +KE N K WR+++ DSLW + V
Sbjct: 406 LNSKK-AKISWNQVCKPKSEGGLGLRSLKEANDVCCLKLVWRIISHGDSLWVKWVEHNLL 464
Query: 358 RVEGGHLCSGGRNESMW-WRNIVALHSEGWFQNHVSRSLGDGSSVLFWTDV*VGELSLRD 416
+ E + N W W+ I L G + +G+G S FW D + SL
Sbjct: 465 KREIFWIVKENANLGSWIWKKI--LKYRGVAKRFCKAEVGNGESTSFWFD----DWSL-- 516
Query: 417 RFSRLYDLSVLKGGV 431
RL D++ ++G +
Sbjct: 517 -LGRLIDVAGIRGTI 530
>At2g17610 putative non-LTR retroelement reverse transcriptase
Length = 773
Score = 210 bits (534), Expect = 2e-54
Identities = 123/399 (30%), Positives = 212/399 (52%), Gaps = 12/399 (3%)
Query: 12 ILDGILVANEVVDDARKRK--KDLLLFKVDFEKAYDSVD*NYLEEVMVKMGFPTLWRKWI 69
I D IL+A+E++ +K + + K+D KA+D ++ ++E +M +MGF W WI
Sbjct: 2 ITDNILIAHELIHSLHTKKLVQPFVATKLDITKAFDKIEWGFIEAIMKQMGFSEKWCNWI 61
Query: 70 KKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEALVVNNLFNG 129
C+ T T S+L+NG P RG+RQGDP+SP+L+LL EG L++A + +G
Sbjct: 62 MTCITTTTYSILINGQPVRRIIPKRGIRQGDPISPYLYLLCTEGLSALIQASIKAKQLHG 121
Query: 130 YKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVNFSKS-LFVGV 188
+K S ++HL FA D+++ C + L +L L+++ SG VNF KS + G
Sbjct: 122 FK-ASRNGPAISHLLFAHDSLVFCKATLEECMTLVNVLKLYEKASGQAVNFQKSAILFGK 180
Query: 189 NVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSRLSSWKSKHLS 248
+ + + +L YLGLP ++ + + ++ ++ +W +K LS
Sbjct: 181 GLDFRTSEQLSQLLGIYKTEGFGRYLGLPEFVGRNKTNAFSFIAQTMDQKMDNWYNKLLS 240
Query: 249 LGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGGGSADHRKIIWVD 308
G+ VL+KS+++++P Y++S F P ++ I S + +FW W H KI WV
Sbjct: 241 PAGKEVLIKSIVTAIPTYSMSCFLLPMRLIHQITSAMR--WFW--WSNTKVKH-KIPWVA 295
Query: 309 WNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVRNRVEGGHLCSGG 368
W+ + +++GGL +R +K+FN+ALL K WR+L + SL RV + + L +
Sbjct: 296 WSKLNDPKKMGGLAIRDLKDFNIALLAKQSWRILQQPFSLMARVFKAKYFPKERLLDAKA 355
Query: 369 RNESMW-WRNIVALHSEGWFQNHVSRSLGDGSSVLFWTD 406
++S + W++I LH + G+G+++ W D
Sbjct: 356 TSQSSYAWKSI--LHGTKLISRGLKYIAGNGNNIQLWKD 392
>At4g09710 RNA-directed DNA polymerase -like protein
Length = 1274
Score = 207 bits (528), Expect = 1e-53
Identities = 142/413 (34%), Positives = 204/413 (49%), Gaps = 27/413 (6%)
Query: 3 QSAFIKGRQILDGILVANEVVDDAR---KRKKDLLLFKVDFEKAYDSVD*NYLEEVMVKM 59
QSAF+ GR I D +L+ +E++ R +K + K D KAYD + N+L+EV++++
Sbjct: 478 QSAFVPGRAIADNVLITHEILHFLRVSGAKKYCSMAIKTDMSKAYDRIKWNFLQEVLMRL 537
Query: 60 GFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLME 119
GF W +W+ +CV T + S L+NGSP RGLRQGDPLSP+LF+L E L
Sbjct: 538 GFHDKWIRWVMQCVCTVSYSFLINGSPQGSVVPSRGLRQGDPLSPYLFILCTEVLSGLCR 597
