

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124215.2 + phase: 0
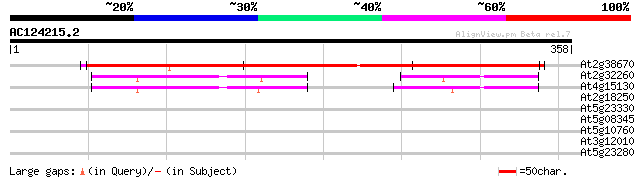
(358 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g38670 putative phospholipid cytidylyltransferase 514 e-146
At2g32260 putative phospholipid cytidylyltransferase 110 9e-25
At4g15130 CTP:phosphorylcholine cytidylyltransferase (AtCCT2) 108 6e-24
At2g18250 hypothetical protein 35 0.083
At5g23330 unknown protein 29 4.5
At5g08345 unknown protein 29 4.5
At5g10760 nucleoid DNA-binding protein cnd41 - like protein 28 5.9
At3g12010 unknown protein 28 5.9
At5g23280 unknown protein 28 7.7
>At2g38670 putative phospholipid cytidylyltransferase
Length = 421
Score = 514 bits (1323), Expect = e-146
Identities = 248/289 (85%), Positives = 268/289 (91%), Gaps = 1/289 (0%)
Query: 50 RNKKPIRVYMDGCFDMMHYGHCNALRQARALGDQLIVGVVSDDEIIANKGPPVTPLHERL 109
R +KP+RVYMDGCFDMMHYGHCNALRQARALGDQL+VGVVSD+EIIANKGPPVTPLHER+
Sbjct: 50 RKRKPVRVYMDGCFDMMHYGHCNALRQARALGDQLVVGVVSDEEIIANKGPPVTPLHERM 109
Query: 110 IMVNAVKWVDEVIPEAPYAITEEFMKKLFDEYNIDYIIHGDDPCVLPDGTDAYAHAKKAG 169
MV AVKWVDEVI +APYAITE+FMKKLFDEY IDYIIHGDDPCVLPDGTDAYA AKKAG
Sbjct: 110 TMVKAVKWVDEVISDAPYAITEDFMKKLFDEYQIDYIIHGDDPCVLPDGTDAYALAKKAG 169
Query: 170 RYKQIKRTEGVSSTDIVGRMLLCVRERNIADTHNHSSLQRQFSNGRGQQKFEDGGVVASG 229
RYKQIKRTEGVSSTDIVGRMLLCVRER+I+DTH+ SSLQRQFS+G KFED G ++G
Sbjct: 170 RYKQIKRTEGVSSTDIVGRMLLCVRERSISDTHSRSSLQRQFSHGHSSPKFED-GASSAG 228
Query: 230 TRVSHFLPTSRRIVQFSNGRSPGPDARIVYIDGAFDLFHAGHVEILRLARDRGDFLLVGI 289
TRVSHFLPTSRRIVQFSNG+ PGPDARI+YIDGAFDLFHAGHVEILR AR+ GDFLLVGI
Sbjct: 229 TRVSHFLPTSRRIVQFSNGKGPGPDARIIYIDGAFDLFHAGHVEILRRARELGDFLLVGI 288
Query: 290 HTDQTVSATRGLHRPIMSLHERSLSVLACRYVDEVIIGAPWEVSKDMVS 338
H DQTVSA RG HRPIM+LHERSLSVLACRYVDEVIIGAPWEVS+D ++
Sbjct: 289 HNDQTVSAKRGAHRPIMNLHERSLSVLACRYVDEVIIGAPWEVSRDTIT 337
Score = 77.4 bits (189), Expect = 1e-14
Identities = 40/106 (37%), Positives = 62/106 (57%), Gaps = 6/106 (5%)
Query: 46 NGRRRNKKPIRVYMDGCFDMMHYGHCNALRQARALGDQLIVGVVSDDEIIANKGP--PVT 103
NG+ +Y+DG FD+ H GH LR+AR LGD L+VG+ +D + A +G P+
Sbjct: 246 NGKGPGPDARIIYIDGAFDLFHAGHVEILRRARELGDFLLVGIHNDQTVSAKRGAHRPIM 305
Query: 104 PLHERLIMVNAVKWVDEVIPEAPYAITEEFMKKLFDEYNIDYIIHG 149
LHER + V A ++VDEVI AP+ ++ + ++I ++HG
Sbjct: 306 NLHERSLSVLACRYVDEVIIGAPWEVSRD----TITTFDISLVVHG 347
Score = 75.5 bits (184), Expect = 4e-14
Identities = 37/84 (44%), Positives = 54/84 (64%), Gaps = 2/84 (2%)
Query: 258 VYIDGAFDLFHAGHVEILRLARDRGDFLLVGIHTDQTVSATRGLHRPIMSLHERSLSVLA 317
VY+DG FD+ H GH LR AR GD L+VG+ +D+ + A +G P+ LHER V A
Sbjct: 57 VYMDGCFDMMHYGHCNALRQARALGDQLVVGVVSDEEIIANKG--PPVTPLHERMTMVKA 114
Query: 318 CRYVDEVIIGAPWEVSKDMVSLIF 341
++VDEVI AP+ +++D + +F
Sbjct: 115 VKWVDEVISDAPYAITEDFMKKLF 138
>At2g32260 putative phospholipid cytidylyltransferase
Length = 332
Score = 110 bits (276), Expect = 9e-25
