

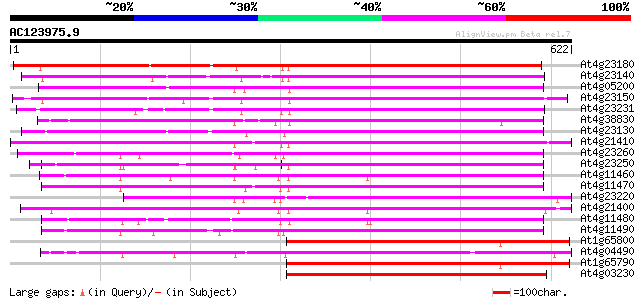
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123975.9 + phase: 0 /pseudo
(622 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g23180 receptor-like protein kinase 4 (RLK4) 498 e-141
At4g23140 receptor-like protein kinase 5 (RLK5) 466 e-131
At4g05200 receptor protein kinase - like protein 458 e-129
At4g23150 serine/threonine kinase - like protein 454 e-128
At4g23231 unknown protein 431 e-121
At4g38830 receptor-like protein kinase - like protein 421 e-118
At4g23130 receptor-like protein kinase 6 (RLK6) 411 e-115
At4g21410 serine/threonine kinase like protein 407 e-114
At4g23260 unknown protein 396 e-110
At4g23250 protein kinase - like protein 395 e-110
At4g11460 serine/threonine kinase-like protein 395 e-110
At4g11470 serine/threonine kinase-like protein 380 e-106
At4g23220 serine/threonine kinase - like protein 379 e-105
At4g21400 serine/threonine protein kinase - like protein 374 e-104
At4g11480 serine/threonine kinase-like protein 369 e-102
At4g11490 serine/threonine kinase-like protein 365 e-101
At1g65800 receptor kinase, putative 363 e-100
At4g04490 putative receptor-like protein kinase 360 e-99
At1g65790 receptor kinase, putative 360 e-99
At4g03230 putative receptor kinase 358 5e-99
>At4g23180 receptor-like protein kinase 4 (RLK4)
Length = 669
Score = 498 bits (1281), Expect = e-141
Identities = 282/628 (44%), Positives = 388/628 (60%), Gaps = 48/628 (7%)
Query: 5 PIVFKFIQFFLVLYTLDFLSYLALADPP---YEICSTRNIYANGSSFDNNLSNLLLSLPF 61
PI+ + FF + S+ A P Y C Y + S+++NNL LL SL
Sbjct: 10 PIMSYYSSFFFLFLFSFLTSFRVSAQDPTYVYHTCQNTANYTSNSTYNNNLKTLLASLSS 69
Query: 62 NDSNSISKFGNTSSGIGLDRVYGLYMCLDFVSNETCLKCVTNAIADTVKLCPQSKEAVVY 121
+++ + F N + G DRV GL+ C VS E C +CV+ A+ DT+ CP KEA +Y
Sbjct: 70 RNASYSTGFQNATVGQAPDRVTGLFNCRGDVSTEVCRRCVSFAVNDTLTRCPNQKEATLY 129
Query: 122 EEFCQVRYSNKNFIGSLNVNGNIGKDNVQNISEPVKFETSVNKLLNGLTKIASFNV-SAN 180
+ C +RYSN+N + +L G + N +N++ + + + +L L + A+ + S+
Sbjct: 130 YDECVLRYSNQNILSTLITTGGVILVNTRNVTSN-QLDLLSDLVLPTLNQAATVALNSSK 188
Query: 181 MYATGEVPFED-KTIYALVQCTRDLAANDCSRCLLSAIGDIPGCCYASIGARVMSRSCYL 239
+ T + F ++ Y LVQCT DL DCSRCL I IP IGAR+++ SC
Sbjct: 189 KFGTRKNNFTALQSFYGLVQCTPDLTRQDCSRCLQLVINQIPT---DRIGARIINPSCTS 245
Query: 240 RYEFYPFYLGE-----------KEQTKSSTNLGGKNNSSKIWMITVIAVGVGLVIIIFIC 288
RYE Y FY S+ GK+ +SK+ +I ++ V + + +++FI
Sbjct: 246 RYEIYAFYTESAVPPPPPPPSISTPPVSAPPRSGKDGNSKVLVIAIV-VPIIVAVLLFIA 304
