

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC124214.6 + phase: 0
(484 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
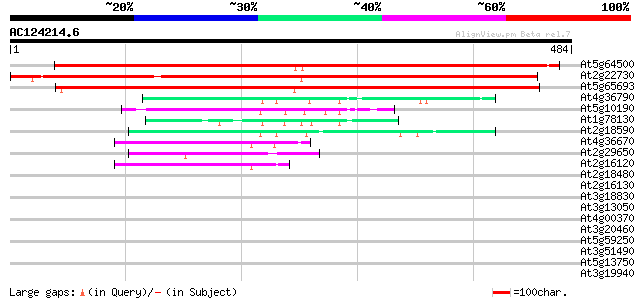
At5g64500 unknown protein 678 0.0
At2g22730 hypothetical protein 622 e-178
At5g65693 unknown protein 576 e-164
At4g36790 unknown protein 54 3e-07
At5g10190 unknown protein 49 5e-06
At1g78130 transporter like protein 49 6e-06
At2g18590 hypothetical protein 45 1e-04
At4g36670 sugar transporter like protein 44 2e-04
At2g29650 Na+-dependent inorganic phosphate cotransporter like p... 42 8e-04
At2g16120 putative sugar transporter 42 8e-04
At2g18480 putative sugar transporter 42 0.001
At2g16130 putative sugar transporter 42 0.001
At3g18830 sugar transporter like protein 41 0.002
At3g13050 putative transporter 41 0.002
At4g00370 unknown protein 40 0.004
At3g20460 sugar transporter, putative 38 0.011
At5g59250 D-xylose-H+ symporter - like protein 37 0.032
At3g51490 sugar transporter-like protein 35 0.12
At5g13750 transporter-like protein 34 0.16
At3g19940 putative monosaccharide transport protein, STP4 34 0.21
>At5g64500 unknown protein
Length = 484
Score = 678 bits (1750), Expect = 0.0
Identities = 325/458 (70%), Positives = 385/458 (83%), Gaps = 23/458 (5%)
Query: 39 IPATSWFTPKRLLAIFCVINMLNYLDRGAIASNGVNGHRGTCTD-GICKSGTGIQGDFNL 97
I SWFTPK+LL +FCV+N++NY+DRGAIASNG+NG RG+CT G C SG+GIQGDFNL
Sbjct: 27 ISEPSWFTPKKLLFVFCVVNLINYIDRGAIASNGINGSRGSCTSSGTCSSGSGIQGDFNL 86
Query: 98 TNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLSFNFWSIT 157
+NF+DGVLSSAFMVGLL+ASPIFASL+KSVNPFRLIGVGLS+WT+A + CGLSF+FWSIT
Sbjct: 87 SNFEDGVLSSAFMVGLLVASPIFASLAKSVNPFRLIGVGLSIWTLAVIGCGLSFDFWSIT 146
Query: 158 VCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGGVVGSHFG 217
+CRM VGVGEASF+SLAAPFIDDNAP QK+ WL++FYMCIP GYA GYVYGGVVGS
Sbjct: 147 ICRMFVGVGEASFVSLAAPFIDDNAPHDQKSAWLAVFYMCIPTGYAFGYVYGGVVGSVLP 206
Query: 218 WRYAFWVEAVLMLPFAILGFVMKPLQLK------------DQLSLF---------LKDMK 256
WR AFW EA+LMLPFA+LGFV+KPL LK D L++ +KD+K
Sbjct: 207 WRAAFWGEAILMLPFAVLGFVIKPLHLKGFAPDDTGKPRTDNLNVLPVGYGFSAVMKDLK 266
Query: 257 ELLSDKVFVVNVLGYIAYNFVIGAYSYWGPKAGYSIYNMTNADMIFGGITIVCGILGTLA 316
LL DKV+V N+LGYIAYNFV+GAYSYWGPKAGY+IY M NADMIFGG+T+VCGI+GTL+
Sbjct: 267 LLLVDKVYVTNILGYIAYNFVLGAYSYWGPKAGYNIYKMENADMIFGGVTVVCGIVGTLS 326
Query: 317 GGLVLDYMTNTLSNAFKLLSLTTLVGGAFCFGAFAFKSMYGFLALFAIGELLVFATQGPV 376
GG++LDYM T+SNAFK+LS++T +G FCF AF FKSMY FLALFA+GELLVFATQGPV
Sbjct: 327 GGVILDYMDATISNAFKVLSVSTFIGAIFCFAAFCFKSMYAFLALFAVGELLVFATQGPV 386
Query: 377 NFVCLHCVKPSLRPLSMAMSTVAIHIFGDVPSAPLVGVVQDHINNWRTTALILTTIFFPA 436
NF+ LHCVKPSLRPL+MAMSTV+IHIFGDVPS+PLVGV+QD++NNWR T+L+LT + FPA