Query: 120 ALVVNNLFNGYKV--GSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLK 177
+ G +V GS + V HL FADDT+ C + AL IL ++ SG
Sbjct: 598 KAQEKGVMVGIRVARGSPQ---VNHLLFADDTMFFCKTNPTCCGALSNILKKYELASGQS 654
Query: 178 VNFSKS--LFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRI 235
+N +KS F ++ N G I YLGLP + + +++RI
Sbjct: 655 INLAKSAITFSSKTPQDIKRRVKLSLRIDNEGGIG-KYLGLPEHFGRRKRDIFSSIVDRI 713
Query: 236 NSRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWG 295
R SW + LS G+ +LLK+VLSS+P YA+ FK P+ + I+S+L F W
Sbjct: 714 RQRSHSWSIRFLSSAGKQILLKAVLSSMPSYAMMCFKLPASLCKQIQSVLTRF-----WW 768
Query: 296 GGSADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAV 355
D RK+ WV W+ + GGL R I+ K WR+L E SL RVL
Sbjct: 769 DSKPDKRKMAWVSWDKLTLPINEGGLGFREIE-------AKLSWRILKEPHSLLSRVLLG 821
Query: 356 RNRVEGGHL-CSGGRN-ESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTD 406
+ + CS + S WR I+A + + S+G G S+ WT+
Sbjct: 822 KYCNTSSFMDCSASPSFASHGWRGILA--GRDLLRKGLGWSIGQGDSINVWTE 872
>At2g16680 putative non-LTR retroelement reverse transcriptase
Length = 1319
Score = 206 bits (525), Expect = 3e-53
Identities = 136/418 (32%), Positives = 209/418 (49%), Gaps = 31/418 (7%)
Query: 3 QSAFIKGRQILDGILVANEVVDDAR---KRKKDLLLFKVDFEKAYDSVD*NYLEEVMVKM 59
QSAF R I D IL+A+E+V R K K+ ++FK D KAYD V+ ++L+E++V +
Sbjct: 489 QSAFFAERLISDNILIAHEIVHSLRTNDKISKEFMVFKTDMSKAYDRVEWSFLQEILVAL 548
Query: 60 GFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEG-FHVLM 118
GF W WI CV + T SVL+NG + RG+RQGDP+SPFLF+L E H+L
Sbjct: 549 GFNDKWISWIMGCVTSVTYSVLINGQHFGHITPERGIRQGDPISPFLFVLCTEALIHILQ 608
Query: 119 EALVVNNLFNGYKVGSHEMVG----VTHLQFADDTIILCDKSWANIRALRAILLLFQELS 174
+A N KV + G V HL F DDT ++C + ++ + L + +S
Sbjct: 609 QA------ENSKKVSGIQFNGSGPSVNHLLFVDDTQLVCRATKSDCEQMMLCLSQYGHIS 662
Query: 175 GLKVNFSK-SLFVGVNVHGS---WLAEAATV-LNCNAGSIPFLYLGLPIGGNASRMVFWK 229
G +N K S+ GV V W+ + + L G YLGLP + S+ +
Sbjct: 663 GQLINVEKSSITFGVKVDEDTKRWIKNRSGIHLEGGTGK----YLGLPENLSGSKQDLFG 718
Query: 230 PLINRINSRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFF 289
+ ++ S LS W K LS GG+ +LLKS+ +LPVY ++ F+ P G+ + + S++ +F
Sbjct: 719 YIKEKLQSHLSGWYDKTLSQGGKEILLKSIALALPVYIMTCFRLPKGLCTKLTSVMMDF- 777
Query: 290 FWGGWGGGSADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLW 349
W KI W+ + + +GG + ++ FN ALL K WR+ ++ S+
Sbjct: 778 ----WWNSMEFSNKIHWIGGKKLTLPKSLGGFGFKDLQCFNQALLAKQAWRLFSDSKSIV 833
Query: 350 FRVLAVRNRVEGGHL-CSGGRNESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTD 406
++ R + L G S WR+I L+ + R +G+G W D