Query: 289 YLCFLRNRQSKLF-------GDCISV----------------NFTDSNKLGEGGFGPVYK 325
CFL R K + GD I+ +F +SNK+G+GGFG VYK
Sbjct: 305 GYCFLTRRARKSYYTPSAFAGDDITTADSLQLDYRTIQTATDDFVESNKIGQGGFGEVYK 364
Query: 326 GILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVKLLGFCVDGEEKLLVYEYL 385
G LSDG EVA+KRLS S QG EF NEV+L+ KLQH+NLV+LLGFC+DGEE++LVYEY+
Sbjct: 365 GTLSDGTEVAVKRLSKSSGQGEVEFKNEVVLVAKLQHRNLVRLLGFCLDGEERVLVYEYV 424
Query: 386 PNGSLDVVLFE--QHAQLDWTKRLDIINGIARGILYLHEDSRLQIIHRDLKASNVLLDND 443
PN SLD LF+ + QLDWT+R II G+ARGILYLH+DSRL IIHRDLKASN+LLD D
Sbjct: 425 PNKSLDYFLFDPAKKGQLDWTRRYKIIGGVARGILYLHQDSRLTIIHRDLKASNILLDAD 484
Query: 444 MNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEGLYSIKSDVFGFGVLLLEII 503
MNPKI+DFGMARIF + E NT+ IVGTYGYM+PEYAM G YS+KSDV+ FGVL+LEII
Sbjct: 485 MNPKIADFGMARIFGLDQTEENTSRIVGTYGYMSPEYAMHGQYSMKSDVYSFGVLVLEII 544
Query: 504 TGIRNAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLLL--CPGDQFLRYMNIGLLCVQE 561
+G +N+ F + L++YAW LW++G+ LEL DP ++ C ++ +R ++IGLLCVQE
Sbjct: 545 SGKKNSSFYQTDGAHDLVSYAWGLWSNGRPLELVDPAIVENCQRNEVVRCVHIGLLCVQE 604
Query: 562 DAFDRPTMSSVVLMLMNESVMLGQPGKP 589
D +RPT+S++VLML + +V L P +P
Sbjct: 605 DPAERPTLSTIVLMLTSNTVTLPVPRQP 632
>At4g23140 receptor-like protein kinase 5 (RLK5)
Length = 674
Score = 466 bits (1198), Expect = e-131
Identities = 274/637 (43%), Positives = 377/637 (59%), Gaps = 63/637 (9%)
Query: 14 FLVLYTLDFLSYLALADPPYEI---CSTRNIYANGSSFDNNLSNLLLSLPFNDSNSISKF 70
FL L++ S+ A A P+ + C Y++ S++ NL LL SL +++ + F
Sbjct: 9 FLFLFSFLTSSFTASAQDPFYLNHYCPNTTTYSSNSTYSTNLRTLLSSLSSRNASYSTGF 68
Query: 71 GNTSSGIGLDRVYGLYMCLDFVSNETCLKCVTNAIADTVKLCPQSKEAVVYEEFCQVRYS 130
N ++G DRV GL++C VS E C CV ++ T+ LCP+ +EAV Y E C +RYS
Sbjct: 69 QNATAGKAPDRVTGLFLCRGDVSPEVCRNCVAFSVNQTLNLCPKVREAVFYYEQCILRYS 128
Query: 131 NKNFIGSLNVN-GNIGKDNVQNISEPVK----FETSVNKLLNGLTKIASFNVSANMYATG 185
+KN + + N G N IS K F + V+ ++ A+ N S +Y
Sbjct: 129 HKNILSTAITNEGEFILSNTNTISPNQKQIDGFTSFVSSTMSEAAGKAA-NSSRKLYTVN 187
Query: 186 EVPFEDKTIYALVQCTRDLAANDCSRCLLSAIGDIPGCCYASIGARVMSRSCYLRYEFYP 245
+ +Y L+QCT DL DC CL S+I G + IGAR+ SC RYE YP
Sbjct: 188 TELTAYQNLYGLLQCTPDLTRADCLSCLQSSIN---GMALSRIGARLYWPSCTARYELYP 244
Query: 246 FYLGEKEQTK---------------------SSTNLGGKNNSSKIWMITVIAVGVGLVII 284
FY +T SS++L GK+ +S + ++ V+ + V L+ I
Sbjct: 245 FYNESAIETPPLPPPPPPPPPRESLVSTPPISSSSLPGKSGNSTVLVVAVVVLAV-LLFI 303