Sbjct: 387 NFIVLHCVKPSLRPLAMAMSTVSIHIFGDVPSSPLVGVLQDYVNNWRVTSLVLTFVLFPA 446
Query: 437 AAIWFIGIFLNSKDKFNEESEHQVSRVEGTTTAPLLEE 474
AAIW IGIFLNS D++NE+SE E +T APLL+E
Sbjct: 447 AAIWSIGIFLNSVDRYNEDSEPDAVTRE-STAAPLLQE 483
>At2g22730 hypothetical protein
Length = 507
Score = 622 bits (1604), Expect = e-178
Identities = 318/496 (64%), Positives = 367/496 (73%), Gaps = 47/496 (9%)
Query: 1 MAQKSEDEPKPATTTSSS--STPNPVPSSVEPNMLPKSTMIPATSWFTPKRLLAIFCVIN 58
M K ED P T T+S ST + P + E + ++ ++S +P LL IFC+IN
Sbjct: 1 MVTKEEDCLPPVTETTSRCYSTSSSTPLA-ELETVRSLEIVESSSSLSPVWLLVIFCIIN 59
Query: 59 MLNYLDRGAIASNGVNGHRGTCTD-GICKSGTGIQGDFNLTNFQDGVLSSAFMVGLLIAS 117
+LNY+DRGAIASNGVNG +C D G C TGIQG FNL+NF+DGVLSS+FMVGLLIAS
Sbjct: 60 LLNYMDRGAIASNGVNGSTRSCNDKGKCTLATGIQGHFNLSNFEDGVLSSSFMVGLLIAS 119
Query: 118 PIFASLSKSVNPFRLIGVGLSVWTVATLCCGLSFNFWSITVCRMLVGVGEASFISLAAPF 177
PIFASL+K RLIGVGL+VWT+A L CG SF FW I +CRM VGVGEASFISLAAPF
Sbjct: 120 PIFASLAK-----RLIGVGLTVWTIAVLGCGSSFAFWFIVLCRMFVGVGEASFISLAAPF 174
Query: 178 IDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGGVVGSHFGWRYAFWVEAVLMLPFAILGF 237
IDDNAP QK WL +FYMCIP G A+GYVYGG VG HF WRYAFW EAVLM PFA+LGF
Sbjct: 175 IDDNAPQEQKAAWLGLFYMCIPSGVALGYVYGGYVGKHFSWRYAFWGEAVLMAPFAVLGF 234
Query: 238 VMKPLQLKDQLSL--------------------------------------FLKDMKELL 259
+MKPLQLK +L F KDMK L
Sbjct: 235 LMKPLQLKGSETLKNNNRLQVDNEIEHDQFEVSIETSKSSYANAVFKSFTGFAKDMKVLY 294
Query: 260 SDKVFVVNVLGYIAYNFVIGAYSYWGPKAGYSIYNMTNADMIFGGITIVCGILGTLAGGL 319
+KVFVVNVLGY++YNFVIGAYSYWGPKAGY+IY M NADMIFG +TI+CGI+GTL+GG
Sbjct: 295 KEKVFVVNVLGYVSYNFVIGAYSYWGPKAGYNIYKMKNADMIFGAVTIICGIVGTLSGGF 354
Query: 320 VLDYMTNTLSNAFKLLSLTTLVGGAFCFGAFAFKSMYGFLALFAIGELLVFATQGPVNFV 379
+LD +T T+ NAFKLLS T +G FCF AF KS+YGF+ALFA+GELLVFATQ PVN+V
Sbjct: 355 ILDRVTATIPNAFKLLSGATFLGAVFCFTAFTLKSLYGFIALFALGELLVFATQAPVNYV 414
Query: 380 CLHCVKPSLRPLSMAMSTVAIHIFGDVPSAPLVGVVQDHINNWRTTALILTTIFFPAAAI 439
CLHCVKPSLRPLSMA+STVAIHIFGDVPS+PLVG+VQDHIN+WR T LILT+I F AAAI
Sbjct: 415 CLHCVKPSLRPLSMAISTVAIHIFGDVPSSPLVGIVQDHINSWRKTTLILTSILFLAAAI 474
Query: 440 WFIGIFLNSKDKFNEE 455
WFIGIF+NS D+FN+E
Sbjct: 475 WFIGIFINSVDRFNQE 490
>At5g65693 unknown protein
Length = 492
Score = 576 bits (1484), Expect = e-164
Identities = 280/457 (61%), Positives = 339/457 (73%), Gaps = 39/457 (8%)
Query: 40 PATS--WFTPKRLLAIFCVINMLNYLDRGAIASNGVNGHRGTC-TDGICKSGTGIQGDFN 96
PAT + TP R + I C+IN++NY+DRG IASNGVNG C G+C +GTGIQG+FN
Sbjct: 17 PATKKRFLTPGRFVTILCIINLINYVDRGVIASNGVNGSSKVCDAKGVCSAGTGIQGEFN 76
Query: 97 LTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLSFNFWSI 156
LTNF+DG+LSSAFMVGLL+ASPIFA LSK NPF+LIGVGL+VWT+A + CG S+NFW I