Sbjct: 834 SQIFKSRYFMNTDFLNARQGTRPSYTWRSI--LYGRELLNGGLKRLIGNGEQTNVWID 889
>At2g31080 putative non-LTR retroelement reverse transcriptase
Length = 1231
Score = 206 bits (524), Expect = 3e-53
Identities = 138/416 (33%), Positives = 211/416 (50%), Gaps = 25/416 (6%)
Query: 3 QSAFIKGRQILDGILVANEVVDDARKRK--KDLLLFKVDFEKAYDSVD*NYLEEVMVKMG 60
Q++FI GR +D I++ E V R++K K +L K+D EKAYD V ++L+E + G
Sbjct: 401 QASFIPGRLSIDNIVLVQEAVHSMRRKKGRKGWMLLKLDLEKAYDRVRWDFLQETLEAAG 460
Query: 61 FPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEA 120
W I V + SVL NG T F RGLRQGDPLSP+LF+L E L+EA
Sbjct: 461 LSEGWTSRIMAGVTDPSMSVLWNGERTDSFVPARGLRQGDPLSPYLFVLCLERLCHLIEA 520
Query: 121 LVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVNF 180
V + V S ++H+ FADD I+ + S A IR +R +L F E SG KV+
Sbjct: 521 SVGKREWKPIAV-SCGGSKLSHVCFADDLILFAEASVAQIRIIRRVLERFCEASGQKVSL 579
Query: 181 SKS-LFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSRL 239
KS +F NV + YLG+PI + ++ R+++RL
Sbjct: 580 EKSKIFFSHNVSREMEQLISEESGIGCTKELGKYLGMPILQKRMNKETFGEVLERVSARL 639
Query: 240 SSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGGGSA 299
+ WK + LSL GR+ L K+VLSS+PV+ +S P + +++ F + G +
Sbjct: 640 AGWKGRSLSLAGRITLTKAVLSSIPVHVMSAILLPVSTLDTLDRYSRTFLW-----GSTM 694
Query: 300 DHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVRNRV 359
+ +K + W +C+ + GG+ +R ++ N AL+ K WR+L +++SLW RV+ + +V
Sbjct: 695 EKKKQHLLSWRKICKPKAEGGIGLRSARDMNKALVAKVGWRLLQDKESLWARVVRKKYKV 754
Query: 360 EGGHLCSGGRNESMW---WRNI------VALHSEGWFQNHVSRSLGDGSSVLFWTD 406
G S + + W WR++ V + GW GDG ++ FW D
Sbjct: 755 GGVQDTSWLKPQPRWSSTWRSVAVGLREVVVKGVGWVP-------GDGCTIRFWLD 803
>At2g11240 pseudogene
Length = 1044
Score = 206 bits (524), Expect = 3e-53
Identities = 131/409 (32%), Positives = 201/409 (49%), Gaps = 13/409 (3%)
Query: 3 QSAFIKGRQILDGILVANEVVDDARK---RKKDLLLFKVDFEKAYDSVD*NYLEEVMVKM 59
QSAF+ R D +L+ +E + + K+ + K + KAYD ++ ++++ VM +M
Sbjct: 363 QSAFVPKRASNDNVLITHEALHYLKSLGAEKRCFMAVKTNMSKAYDRIEWDFIKLVMQEM 422
Query: 60 GFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLME 119
GF W WI +C+ T + S L+NGS + RGLRQGDPLSPFLF++ +E L
Sbjct: 423 GFHQTWISWILQCITTVSYSFLLNGSAQGAVTPERGLRQGDPLSPFLFIICSEVLSGLCR 482
Query: 120 ALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVN 179
++ G +V S V HL FADDTI C + + IL ++E SG +N
Sbjct: 483 KAQLDGSLLGLRV-SKGNPRVNHLLFADDTIFFCRSDLKSCKTFLCILKKYEEASGQMIN 541
Query: 180 FSKSLFVGVNVHGSWL-AEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSR 238
SKS + EA +L YLGLP + + +++RI R