Query: 285 IFICYLCFLRNRQSKLF--------GDCISV----------------NFTDSNKLGEGGF 320
+ Y CFL ++ K F GD ++ +F +SNK+G GGF
Sbjct: 304 ALVGY-CFLAKKKKKTFDTASASEVGDDMATADSLQLDYRTIQTATNDFAESNKIGRGGF 362
Query: 321 GPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVKLLGFCVDGEEKLL 380
G VYKG S+G+EVA+KRLS S QG EF EV+++ KLQH+NLV+LLGF + GEE++L
Sbjct: 363 GEVYKGTFSNGKEVAVKRLSKNSRQGEAEFKTEVVVVAKLQHRNLVRLLGFSLQGEERIL 422
Query: 381 VYEYLPNGSLDVVLFE--QHAQLDWTKRLDIINGIARGILYLHEDSRLQIIHRDLKASNV 438
VYEY+PN SLD +LF+ + QLDW +R +II GIARGILYLH+DSRL IIHRDLKASN+
Sbjct: 423 VYEYMPNKSLDCLLFDPTKQIQLDWMQRYNIIGGIARGILYLHQDSRLTIIHRDLKASNI 482
Query: 439 LLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEGLYSIKSDVFGFGVL 498
LLD D+NPKI+DFGMARIF + + NT+ IVGTYGYMAPEYAM G +S+KSDV+ FGVL
Sbjct: 483 LLDADINPKIADFGMARIFGLDQTQDNTSRIVGTYGYMAPEYAMHGQFSMKSDVYSFGVL 542
Query: 499 LLEIITGIRNAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLLL--CPGDQFLRYMNIGL 556
+LEII+G +N+ F S LL +AW LW + K L+L DPL+ C + +R ++IGL
Sbjct: 543 VLEIISGRKNSSFGESDGAQDLLTHAWRLWTNKKALDLVDPLIAENCQNSEVVRCIHIGL 602
Query: 557 LCVQEDAFDRPTMSSVVLMLMNESVMLGQPGKPPFSV 593
LCVQED RP +S+V +ML + +V L P +P F +
Sbjct: 603 LCVQEDPAKRPAISTVFMMLTSNTVTLPVPRQPGFFI 639
>At4g05200 receptor protein kinase - like protein
Length = 675
Score = 458 bits (1178), Expect = e-129
Identities = 267/605 (44%), Positives = 363/605 (59%), Gaps = 48/605 (7%)
Query: 33 YEICSTRNIYANGSSFDNNLSNLLLSLPFNDSNSISKFGNTSSGIGLD--RVYGLYMCLD 90
Y IC Y+ SS+ NL +L SL ++ S F N ++G D RVYG+++C
Sbjct: 31 YHICPNTTTYSRNSSYLTNLRTVLSSLSSPNAAYASLFDNAAAGEENDSNRVYGVFLCRG 90
Query: 91 FVSNETCLKCVTNAIADTVKLCPQSKEAVVYEEFCQVRYSNKNFIGSLNVNGNIGKDNVQ 150
VS E C CV A +T++ CP+ K AV++ + C VRYSN++ +G + + + N Q
Sbjct: 91 DVSAEICRDCVAFAANETLQRCPREKVAVIWYDECMVRYSNQSIVGQMRIRPGVFLTNKQ 150
Query: 151 NISEPV--KFETSVNKLLNGLTKIASFNVSANMYATGEVPFED-KTIYALVQCTRDLAAN 207
NI+E +F S+ LL + A+ +S+ +AT + F +TIY+LVQCT DL
Sbjct: 151 NITENQVSRFNESLPALLIDVAVKAA--LSSRKFATEKANFTVFQTIYSLVQCTPDLTNQ 208
Query: 208 DCSRCLLSAIGDIPGCCYASIGARVMSRSCYLRYEFYPFY-----LGEKEQTKSST---- 258
DC CL I +P CC S+G RV++ SC RYE YPFY SST
Sbjct: 209 DCESCLRQVINYLPRCCDRSVGGRVIAPSCSFRYELYPFYNETIAAAPMAPPPSSTVTAP 268
Query: 259 --NLGGKNNSSKIWMITVIAVGVGL-VIIIFICYLCFL--RNRQSKLFGDCISVN----- 308
N+ + K + V A+ V + V ++ + +C+L R R +KL + ++
Sbjct: 269 PLNIPSEKGKGKNLTVIVTAIAVPVSVCVLLLGAMCWLLARRRNNKLSAETEDLDEDGIT 328