Sbjct: 77 LTNFEDGLLSSAFMVGLLVASPIFAGLSKRFNPFKLIGVGLTVWTIAVIGCGFSYNFWMI 136
Query: 157 TVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGGVVGSHF 216
V RM VGVGEASFISLAAP+IDD+AP ++K WL +FYMCIP G A+GYV+GG +G+H
Sbjct: 137 AVFRMFVGVGEASFISLAAPYIDDSAPVARKNFWLGLFYMCIPAGVALGYVFGGYIGNHL 196
Query: 217 GWRYAFWVEAVLMLPFAILGFVMKPLQL-------------------------------- 244
GWR+AF++EA+ M F IL F +KP Q
Sbjct: 197 GWRWAFYIEAIAMAVFVILSFCIKPPQQLKGFADKDSKKPSTSIETVAPTDAEASQIKTK 256
Query: 245 ----KDQLSLFLKDMKELLSDKVFVVNVLGYIAYNFVIGAYSYWGPKAGYSIYNMTNADM 300
K+ + LF KD+K L S+KVF+VNVLGYI YNFVIGAYSYWGPKAG+ IY M NADM
Sbjct: 257 TPKSKNLVVLFGKDLKALFSEKVFIVNVLGYITYNFVIGAYSYWGPKAGFGIYKMKNADM 316
Query: 301 IFGGITIVCGILGTLAGGLVLDYMTNTLSNAFKLLSLTTLVGGAFCFGAFAFKSMYGFLA 360
IFGG+TI+CGI+GTL G VLD + TLSN FKLL+ +TL+G AFCF AF K+MY F+A
Sbjct: 317 IFGGLTIICGIIGTLGGSYVLDRINATLSNTFKLLAASTLLGAAFCFTAFLMKNMYAFIA 376
Query: 361 LFAIGELLVFATQGPVNFVCLHCVKPSLRPLSMAMSTVAIHIFGDVPSAPLVGVVQDHIN 420
LFA+GE+L+FA Q PVNFVCLHCV+P+LRPLSMA STV IHI GDVPS+PL G +QDH+
Sbjct: 377 LFAVGEILIFAPQAPVNFVCLHCVRPNLRPLSMASSTVLIHILGDVPSSPLYGKMQDHLK 436
Query: 421 NWRTTALILTTIFFPAAAIWFIGIFLNSKDKFNEESE 457
NWR + LI+T+I F AA IW IGIF+NS D+ NE SE
Sbjct: 437 NWRKSTLIITSILFLAAIIWGIGIFMNSVDRSNEVSE 473
>At4g36790 unknown protein
Length = 489
Score = 53.5 bits (127), Expect = 3e-07
Identities = 74/336 (22%), Positives = 128/336 (38%), Gaps = 36/336 (10%)
Query: 115 IASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLSFNFWSITVCRMLVGVGEASFISLA 174
+ASP+ L + + ++ +G W ++T G S F + + R + G G A I
Sbjct: 93 LASPLAGVLVITYDRPIVLAIGTFCWALSTAAVGASSYFIQVALWRAVNGFGLAIVIPAL 152
Query: 175 APFIDDNAPASQKTVWLSIFYMC-IPGGYAIGYVYGGVVGSHF----GWRYAFWVEAVL- 228
FI D+ + + + GG G V + GS F GWR AF + A L
Sbjct: 153 QSFIADSYKDGARGAGFGMLNLIGTIGGIGGGVVATVMAGSEFWGIPGWRCAFIMMAALS 212
Query: 229 -MLPFAILGFVMKPLQLKDQLSLFLKDMKE--------LLSDKVFVVNVLGYIAYNFVIG 279
++ + FV+ P + ++ L M + V V+ I +IG
Sbjct: 213 AVIGLLVFLFVVDPRKNIEREELMAHKMNSNSVWNDSLAAAKSVVKVSTFQIIVAQGIIG 272
Query: 280 AYSY--------WGPKAGYSIYNMTNADMIFGGITIVCGILGTLAGGLVLDYMTNTLSNA 331
++ + W G+ +N T A + G+ G +GTL GG++ D M+ N+
Sbjct: 273 SFPWTAMVFFTMWFELIGFD-HNQTAALL---GVFATGGAIGTLMGGIIADKMSRIYPNS 328
Query: 332 FKLLSLTTLVGGAFCFGAFAFK------SMYGF--LALFAIGELLVFATQGPVNFVCLHC 383
+++ F K S Y + LF +G + + +
Sbjct: 329 GRVMCAQFSAFMGIPFSIILLKVIPQSTSSYSIFSITLFLMGLTITWCGSAVNAPMFAEV 388
Query: 384 VKPSLRPLSMAMSTVAIHIFGDVPSAPLVGVVQDHI 419
V P R + A F +APLVG++ + +
Sbjct: 389 VPPRHRTMIYAFDRAFEGSFSSF-AAPLVGILSEKL 423
>At5g10190 unknown protein
Length = 488
Score = 49.3 bits (116), Expect = 5e-06
Identities = 64/259 (24%), Positives = 106/259 (40%), Gaps = 39/259 (15%)