Sbjct: 542 KSKSAITFSRKTPDHIKTEAQQILGIQLVGGLGKYLGLPKMFGRKKRDLFNQIVDRIRQR 601
Query: 239 LSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGGGS 298
SW S+ LS G+ +LKSVL+S+P Y +S FK + I+S L +F W S
Sbjct: 602 SLSWSSRFLSTAGKTTMLKSVLASMPTYTMSCFKLLVSLCKRIQSALTHF-----WWDSS 656
Query: 299 ADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVRNR 358
AD +K+ W+ W+ + ++++ GGL + I FN ALL K WR++ + R+L +
Sbjct: 657 ADKKKMCWIAWSKMAKNKKEGGLGFKDITNFNDALLAKLSWRIVQSPSCVLVRILLGKYC 716
Query: 359 VEGGHL-CSGGRNESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTD 406
L CS S WR I + ++ + + +G G L W +
Sbjct: 717 RTSSFLDCSVTAASSHGWRGICT--GKDLIKSQLGKVIGSGLDTLVWNE 763
>At2g01840 putative non-LTR retroelement reverse transcriptase
Length = 1715
Score = 203 bits (517), Expect = 2e-52
Identities = 130/413 (31%), Positives = 209/413 (50%), Gaps = 35/413 (8%)
Query: 1 DVQSAFIKGRQILDGILVANEVVDDARKRKK---DLLLFKVDFEKAYDSVD*NYLEEVMV 57
D Q+AF+ G+ I D +LVA+E++ + R++ + K D KAYD V+ N+LE+VM+
Sbjct: 897 DSQAAFVPGQNISDNVLVAHELLHSLKSRRECQSGYVAVKTDISKAYDRVEWNFLEKVMI 956
Query: 58 KMGFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVL 117
++GF W KWI CV + + VL+NGSP + RG+RQGDPLSP+LFL AE +
Sbjct: 957 QLGFAPRWVKWIMTCVTSVSYEVLINGSPYGKIFPSRGIRQGDPLSPYLFLFCAEVLSNM 1016
Query: 118 MEALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLK 177
+ VN +G K+ + + + ++HL FADD++ C S NI L I ++E SG K
Sbjct: 1017 LRKAEVNKQIHGMKI-TKDCLAISHLLFADDSLFFCRASNQNIEQLALIFKKYEEASGQK 1075
Query: 178 VNFSK-SLFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRIN 236
+N++K S+ G + +L + YLGLP ++ ++ ++ ++
Sbjct: 1076 INYAKSSIIFGQKIPTMRRQRLHRLLGIDNVRGGGKYLGLPEQLGRRKVELFEYIVTKVK 1135
Query: 237 SRLSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGG 296
R W +LS G+ +++K++ +LPVY+++ F P+ I + I S++ F+WG
Sbjct: 1136 ERTEGWAYNYLSPAGKEIVIKAIAMALPVYSMNCFLLPTLICNEINSLI-TAFWWG---- 1190
Query: 297 GSADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVR 356
+ G L + + +FN ALL K WR+LT SL R+
Sbjct: 1191 ------------------KENEGDLGFKDLHQFNRALLAKQAWRILTNPQSLLARLYKGL 1232
Query: 357 NRVEGGHL-CSGGRNESMWWRNIVALHSEG--WFQNHVSRSLGDGSSVLFWTD 406
+L + G + S W +I EG Q + LGDG + W D
Sbjct: 1233 YYPNTTYLRANKGGHASYGWNSI----QEGKLLLQQGLRVRLGDGQTTKIWED 1281
>At2g28980 putative non-LTR retroelement reverse transcriptase
Length = 1529
Score = 201 bits (510), Expect = 1e-51
Identities = 131/408 (32%), Positives = 206/408 (50%), Gaps = 14/408 (3%)
Query: 3 QSAFIKGRQILDGILVANEVVDDARKRK-KDLLLFKVDFEKAYDSVD*NYLEEVMVKMGF 61
QSAF+K R +++ +L+A E+V D K K+D KA+DSV +L + + F