Identities = 56/143 (39%), Positives = 84/143 (58%), Gaps = 9/143 (6%)
Query: 53 KPIRVYMDGCFDMMHYGHCNALRQARAL---GDQLIVGVVSDDEIIANKGPPVTPLHERL 109
+P+RVY DG +D+ H+GH +L QA+ L+VG +D+ KG V ER
Sbjct: 33 RPVRVYADGIYDLFHFGHARSLEQAKLAFPNNTYLLVGCCNDETTHKYKGRTVMTAEERY 92
Query: 110 IMVNAVKWVDEVIPEAPYAITEEFMKKLFDEYNIDYIIHGDDPCVLPDGT--DAYAHAKK 167
+ KWVDEVIP+AP+ + +EF+ D++ IDY+ H P G D Y KK
Sbjct: 93 ESLRHCKWVDEVIPDAPWVVNQEFL----DKHQIDYVAHDSLPYADSSGAGKDVYEFVKK 148
Query: 168 AGRYKQIKRTEGVSSTDIVGRML 190
GR+K+ +RTEG+S++DI+ R++
Sbjct: 149 VGRFKETQRTEGISTSDIIMRIV 171
Score = 64.3 bits (155), Expect = 1e-10
Identities = 38/92 (41%), Positives = 53/92 (57%), Gaps = 6/92 (6%)
Query: 250 SPGPDARI-VYIDGAFDLFHAGHVEIL---RLARDRGDFLLVGIHTDQTVSATRGLHRPI 305
SP D + VY DG +DLFH GH L +LA +LLVG D+T +G R +
Sbjct: 28 SPPTDRPVRVYADGIYDLFHFGHARSLEQAKLAFPNNTYLLVGCCNDETTHKYKG--RTV 85
Query: 306 MSLHERSLSVLACRYVDEVIIGAPWEVSKDMV 337
M+ ER S+ C++VDEVI APW V+++ +
Sbjct: 86 MTAEERYESLRHCKWVDEVIPDAPWVVNQEFL 117
>At4g15130 CTP:phosphorylcholine cytidylyltransferase (AtCCT2)
Length = 300
Score = 108 bits (269), Expect = 6e-24
Identities = 57/142 (40%), Positives = 83/142 (58%), Gaps = 8/142 (5%)
Query: 53 KPIRVYMDGCFDMMHYGHCNALRQARAL--GDQLIVGVVSDDEIIANKGPPVTPLHERLI 110
+P+RVY DG FD+ H+GH A+ QA+ L+VG +D+ KG V ER
Sbjct: 19 RPVRVYADGIFDLFHFGHARAIEQAKKSFPNTYLLVGCCNDEITNKFKGKTVMTESERYE 78
Query: 111 MVNAVKWVDEVIPEAPYAITEEFMKKLFDEYNIDYIIHGDDPCVLPD--GTDAYAHAKKA 168
+ KWVDEVIP+AP+ +T EF+ D++ IDY+ H P G D Y K
Sbjct: 79 SLRHCKWVDEVIPDAPWVLTTEFL----DKHKIDYVAHDALPYADTSGAGNDVYEFVKSI 134
Query: 169 GRYKQIKRTEGVSSTDIVGRML 190
G++K+ KRTEG+S++DI+ R++
Sbjct: 135 GKFKETKRTEGISTSDIIMRIV 156
Score = 58.9 bits (141), Expect = 4e-09
Identities = 35/95 (36%), Positives = 52/95 (53%), Gaps = 5/95 (5%)
Query: 246 SNGRSPGPDARI-VYIDGAFDLFHAGHVEILRLARDR--GDFLLVGIHTDQTVSATRGLH 302
S G S D + VY DG FDLFH GH + A+ +LLVG D+ + +G
Sbjct: 10 SGGDSSSSDRPVRVYADGIFDLFHFGHARAIEQAKKSFPNTYLLVGCCNDEITNKFKG-- 67
Query: 303 RPIMSLHERSLSVLACRYVDEVIIGAPWEVSKDMV 337
+ +M+ ER S+ C++VDEVI APW ++ + +
Sbjct: 68 KTVMTESERYESLRHCKWVDEVIPDAPWVLTTEFL 102
>At2g18250 hypothetical protein
Length = 176
Score = 34.7 bits (78), Expect = 0.083
Identities = 32/107 (29%), Positives = 48/107 (43%), Gaps = 10/107 (9%)
Query: 57 VYMDGCFDMMHYGHCNALRQARALGDQLIVGVVSDDEIIANK--GPPVTPLHERLIMVNA 114
V + G FD +H GH L+ A L IV V D ++ K + P+ ER M N
Sbjct: 18 VVLGGTFDRLHDGHRMFLKAAAELARDRIVVGVCDGPMLTKKQFSDMIQPIEER--MRNV 75
Query: 115 VKWVDEVIPE---APYAITEEFMKKLFDEYNIDYIIHGDDPCVLPDG 158
+V + PE IT+ + + DE N++ I+ + LP G
Sbjct: 76 ETYVKSIKPELVVQAEPITDPYGPSIVDE-NLEAIVVSKE--TLPGG 119
>At5g23330 unknown protein
Length = 354
Score = 28.9 bits (63), Expect = 4.5
Identities = 22/101 (21%), Positives = 44/101 (42%), Gaps = 10/101 (9%)
Query: 61 GCFDMMHYGHCNALRQARALGDQLIVGVVSDDEIIANK--GPPVTPLHERLIMVNAVKWV 118