Query: 97 LTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLSFNFWSI 156
LT F+ V SS + P+ A LS N +I +G +W AT +S F+ +
Sbjct: 46 LTLFRSIVQSSCY--------PLAAYLSSRHNRAHVIALGAFLWATATFLVAVSTTFFQV 97
Query: 157 TVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGGVVGSH- 215
V R L G+G A + D+ + + G +GYV + S
Sbjct: 98 AVSRGLNGIGLAIVTPAIQSLVADSTDDYNRGMAFGWLGFTSNIGSILGYVCSILFASKS 157
Query: 216 ----FGWRYAFWVEAVLMLPFAILG--FVMKPLQLKDQLSLFLK------DMKELLSDKV 263
GWR AF + AV+ + IL F P +++ +K D+++LL +
Sbjct: 158 FNGVAGWRIAFLLVAVVSVIVGILVRLFATDPHYSDRKITKHVKDKPFWSDIRDLLKEAK 217
Query: 264 FVVNVLGY-------IAYNFVIGAYSY---WGPKAGYSIYNMTNADMIFGGITIVCGILG 313
V+ + + ++ +F A ++ W G+S + T A ++ TI C +
Sbjct: 218 MVIKIPSFQIFVAQGVSGSFPWSALAFAPLWLELIGFS--HKTTA-VLVTLFTISCSL-- 272
Query: 314 TLAGGLVLDYMTNTLSNAF 332
GGL YM +TL+ F
Sbjct: 273 ---GGLFGGYMGDTLAKKF 288
>At1g78130 transporter like protein
Length = 490
Score = 48.9 bits (115), Expect = 6e-06
Identities = 58/255 (22%), Positives = 101/255 (38%), Gaps = 52/255 (20%)
Query: 118 PIFASLSKSVNPFRLIGVGLSVWTVATLCCGLSFNFWSITVCRMLVGVGEASFISLAAPF 177
P+ A ++ N +I +G +W+ AT S F+ + V R L G+G ++L AP
Sbjct: 59 PLAAYMAIRHNRAHVIALGAFLWSAATFLVAFSSTFFQVAVSRALNGIG----LALVAPA 114
Query: 178 ID-------DNAPASQKTVWLSIFYMCIPGGYAIGYVYGGVVGSHF---------GWRYA 221
I D+A WL + IG + GG+ GWR A
Sbjct: 115 IQSLVADSTDDANRGTAFGWLQL-------TANIGSILGGLCSVLIAPLTFMGIPGWRVA 167
Query: 222 FWVEAVLMLPFAIL--GFVMKPLQLKDQLSL---------FLKDMKELL--SDKVFVVNV 268
F + V+ + +L F P +KD + + F ++K+L+ +D V +
Sbjct: 168 FHIVGVISVIVGVLVRVFANDPHFVKDGVDVSNQPGSRKPFCTEVKDLVREADTVIKIRS 227
Query: 269 LGYIAYNFVIGAYSY--------WGPKAGYSIYNMTNADMIFGGITIVCGILGTLAGGLV 320
I V G++ + W G+S G+ + LG L GG +
Sbjct: 228 FQIIVAQGVTGSFPWSALSFAPMWLELIGFS----HGKTAFLMGLFVAASSLGGLFGGKM 283
Query: 321 LDYMTNTLSNAFKLL 335
D+++ L N+ +++
Sbjct: 284 GDFLSTRLPNSGRII 298
>At2g18590 hypothetical protein
Length = 433
Score = 44.7 bits (104), Expect = 1e-04
Identities = 76/345 (22%), Positives = 131/345 (37%), Gaps = 33/345 (9%)
Query: 103 GVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLSFNFWSITVCRML 162
G+LS + +ASP+ + S + + G W +T+ G+S F +T+
Sbjct: 29 GLLSFIRNIVQGLASPLAGLFAISYDRPTVFAFGSFFWVSSTVATGVSRYFIQVTLGVAF 88
Query: 163 VGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGGVVGSH-----FG 217
GVG A + I D+ S + ++ + G G V V+ H G
Sbjct: 89 NGVGHAIVYPVLQSIIADSFKESSRGFGFGLWNLIGTVGGIGGTVVPTVMAGHDFFGISG 148
Query: 218 WRYAFWVEAVL--MLPFAILGFVMKPLQLKDQLSLFLKD------------MKELLSDKV 263
WR AF + A L ++ + FV P + K + D M E S V
Sbjct: 149 WRCAFILSATLSTIVGILVFFFVSDPREKKTSSVIVHHDDQHERDENNGGTMMESPSSSV 208