Query: 309 ------------------FTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEF 350
F++SNKLG GGFG VYKG L G+ VAIKRLS S QG+EEF
Sbjct: 329 STETLQFQFSAIEAATNKFSESNKLGHGGFGEVYKGQLITGETVAIKRLSQGSTQGAEEF 388
Query: 351 INEVMLILKLQHKNLVKLLGFCVDGEEKLLVYEYLPNGSLDVVLF--EQHAQLDWTKRLD 408
NEV ++ KLQH+NL KLLG+C+DGEEK+LVYE++PN SLD LF E+ LDW +R
Sbjct: 389 KNEVDVVAKLQHRNLAKLLGYCLDGEEKILVYEFVPNKSLDYFLFDNEKRRVLDWQRRYK 448
Query: 409 IINGIARGILYLHEDSRLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTT 468
II GIARGILYLH DSRL IIHRDLKASN+LLD DM+PKISDFGMARIF + +ANT
Sbjct: 449 IIEGIARGILYLHRDSRLTIIHRDLKASNILLDADMHPKISDFGMARIFGVDQTQANTKR 508
Query: 469 IVGTYGYMAPEYAMEGLYSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHLW 528
IVGTYGYM+PEYA+ G YS+KSDV+ FGVL+LE+ITG +N+ F L+ Y W LW
Sbjct: 509 IVGTYGYMSPEYAIHGKYSVKSDVYSFGVLVLELITGKKNSSFYEEDGLGDLVTYVWKLW 568
Query: 529 NDGKGLELRDPLLL--CPGDQFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNESVMLGQP 586
+ LEL D + ++ +R ++I LLCVQED+ +RP+M +++M+ + +V L P
Sbjct: 569 VENSPLELVDEAMRGNFQTNEVIRCIHIALLCVQEDSSERPSMDDILVMMNSFTVTLPIP 628
Query: 587 GKPPF 591
+ F
Sbjct: 629 KRSGF 633
>At4g23150 serine/threonine kinase - like protein
Length = 659
Score = 454 bits (1168), Expect = e-128
Identities = 274/658 (41%), Positives = 385/658 (57%), Gaps = 58/658 (8%)
Query: 4 YPIVFKFIQFFLVLYTLDFLSYLALADPPYEI---CSTRNIYANGSSFDNNLSNLLLSLP 60
+P +F F+ FL S+ A A P + C Y++ S++ NL LL SL
Sbjct: 5 FPFIFLFLFSFLT-------SFRASAQDPRFLAYYCPNATTYSSNSTYLTNLKTLLSSLS 57
Query: 61 FNDSNSISKFGNTSSGIGLDRVYGLYMCLDFVSNETCLKCVTNAIADTVKLCPQSKEAVV 120
+++ + F N + G LDRV GL++C VS E C CVT A+ +T CP +EAV
Sbjct: 58 SRNASYSTGFQNATVGQALDRVTGLFLCRGDVSPEVCRNCVTFAVNNTFSRCPNQREAVF 117
Query: 121 YEEFCQVRYSNKNFIGSLNVN-GNIGKDNVQNISEPVKFETS--VNKLLNGLTKIASFNV 177
Y E C +RYS+KN + + N G N +IS P++ + + N +L+ + +IA
Sbjct: 118 YYEECILRYSHKNILSTAITNEGEFILRNPNHIS-PIQNQINQFTNLVLSNMNQIA-IEA 175
Query: 178 SANMYATGEVPFED---KTIYALVQCTRDLAANDCSRCLLSAIGDIPGCCYASIGARVMS 234
+ N + E +T Y LVQCT DL+ +C CL S+I +P ++ IGAR
Sbjct: 176 ADNPRKFSTIKTELTALQTFYGLVQCTPDLSRQNCMNCLTSSINRMP---FSRIGARQFW 232
Query: 235 RSCYLRYEFYPFYLGEKEQTK-------SSTNLGGKNNSSKIWMITVIAVGVGLVIIIFI 287
SC RYE Y FY T +S +L K+ +S + ++ V+ V + + ++IFI
Sbjct: 233 PSCNSRYELYDFYNETAIGTPPPPLPPLASPSLSDKSGNSNVVVVAVV-VPIIVAVLIFI 291
Query: 288 CYLCFLRNRQSKLFGDCISVN-----------------------FTDSNKLGEGGFGPVY 324
CF R K +G +++ F+++NK+G GGFG VY