Sbjct: 859 QSAFVKERLLMENVLLATELVKDYHKESVTPRCAMKIDISKAFDSVQWQFLLNTLEALNF 918
Query: 62 PTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGF-HVLMEA 120
P +R WIK C+ TAT SV VNG F RGLRQG LSP+LF++ H++ EA
Sbjct: 919 PETFRHWIKLCISTATFSVQVNGELAGFFGSSRGLRQGCALSPYLFVICMNVLSHMIDEA 978
Query: 121 LVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVNF 180
V N+ GY E +G+THL FADD ++ D +I + + F SGL+++
Sbjct: 979 AVHRNI--GYHPKC-EKIGLTHLCFADDLMVFVDGHQWSIEGVINVFKEFAGRSGLQISL 1035
Query: 181 SKSLFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSRLS 240
KS V S + + G +P YLGLP+ + PLI + +++S
Sbjct: 1036 EKSTIYLAGVSASDRVQTLSSFPFANGQLPVRYLGLPLLTKQMTTADYSPLIEAVKTKIS 1095
Query: 241 SWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGGGSAD 300
SW ++ LS GRL LL SV+ S+ + +S ++ P+G + IE + + F W G +
Sbjct: 1096 SWTARSLSYAGRLALLNSVIVSIANFWMSAYRLPAGCIREIEKLCSAFL----WSGPVLN 1151
Query: 301 HRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVRNRVE 360
+K + W+S+C+ ++ GGL ++ + E N K WR+L+ + SLW + +
Sbjct: 1152 PKK-AKIAWSSICQPKKEGGLGIKSLAEANKVSCLKLIWRLLSTQPSLWVTWIWTFIIRK 1210
Query: 361 GGHLCSGGRNE--SMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTD 406
G + R+ S W+ ++ + V + +GSS FW D
Sbjct: 1211 GTFWSANERSSLGSWMWKKLLKYRELAKSMHKV--EVRNGSSTSFWYD 1256
>At2g05200 putative non-LTR retroelement reverse transcriptase
Length = 1229
Score = 200 bits (508), Expect = 2e-51
Identities = 136/414 (32%), Positives = 201/414 (47%), Gaps = 23/414 (5%)
Query: 3 QSAFIKGRQILDGILVANEVVDDARK---RKKDLLLFKVDFEKAYDSVD*NYLEEVMVKM 59
QSAF+ GR I D +L+ +EV+ R +K + K D KAYD V+ ++L++V+ +
Sbjct: 432 QSAFVPGRVISDNVLITHEVLHFLRTSSAKKHCSMAVKTDMSKAYDRVEWDFLKKVLQRF 491
Query: 60 GFPTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLME 119
GF ++W W+ +CV + + S L+NG+P + RGLRQGDPLSP LF+L E L
Sbjct: 492 GFHSIWIDWVLECVTSVSYSFLINGTPQGKVVPTRGLRQGDPLSPCLFILCTEVLSGLCT 551
Query: 120 ALVVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVN 179
G +V S V HL FADDT+ + L IL + + SG +N
Sbjct: 552 RAQRLRQLPGVRV-SINGPRVNHLLFADDTMFFSKSDPESCNKLSEILSRYGKASGQSIN 610
Query: 180 FSK-SLFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSR 238
F K S+ S + +L YLGLP + + +I++I +
Sbjct: 611 FHKSSVTFSSKTPRSVKGQVKRILKIRKEGGTGKYLGLPEHFGRRKRDIFGAIIDKIRQK 670
Query: 239 LSSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGGGS 298
SW S+ LS G+ V+LK+VL+S+P+Y++S FK PS + I+S+L F W
Sbjct: 671 SHSWASRFLSQAGKQVMLKAVLASMPLYSMSCFKLPSALCRKIQSLLTRF-----WWDTK 725
Query: 299 ADHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVRNR 358