Query: 264 FVVNVLGYIAYNFVIGAYSYWGPKAGYSIYNMTNADMIFGGITIVCGILGTLAGGLVLDY 323
+ + ++A V ++ + N + GI +G+L GG++ D
Sbjct: 209 WKES---WVAIKDVTKLRTFQIIVLQGIVGFDHNQAALLNGIFATGQAIGSLVGGIIADK 265
Query: 324 MTNTLSNAFKLL--SLTTLVGGAFCFGAF-----AFKSMYGFL-ALFAIGELLVFATQGP 375
M+ N+ +L+ + +G F + S Y FL LF +G L GP
Sbjct: 266 MSRVFPNSGRLICAQFSVFMGAMFSIVLLRMIPQSVNSFYIFLVTLFLMG--LTITWCGP 323
Query: 376 -VNFVCLHCVKPSLRPLSMAMSTVAIHIFGDVPSAPLVGVVQDHI 419
+N L + P+ + A+ + APLVG++ + +
Sbjct: 324 AINSPILAEIVPAKHRTMVYAFDRALEVTFSSFGAPLVGIMSEKL 368
>At4g36670 sugar transporter like protein
Length = 493
Score = 44.3 bits (103), Expect = 2e-04
Identities = 43/174 (24%), Positives = 76/174 (42%), Gaps = 7/174 (4%)
Query: 91 IQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLS 150
I+ D + Q VL+ + L+ S + S + I + ++ + ++ G
Sbjct: 45 IEEDLKTNDVQIEVLTGILNLCALVGSLLAGRTSDIIGRRYTIVLASILFMLGSILMGWG 104
Query: 151 FNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYV--- 207
N+ + R G+G + +A + + A AS + + S+ ++CI G +GY+
Sbjct: 105 PNYPVLLSGRCTAGLGVGFALMVAPVYSAEIATASHRGLLASLPHLCISIGILLGYIVNY 164
Query: 208 YGGVVGSHFGWRYAFWVEAV--LMLPFAILGFVMKPLQLKDQLSLFLKDMKELL 259
+ + H GWR + AV L+L F IL P L Q LK+ KE+L
Sbjct: 165 FFSKLPMHIGWRLMLGIAAVPSLVLAFGILKMPESPRWLIMQGR--LKEGKEIL 216
>At2g29650 Na+-dependent inorganic phosphate cotransporter like
protein
Length = 512
Score = 42.0 bits (97), Expect = 8e-04
Identities = 37/167 (22%), Positives = 69/167 (41%), Gaps = 9/167 (5%)
Query: 103 GVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLS--FNFWSITVCR 160
G++ S+F G L+ + +V R++G G+ W++AT+ ++ + V R
Sbjct: 142 GLIQSSFFWGYLLTQIAGGIWADTVGGKRVLGFGVIWWSIATILTPVAAKLGLPYLLVVR 201
Query: 161 MLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGGVVGSHFGWRY 220
+GVGE + + P +++ L++ Y + G G + + FGW
Sbjct: 202 AFMGVGEGVAMPAMNNILSKWVPVQERSRSLALVYSGMYLGSVTGLAFSPFLIHQFGWPS 261
Query: 221 AFWVEAVLMLPFAILGFVMKPLQLKDQLSLFLKDMKELLSDKVFVVN 267
F+ F LG V L L S L+D L ++ + +
Sbjct: 262 VFY-------SFGSLGTVWLTLWLTKAESSPLEDPTLLPEERKLIAD 301
>At2g16120 putative sugar transporter
Length = 511
Score = 42.0 bits (97), Expect = 8e-04
Identities = 38/154 (24%), Positives = 65/154 (41%), Gaps = 4/154 (2%)
Query: 91 IQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLS 150
I+ D L++ Q +L + L+ S S + I + + + L G +
Sbjct: 54 IKDDLKLSDVQLEILMGILNIYSLVGSGAAGRTSDWLGRRYTIVLAGAFFFCGALLMGFA 113
Query: 151 FNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYV--- 207
N+ I V R + G+G + +A + + APAS + S + I G +GYV
Sbjct: 114 TNYPFIMVGRFVAGIGVGYAMMIAPVYTAEVAPASSRGFLTSFPEIFINIGILLGYVSNY 173
Query: 208 YGGVVGSHFGWRYAFWVEAVLMLPFAILGFVMKP 241
+ + H GWR+ V AV + F +G + P
Sbjct: 174 FFSKLPEHLGWRFMLGVGAVPSV-FLAIGVLAMP 206