Sbjct: 292 AGYCFFAKRAKKTYGTTPALDEDDKTTIESLQLDYRAIQAATNDFSENNKIGRGGFGDVY 351
Query: 325 KGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVKLLGFCVDGEEKLLVYEY 384
KG S+G EVA+KRLS SEQG EF NEV+++ L+HKNLV++LGF ++ EE++LVYEY
Sbjct: 352 KGTFSNGTEVAVKRLSKTSEQGDTEFKNEVVVVANLRHKNLVRILGFSIEREERILVYEY 411
Query: 385 LPNGSLDVVLFE--QHAQLDWTKRLDIINGIARGILYLHEDSRLQIIHRDLKASNVLLDN 442
+ N SLD LF+ + QL WT+R II GIARGILYLH+DSRL IIHRDLKASN+LLD
Sbjct: 412 VENKSLDNFLFDPAKKGQLYWTQRYHIIGGIARGILYLHQDSRLTIIHRDLKASNILLDA 471
Query: 443 DMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEGLYSIKSDVFGFGVLLLEI 502
DMNPKI+DFGMARIF + + NT+ IVGTYGYM+PEYAM G +S+KSDV+ FGVL+LEI
Sbjct: 472 DMNPKIADFGMARIFGMDQTQQNTSRIVGTYGYMSPEYAMRGQFSMKSDVYSFGVLVLEI 531
Query: 503 ITGIRNAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLLL--CPGDQFLRYMNIGLLCVQ 560
I+G +N F + L+ +AW LW +G L+L DP + C + +R +IGLLCVQ
Sbjct: 532 ISGRKNNSFIETDDAQDLVTHAWRLWRNGTALDLVDPFIADSCRKSEVVRCTHIGLLCVQ 591
Query: 561 EDAFDRPTMSSVVLMLMNESVMLGQPGKPPFSVGRLNFIDQNELDLEEEYSVNFLTVS 618
ED RP MS++ +ML + ++ L P +P F V + N LD ++ + +TVS
Sbjct: 592 EDPVKRPAMSTISVMLTSNTMALPAPQQPGFFV--RSRPGTNRLDSDQSTTNKSVTVS 647
>At4g23231 unknown protein
Length = 627
Score = 431 bits (1108), Expect = e-121
Identities = 251/631 (39%), Positives = 367/631 (57%), Gaps = 57/631 (9%)
Query: 8 FKFIQFFLVLYTLDFLSYLALADPPYEICSTRNIYANGSSFDNNLSNLLLSLPFNDSNSI 67
F F+ FFL +T + + IC Y+ S + NL LL SL +++
Sbjct: 7 FIFLFFFLTSFTASVENVFYIK----HICPNTTTYSRNSPYLTNLRTLLSSLSAPNASYS 62
Query: 68 SKFGNTSSGIGLDRVYGLYMCLDFVSNETCLKCVTNAIADTVKLCPQSKEAVVYEEFCQV 127
+ F + +G DRV GL++C VS C CV +I DT+ CP +++V Y + C +
Sbjct: 63 TGFQSARAGQAPDRVTGLFLCRGDVSPAVCRNCVAFSINDTLVQCPSERKSVFYYDECML 122
Query: 128 RYSNKNFIGSL-------NVNGNIGKDNVQNISEPVKFETSVNKLLNGLTKIASFNVSAN 180
RYS++N + +L +NGNI D ++ +F+ V+ +N + + +
Sbjct: 123 RYSDQNILSTLAYDGAWIRMNGNISIDQ----NQMNRFKDFVSSTMNQAA-VKAASSPRK 177
Query: 181 MYATGEVPFEDKTIYALVQCTRDLAANDCSRCLLSAIGDIPGCCYASIGARVMSRSCYLR 240
Y +T+Y LVQCT DL DC CL S+I +P G R + SC R
Sbjct: 178 FYTVKATWTALQTLYGLVQCTPDLTRQDCFSCLESSIKLMP---LYKTGGRTLYSSCNSR 234
Query: 241 YEFYPFYLGEKEQTK--------------SSTNLGGKNNSSKIWMITVIA--VGVGLVII 284
YE + FY +T+ +S +L GK+ +S + ++ ++ + L++I
Sbjct: 235 YELFAFYNETTVRTQQAPPPLPPSSTPLVTSPSLPGKSWNSNVLVVAIVLTILVAALLLI 294
Query: 285 IFICYLCFLRNRQSK---LFGDCISVN---------------FTDSNKLGEGGFGPVYKG 326
C+ ++N GD I+ F+++NK+G+GGFG VYKG