G FD +H GH QA +G ++ V E++ K P V + ++ + +
Sbjct: 90 GKFDALHIGHRELAIQAARIGTPYLLSFVGLAEVLGWKPRAPIVAKCDRKRVLSSWASYC 149
Query: 119 DEVIP---EAPYAIT-----EEFMKKLFDEYNIDYIIHGDD 151
+ P E +A ++F++KL E + ++ G++
Sbjct: 150 GNIAPVEFEIEFASVRHLNPQQFVEKLSRELRVCGVVAGEN 190
>At5g08345 unknown protein
Length = 367
Score = 28.9 bits (63), Expect = 4.5
Identities = 20/101 (19%), Positives = 42/101 (40%), Gaps = 10/101 (9%)
Query: 61 GCFDMMHYGHCNALRQARALGDQLIVGVVSDDEIIA--NKGPPVTPLHERLIMVNAVKWV 118
G FD +H GH QA +G ++ V E++ + P V + ++ + +
Sbjct: 106 GKFDALHIGHRELTIQASRIGAPYLLSFVGMAEVLGWEPRAPIVAKCDRQRVLTSWASYC 165
Query: 119 DEVIPE--------APYAITEEFMKKLFDEYNIDYIIHGDD 151
+ PE + +F++KL E + ++ G++
Sbjct: 166 GDRAPEEYEIEFASVRHLTPRQFVEKLSKELRVCGVVAGEN 206
>At5g10760 nucleoid DNA-binding protein cnd41 - like protein
Length = 464
Score = 28.5 bits (62), Expect = 5.9
Identities = 34/133 (25%), Positives = 57/133 (42%), Gaps = 15/133 (11%)
Query: 216 GQQKFEDGGVVASGT-RVSHFLPTSRRIVQFSNGRSPGPDARIVYIDGAFDLF----HAG 270
G +K + G V+ S T +VS P+S V S + R+V++ GA
Sbjct: 25 GAEKSDSGKVLDSYTIQVSSLFPSSSSCVPSSKASNTKSSLRVVHMHGACSHLSSDARVD 84
Query: 271 HVEILR--LARDRGDFLLVGIHTDQTVSATRGLHRPIMSLHERSLSVLACRYVDEVIIGA 328
H EI+R AR + + ++ VS + P S +++ + Y+ + IG
Sbjct: 85 HDEIIRRDQARVESIYSKLSKNSANEVSEAKSTELPAKS----GITLGSGNYIVTIGIGT 140
Query: 329 PWEVSKDMVSLIF 341
P K +SL+F
Sbjct: 141 P----KHDLSLVF 149
>At3g12010 unknown protein
Length = 685
Score = 28.5 bits (62), Expect = 5.9
Identities = 17/60 (28%), Positives = 29/60 (48%), Gaps = 1/60 (1%)
Query: 161 AYAHAKKAGRYKQIKRTEGVSSTDIVGRMLLCVRERNIADTHNHSSLQRQFSNGRGQQKF 220
AY+++ KAG YK+IK +ST G R R + ++ ++ S+G + F
Sbjct: 424 AYSYSVKAGTYKEIKNENATASTPTPGASPDSERHRQ-SGSNIDEEVEMNLSSGSQENMF 482
>At5g23280 unknown protein
Length = 250
Score = 28.1 bits (61), Expect = 7.7
Identities = 20/63 (31%), Positives = 31/63 (48%), Gaps = 4/63 (6%)
Query: 194 RERNIADTHNHSSLQRQFSNGRGQQKFEDGGVVASGTRVSHFLPTSRRIVQFSNGRSPGP 253
+E + H+H Q F + + QQ+ G AS RV ++LP ++ +G SPG
Sbjct: 187 QEMFLQQQHHHQ--QPLFVHQQQQQQAAMGE--ASAARVGNYLPGHLNLLASLSGGSPGS 242
Query: 254 DAR 256
D R
Sbjct: 243 DRR 245
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.141 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,247,248
Number of Sequences: 26719
Number of extensions: 361376
Number of successful extensions: 661
Number of sequences better than 10.0: 9
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 640
Number of HSP's gapped (non-prelim): 16
length of query: 358
length of database: 11,318,596
effective HSP length: 100
effective length of query: 258
effective length of database: 8,646,696
effective search space: 2230847568
effective search space used: 2230847568
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 61 (28.1 bits)
Medicago: description of AC124215.2