>At2g18480 putative sugar transporter
Length = 508
Score = 41.6 bits (96), Expect = 0.001
Identities = 36/149 (24%), Positives = 63/149 (42%), Gaps = 5/149 (3%)
Query: 91 IQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLS 150
I+ D + + Q VL+ + L+ S S + I + ++ V ++ G
Sbjct: 50 IRDDLKINDTQIEVLAGILNLCALVGSLTAGKTSDVIGRRYTIALSAVIFLVGSVLMGYG 109
Query: 151 FNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYG- 209
N+ + V R + GVG + +A + + + AS + S+ +CI G +GYV
Sbjct: 110 PNYPVLMVGRCIAGVGVGFALMIAPVYSAEISSASHRGFLTSLPELCISLGILLGYVSNY 169
Query: 210 --GVVGSHFGWRYAFWVEAV--LMLPFAI 234
G + GWR + A L+L F I
Sbjct: 170 CFGKLTLKLGWRLMLGIAAFPSLILAFGI 198
>At2g16130 putative sugar transporter
Length = 511
Score = 41.6 bits (96), Expect = 0.001
Identities = 38/154 (24%), Positives = 64/154 (40%), Gaps = 4/154 (2%)
Query: 91 IQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLS 150
I+ D L++ Q +L + LI S S + I + + L G +
Sbjct: 54 IKDDLKLSDVQLEILMGILNIYSLIGSGAAGRTSDWIGRRYTIVLAGFFFFCGALLMGFA 113
Query: 151 FNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYV--- 207
N+ I V R + G+G + +A + + APAS + S + I G +GYV
Sbjct: 114 TNYPFIMVGRFVAGIGVGYAMMIAPVYTTEVAPASSRGFLSSFPEIFINIGILLGYVSNY 173
Query: 208 YGGVVGSHFGWRYAFWVEAVLMLPFAILGFVMKP 241
+ + H GWR+ + AV + F +G + P
Sbjct: 174 FFAKLPEHIGWRFMLGIGAVPSV-FLAIGVLAMP 206
>At3g18830 sugar transporter like protein
Length = 539
Score = 40.8 bits (94), Expect = 0.002
Identities = 34/147 (23%), Positives = 62/147 (42%), Gaps = 3/147 (2%)
Query: 91 IQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLS 150
I+ D + + Q G+L+ + + LI S S + I + +++ + GLS
Sbjct: 64 IKRDLKINDLQIGILAGSLNIYSLIGSCAAGRTSDWIGRRYTIVLAGAIFFAGAILMGLS 123
Query: 151 FNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGG 210
N+ + R + G+G + +A + + +PAS + S + I G +GYV
Sbjct: 124 PNYAFLMFGRFIAGIGVGYALMIAPVYTAEVSPASSRGFLNSFPEVFINAGIMLGYVSNL 183
Query: 211 VVGS---HFGWRYAFWVEAVLMLPFAI 234
+ GWR + AV + AI
Sbjct: 184 AFSNLPLKVGWRLMLGIGAVPSVILAI 210
>At3g13050 putative transporter
Length = 500
Score = 40.8 bits (94), Expect = 0.002
Identities = 35/148 (23%), Positives = 64/148 (42%), Gaps = 4/148 (2%)
Query: 88 GTGIQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCC 147
G +Q +NL+ Q+ +++S G+LI + + +S + + V VA
Sbjct: 48 GPAVQSLWNLSARQESLITSVVFAGMLIGAYSWGIVSDKHGRRKGFIITAVVTFVAGFLS 107
Query: 148 GLSFNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYV 207
S N+ + + R LVG+G LA+ ++ + PA + W+ +F G
Sbjct: 108 AFSPNYMWLIILRCLVGLGLGGGPVLASWYL-EFIPAPSRGTWMVVFSAFWTVGTIFEAS 166
Query: 208 YGGVVGSHFGWRYAFWVEAVLMLPFAIL 235
+V GWR W+ A +P ++L
Sbjct: 167 LAWLVMPRLGWR---WLLAFSSVPSSLL 191