Sbjct: 295 AGYCFAKRVKNSSDNAPAFDGDDITTESLQLDYRMIRAATNKFSENNKIGQGGFGEVYKG 354
Query: 327 ILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVKLLGFCVDGEEKLLVYEYLP 386
S+G EVA+KRLS S QG EF NEV+++ KLQH+NLV+LLGF + G E++LVYEY+P
Sbjct: 355 TFSNGTEVAVKRLSKSSGQGDTEFKNEVVVVAKLQHRNLVRLLGFSIGGGERILVYEYMP 414
Query: 387 NGSLDVVLFE--QHAQLDWTKRLDIINGIARGILYLHEDSRLQIIHRDLKASNVLLDNDM 444
N SLD LF+ + QLDWT+R +I GIARGILYLH+DSRL IIHRDLKASN+LLD DM
Sbjct: 415 NKSLDYFLFDPAKQNQLDWTRRYKVIGGIARGILYLHQDSRLTIIHRDLKASNILLDADM 474
Query: 445 NPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEGLYSIKSDVFGFGVLLLEIIT 504
NPK++DFG+ARIF + + NT+ IVGT+GYMAPEYA+ G +S+KSDV+ FGVL+LEII+
Sbjct: 475 NPKLADFGLARIFGMDQTQENTSRIVGTFGYMAPEYAIHGQFSVKSDVYSFGVLVLEIIS 534
Query: 505 GIRNAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLLL--CPGDQFLRYMNIGLLCVQED 562
G +N F + L+ +AW LW++G L+L DP+++ C + +R ++I LLCVQED
Sbjct: 535 GKKNNSFYETDGAHDLVTHAWRLWSNGTALDLVDPIIIDNCQKSEVVRCIHICLLCVQED 594
Query: 563 AFDRPTMSSVVLMLMNESVMLGQPGKPPFSV 593
+RP +S++ +ML + +V L P +P F V
Sbjct: 595 PAERPILSTIFMMLTSNTVTLPVPLQPGFPV 625
>At4g38830 receptor-like protein kinase - like protein
Length = 665
Score = 421 bits (1082), Expect = e-118
Identities = 254/612 (41%), Positives = 352/612 (57%), Gaps = 55/612 (8%)
Query: 31 PPYEICS--TRNIYANGSSFDNNLSNLLLSLPFNDSNSISKFGNTSSGIGLDRVYGLYMC 88
P +ICS T N N + + NL L+ SL N ++ F N S G ++V + C
Sbjct: 26 PLNQICSNVTGNFTVN-TPYAVNLDRLISSLSSLRRN-VNGFYNISVGDSDEKVNSISQC 83
Query: 89 LDFVSNETCLKCVTNAIADTVKLCPQSKEAVVYEEFCQVRYSNKNFIGSLNVNGNIGKDN 148
V E C+ C+ A V LCP KEA+++ + C RYSN+ L ++ +
Sbjct: 84 RGDVKLEVCINCIAMAGKRLVTLCPVQKEAIIWYDKCTFRYSNRTIFNRLEISPHTSITG 143
Query: 149 VQNIS-EPVKFETSVNKLLNGLTKIAS-FNVSANMYATGEVPFED-KTIYALVQCTRDLA 205
+N + + +E S+ LL GL AS S + GE +T++ LVQCT D++
Sbjct: 144 TRNFTGDRDSWEKSLRGLLEGLKNRASVIGRSKKNFVVGETSGPSFQTLFGLVQCTPDIS 203
Query: 206 ANDCSRCLLSAIGDIPGCCYASIGARVMSRSCYLRYEFYPFY----------------LG 249
DCS CL I IP CC +G+ VMS SC L Y + FY
Sbjct: 204 EEDCSYCLSQGIAKIPSCCDMKMGSYVMSPSCMLAYAPWRFYDPVDTDDPSSVPATPSRP 263
Query: 250 EKEQTKSSTNLGGKNNSSKIWMITVIAVGVGLVIIIFICYLCFL--------RNRQSKLF 301
K +T+S T G KN +I A V +V++ + + FL RN ++K
Sbjct: 264 PKNETRSVTQ-GDKNRGVPKALIFASA-SVAIVVLFIVLLVVFLKLRRKENIRNSENKHE 321
Query: 302 GDCISVN---------------FTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQG 346
+ IS + F+ NKLGEGGFG VYKG+LSDGQ++A+KRLS ++QG