D RK WV W+ + + GGL R I+ N +LL K WR+L +SL R+L +
Sbjct: 726 PDVRKTSWVAWSKLTNPKNAGGLGFRDIERCNDSLLAKLGWRLLNSPESLLSRILLGKYC 785
Query: 359 VEGGHL-CSGGRNESMWWRNIVA-----LHSEGWFQNHVSRSLGDGSSVLFWTD 406
+ C S WR+I+A GW + +G V W D
Sbjct: 786 HSSSFMECKLPSQPSHGWRSIIAGREILKEGLGWL-------ITNGEKVSIWND 832
>At1g25430 hypothetical protein
Length = 1213
Score = 199 bits (506), Expect = 4e-51
Identities = 128/408 (31%), Positives = 207/408 (50%), Gaps = 16/408 (3%)
Query: 3 QSAFIKGRQILDGILVANEVVDDAR-KRKKDLLLFKVDFEKAYDSVD*NYLEEVMVKMGF 61
QSAF+ GR + + +L+A ++V + KVD +KA+DSV ++ + +
Sbjct: 556 QSAFLPGRSLAENVLLATDLVHGYNWSNISPRGMLKVDLKKAFDSVRWEFVIAALRALAI 615
Query: 62 PTLWRKWIKKCVGTATASVLVNGSPTYEFSLHRGLRQGDPLSPFLFLLAAEGFHVLMEAL 121
P + WI +C+ T T +V +NG F +GLRQGDPLSP+LF+LA E F L+ +
Sbjct: 616 PEKFINWISQCISTPTFTVSINGGNGGFFKSTKGLRQGDPLSPYLFVLAMEAFSNLLHSR 675
Query: 122 VVNNLFNGYKVGSHEMVGVTHLQFADDTIILCDKSWANIRALRAILLLFQELSGLKVNFS 181
+ L + + S+ + ++HL FADD +I D ++ + L F SGLKVN
Sbjct: 676 YESGLIHYHPKASN--LSISHLMFADDVMIFFDGGSFSLHGICETLDDFASWSGLKVNKD 733
Query: 182 KS--LFVGVNVHGSWLAEAATVLNCNAGSIPFLYLGLPIGGNASRMVFWKPLINRINSRL 239
KS G+N S A G++P YLGLP+ R+ ++PL+ +I +R
Sbjct: 734 KSHLYLAGLNQLES---NANAAYGFPIGTLPIRYLGLPLMNRKLRIAEYEPLLEKITARF 790
Query: 240 SSWKSKHLSLGGRLVLLKSVLSSLPVYALSFFKAPSGIVSSIESILNNFFFWGGWGGGSA 299
SW +K LS GR+ L+ SV+ + +S F P G + IES+ + F + G+
Sbjct: 791 RSWVNKCLSFAGRIQLISSVIFGSINFWMSTFLLPKGCIKRIESLCSRFLW-----SGNI 845
Query: 300 DHRKIIWVDWNSVCRSQEVGGLEVRRIKEFNLALLGKWCWRVLTERDSLWFRVLAVRNRV 359
+ K I V W ++C + GGL +RR+ E+N L + WR+ +DSLW + +
Sbjct: 846 EQAKGIKVSWAALCLPKSEGGLGLRRLLEWNKTLSMRLIWRLFVAKDSLWADWQHLHHLS 905
Query: 360 EGGH-LCSGGRNESMWWRNIVALHSEGWFQNHVSRSLGDGSSVLFWTD 406
G GG+++S W+ +++L + +G+G +W D
Sbjct: 906 RGSFWAVEGGQSDSWTWKRLLSLRPLA--HQFLVCKVGNGLKADYWYD 951
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.340 0.151 0.516
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,481,962
Number of Sequences: 26719
Number of extensions: 608595
Number of successful extensions: 2126
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 64
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 1852
Number of HSP's gapped (non-prelim): 99
length of query: 679
length of database: 11,318,596
effective HSP length: 106
effective length of query: 573
effective length of database: 8,486,382
effective search space: 4862696886
effective search space used: 4862696886
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC124215.6