>At4g00370 unknown protein
Length = 541
Score = 39.7 bits (91), Expect = 0.004
Identities = 29/136 (21%), Positives = 60/136 (43%), Gaps = 10/136 (7%)
Query: 94 DFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCC------ 147
++N ++ G++ S+F G L+ + + ++G G+ W+ AT+
Sbjct: 162 EYNWSSATVGLIQSSFFWGYLLTQILGGIWADKFGGKVVLGFGVVWWSFATIMTPIAARL 221
Query: 148 GLSFNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYV 207
GL F + V R +G+GE + + P S+++ L++ Y + G G
Sbjct: 222 GLPF----LLVVRAFMGIGEGVAMPAMNNMLSKWIPVSERSRSLALVYSGMYLGSVTGLA 277
Query: 208 YGGVVGSHFGWRYAFW 223
+ ++ + FGW F+
Sbjct: 278 FSPMLITKFGWPSVFY 293
>At3g20460 sugar transporter, putative
Length = 488
Score = 38.1 bits (87), Expect = 0.011
Identities = 28/135 (20%), Positives = 53/135 (38%)
Query: 78 GTCTDGICKSGTGIQGDFNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGL 137
GT + TGI NL+ + + +G L+ + + L+ +GV
Sbjct: 68 GTAAGFTSPAQTGIMAGLNLSLAEFSFFGAVLTIGGLVGAAMSGKLADVFGRRGALGVSN 127
Query: 138 SVWTVATLCCGLSFNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMC 197
S L S WS+ + R+ +GV + +I + AP + + +I +
Sbjct: 128 SFCMAGWLMIAFSQATWSLDIGRLFLGVAAGVASYVVPVYIVEIAPKKVRGTFSAINSLV 187
Query: 198 IPGGYAIGYVYGGVV 212
+ A+ Y+ G V+
Sbjct: 188 MCASVAVTYLLGSVI 202
>At5g59250 D-xylose-H+ symporter - like protein
Length = 558
Score = 36.6 bits (83), Expect = 0.032
Identities = 85/373 (22%), Positives = 146/373 (38%), Gaps = 71/373 (19%)
Query: 95 FNLTNFQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLSFNFW 154
FN + Q G++ S + G L+ S ++ + R + + ++ + +L G + +
Sbjct: 137 FNFSPVQLGLVVSGSLYGALLGSISVYGVADFLGRRRELIIAAVLYLLGSLITGCAPDLN 196
Query: 155 SITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYVYGG---- 210
+ V R+L G G + A +I + P+ + +S+ + I G +G+ G
Sbjct: 197 ILLVGRLLYGFGIGLAMHGAPLYIAETCPSQIRGTLISLKELFIVLGILLGFSVGSFQID 256
Query: 211 VVGSHFGWRYAFWV---EAVLM------LPFAILGFVMKPLQLKDQLSLFLKDMKELLSD 261
VVG GWRY + A+LM LP + +++ +Q K QL + K+ L
Sbjct: 257 VVG---GWRYMYGFGTPVALLMGLGMWSLPASPRWLLLRAVQGKGQLQEY-KEKAMLALS 312
Query: 262 KVFVVNVLGYIAYNFVIGAY-----SYWGPKAGYSIYNM-----TNADMIFGGITIVCGI 311
K+ I+ V AY +Y K+G + + A I GG+ + I
Sbjct: 313 KLRGRPPGDKISEKLVDDAYLSVKTAYEDEKSGGNFLEVFQGPNLKALTIGGGLVLFQQI 372
Query: 312 LGTLAGGLVLDYMTNTLSNA-----------------FKLLSL-------------TTLV 341
G + VL Y + L A FKLL L+
Sbjct: 373 TGQPS---VLYYAGSILQTAGFSAAADATRVSVIIGVFKLLMTWVAVAKVDDLGRRPLLI 429
Query: 342 GG--AFCFGAFAFKSMYGFLA---LFAIGELLVFA-----TQGPVNFVCLHCVKPSLRPL 391
GG F + Y FL L A+G LL++ + GP++++ + + P LR
Sbjct: 430 GGVSGIALSLFLLSAYYKFLGGFPLVAVGALLLYVGCYQISFGPISWLMVSEIFP-LRTR 488
Query: 392 SMAMSTVAIHIFG 404
+S + FG
Sbjct: 489 GRGISLAVLTNFG 501