Sbjct: 322 NENISTDSMKFDFSVLQDATSHFSLENKLGEGGFGAVYKGVLSDGQKIAVKRLSKNAQQG 381
Query: 347 SEEFINEVMLILKLQHKNLVKLLGFCVDGEEKLLVYEYLPNGSLDVVLFE--QHAQLDWT 404
EF NE +L+ KLQH+NLVKLLG+ ++G E+LLVYE+LP+ SLD +F+ Q +L+W
Sbjct: 382 ETEFKNEFLLVAKLQHRNLVKLLGYSIEGTERLLVYEFLPHTSLDKFIFDPIQGNELEWE 441
Query: 405 KRLDIINGIARGILYLHEDSRLQIIHRDLKASNVLLDNDMNPKISDFGMARIF-AGSEGE 463
R II G+ARG+LYLH+DSRL+IIHRDLKASN+LLD +M PKI+DFGMAR+F +
Sbjct: 442 IRYKIIGGVARGLLYLHQDSRLRIIHRDLKASNILLDEEMTPKIADFGMARLFDIDHTTQ 501
Query: 464 ANTTTIVGTYGYMAPEYAMEGLYSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAY 523
T IVGT+GYMAPEY M G +S K+DV+ FGVL+LEII+G +N+GF + L+++
Sbjct: 502 RYTNRIVGTFGYMAPEYVMHGQFSFKTDVYSFGVLVLEIISGKKNSGFSSEDSMGDLISF 561
Query: 524 AWHLWNDGKGLELRDPLLLC----PGDQFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNE 579
AW W +G L L D +L+ + +R +NIGLLCVQE +RP+M+SVVLML
Sbjct: 562 AWRNWKEGVALNLVDKILMTMSSYSSNMIMRCINIGLLCVQEKVAERPSMASVVLMLDGH 621
Query: 580 SVMLGQPGKPPF 591
++ L +P KP F
Sbjct: 622 TIALSEPSKPAF 633
>At4g23130 receptor-like protein kinase 6 (RLK6)
Length = 659
Score = 411 bits (1056), Expect = e-115
Identities = 251/624 (40%), Positives = 358/624 (57%), Gaps = 53/624 (8%)
Query: 14 FLVLYTLDFLSYLA-LADPPY--EICSTRNIYANGSSFDNNLSNLLLSLPFNDSNSISKF 70
FL+ + + L A L DP Y +C+ R + S + +NL LL SL N++
Sbjct: 11 FLLTFFIGSLRVSAQLQDPTYVGHVCTNR--ISRNSIYFSNLQTLLTSLSSNNAYFSLGS 68
Query: 71 GNTSSGIGLDRVYGLYMCLDFVSNETCLKCVTNAIADTVKLCPQSKEAVVYEEFCQVRYS 130
+ + G D V+GLY+C +S E+C +CV A DT CP KE ++ + C + YS
Sbjct: 69 HSLTKGQNSDMVFGLYLCKGDLSPESCRECVIFAAKDTRSRCPGGKEFLIQYDECMLGYS 128
Query: 131 NKNFIGSLNVNGNIGKDNVQNIS--EPVKFETSVNKLLNGLTKIASFNVSANMYATGEVP 188
++N I N Q ++ + +F +V L+ + A+ N ++ +A +
Sbjct: 129 DRNIFMDTVTTTTIITWNTQKVTADQSDRFNDAVLSLMKKSAEEAA-NSTSKKFAVKKSD 187
Query: 189 FED-KTIYALVQCTRDLAANDCSRCLLSAIGDIPGCCYASIGARVMSRSCYLRYEFYPFY 247
F +++YA VQC DL + DC CL +I ++ + +G R + SC RYE YPFY
Sbjct: 188 FSSSQSLYASVQCIPDLTSEDCVMCLQQSIKEL---YFNKVGGRFLVPSCNSRYEVYPFY 244
Query: 248 LGEKEQTKSSTNLG--------------GKNNSSKIWMITVIAVGVGLVIIIFICYLCFL 293
E T + GK +S + +I ++ V V + ++I + F
Sbjct: 245 KETIEGTVLPPPVSAPPLPLVSTPSFPPGKGKNSTVIIIAIV-VPVAISVLICVAVFSFH 303
Query: 294 RNRQSKLFGD---------------------CISVNFTDSNKLGEGGFGPVYKGILSDGQ 332
++++K D + F+ NKLG+GGFG VYKG L +G
Sbjct: 304 ASKRAKKTYDTPEEDDITTAGSLQFDFKVIEAATDKFSMCNKLGQGGFGQVYKGTLPNGV 363
Query: 333 EVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVKLLGFCVDGEEKLLVYEYLPNGSLDV 392