>At3g51490 sugar transporter-like protein
Length = 729
Score = 34.7 bits (78), Expect = 0.12
Identities = 25/142 (17%), Positives = 62/142 (43%), Gaps = 2/142 (1%)
Query: 102 DGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLSFNFWSITVCRM 161
+G++ + ++G + + +S V ++ + ++ ++++ S N + + R+
Sbjct: 45 EGLIVAMSLIGATLITTFSGPVSDKVGRRSMLILSSVLYFLSSIVMFWSPNVYVLLFARL 104
Query: 162 LVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGY--VYGGVVGSHFGWR 219
L G G ++L +I + AP+ + + + C GG + Y V+G + WR
Sbjct: 105 LDGFGIGLAVTLVPIYISETAPSEIRGLLNTFPQFCGSGGMFLSYCLVFGMSLQESPSWR 164
Query: 220 YAFWVEAVLMLPFAILGFVMKP 241
V ++ + + +L P
Sbjct: 165 LMLGVLSIPSIAYFVLAAFFLP 186
>At5g13750 transporter-like protein
Length = 478
Score = 34.3 bits (77), Expect = 0.16
Identities = 30/126 (23%), Positives = 49/126 (38%), Gaps = 11/126 (8%)
Query: 94 DFNLTN------FQDGVLSSAFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCC 147
DFN+ F G + +FM+G S + ++ +I +G + V
Sbjct: 65 DFNIAKKEEDIGFYAGFVGCSFMLGRAFTSVAWGLVADRYGRKPVILIGTASVVVFNTLF 124
Query: 148 GLSFNFWSITVCRMLVGVGEASFISLAAPFIDDNAPASQKTVWLSIFYMCIPGGYAIGYV 207
GLS NFW + R +G SF L P I A + + + + + IG +
Sbjct: 125 GLSLNFWMAIITRFCLG----SFNGLLGP-IKAYAMEIFRDEYQGLALSAVSTAWGIGLI 179
Query: 208 YGGVVG 213
G +G
Sbjct: 180 IGPAIG 185
>At3g19940 putative monosaccharide transport protein, STP4
Length = 514
Score = 33.9 bits (76), Expect = 0.21
Identities = 33/135 (24%), Positives = 59/135 (43%), Gaps = 8/135 (5%)
Query: 108 AFMVGLLIASPIFASLSKSVNPFRLIGVGLSVWTVATLCCGLSFNFWSITVCRMLVGVGE 167
A +V +AS I + V+ F +G + + L + N + + R+L+GVG
Sbjct: 94 AALVASFMASVITRKHGRKVSMF----IGGLAFLIGALFNAFAVNVSMLIIGRLLLGVG- 148
Query: 168 ASFISLAAP-FIDDNAPASQKTVWLSIFYMCIPGGYAIGYV--YGGVVGSHFGWRYAFWV 224
F + + P ++ + APA + F M I G + + YG + GWR + +
Sbjct: 149 VGFANQSTPVYLSEMAPAKIRGALNIGFQMAITIGILVANLINYGTSKMAQHGWRVSLGL 208
Query: 225 EAVLMLPFAILGFVM 239
AV + I F++
Sbjct: 209 AAVPAVVMVIGSFIL 223
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.139 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,992,223
Number of Sequences: 26719
Number of extensions: 483178
Number of successful extensions: 2278
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 26
Number of HSP's that attempted gapping in prelim test: 2213
Number of HSP's gapped (non-prelim): 77
length of query: 484
length of database: 11,318,596
effective HSP length: 103
effective length of query: 381
effective length of database: 8,566,539
effective search space: 3263851359
effective search space used: 3263851359
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 62 (28.5 bits)
Medicago: description of AC124214.6