+VA+KRLS S QG +EF NEV+++ KLQH+NLVKLLGFC++ EEK+LVYE++ N SLD
Sbjct: 364 QVAVKRLSKTSGQGEKEFKNEVVVVAKLQHRNLVKLLGFCLEREEKILVYEFVSNKSLDY 423
Query: 393 VLFEQH--AQLDWTKRLDIINGIARGILYLHEDSRLQIIHRDLKASNVLLDNDMNPKISD 450
LF+ +QLDWT R II GIARGILYLH+DSRL IIHRDLKA N+LLD DMNPK++D
Sbjct: 424 FLFDSRMQSQLDWTTRYKIIGGIARGILYLHQDSRLTIIHRDLKAGNILLDADMNPKVAD 483
Query: 451 FGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEGLYSIKSDVFGFGVLLLEIITGIRNAG 510
FGMARIF + EA+T +VGTYGYM+PEYAM G +S+KSDV+ FGVL+LEII+G +N+
Sbjct: 484 FGMARIFEIDQTEAHTRRVVGTYGYMSPEYAMYGQFSMKSDVYSFGVLVLEIISGRKNSS 543
Query: 511 -FCYSKTTPSLLAYAWHLWNDGKGLELRDPLL--LCPGDQFLRYMNIGLLCVQEDAFDRP 567
+ + +L+ Y W LW+DG L+L D ++ +R ++I LLCVQED +RP
Sbjct: 544 LYQMDASFGNLVTYTWRLWSDGSPLDLVDSSFRDSYQRNEIIRCIHIALLCVQEDTENRP 603
Query: 568 TMSSVVLMLMNESVMLGQPGKPPF 591
TMS++V ML S+ L P P F
Sbjct: 604 TMSAIVQMLTTSSIALAVPQPPGF 627
>At4g21410 serine/threonine kinase like protein
Length = 679
Score = 407 bits (1047), Expect = e-114
Identities = 262/681 (38%), Positives = 359/681 (52%), Gaps = 62/681 (9%)
Query: 1 MFHYPIVFKFIQFFLVLYTLDFLSYLALADPPYEICSTRNIYANGSSFDNNLSNLLLSLP 60
M H ++F F F + F + P C R + S+F NL+ L+ SL
Sbjct: 1 MEHVRVIFFFACFLTLAPFHAFAQVDSYEFDPDFNCVDRGNFTANSTFAGNLNRLVSSLS 60
Query: 61 FNDSNSISKFGNTSSGIGLDRVYGLYMCLDFVSNETCLKCVTNAIADTVKLCPQSKEAVV 120
S + + +S +R Y + +C V + C+ C+ A + K CP +K+AVV
Sbjct: 61 SLKSQAYGFYNLSSGDSSGERAYAIGLCRREVKRDDCVSCIQTAARNLTKQCPLTKQAVV 120
Query: 121 YEEFCQVRYSNKNFIGSLNVNGNIGKDNVQNIS-EPVKFETSVNKLLNGLTKIASFNVSA 179
+ C RYSN+ G N + IS FE LL+ L IA+
Sbjct: 121 WYTHCMFRYSNRTIYGRKETNPTKAFIAGEEISANRDDFERLQRGLLDRLKGIAAAGGPN 180
Query: 180 NMYA--TGEVPFEDKTIYALVQCTRDLAANDCSRCLLSAIGDIPGCCYASIGARVMSRSC 237
YA G + Y VQCT DL+ DC+ CL+ +IP CC A IG R S SC
Sbjct: 181 RKYAQGNGSASAGYRRFYGTVQCTPDLSEQDCNDCLVFGFENIPSCCDAEIGLRWFSPSC 240
Query: 238 YLRYEFYPFY--------------LGEKEQTKSSTNLGGK-NNSSKIWMITVIAVGVGLV 282
R+E + FY + Q+ + T GK SK+ I I + + LV
Sbjct: 241 NFRFETWRFYEFDADLEPDPPAIQPADSPQSAARTERTGKGKGGSKV--IIAIVIPILLV 298
Query: 283 IIIFICYLCFLRNRQSKL--------------------FGDCISV------------NFT 310
++ IC L+ R++K F + S+ NF+
Sbjct: 299 ALLAICLCLVLKWRKNKSGYKNKVLGKSPLSGSIAEDEFSNTESLLVHFETLKTATDNFS 358
Query: 311 DSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVKLLG 370
N+LG GGFG VYKG+ GQE+A+KRLS S QG EF NE++L+ KLQH+NLV+L+G
Sbjct: 359 SENELGRGGFGSVYKGVFPQGQEIAVKRLSGNSGQGDNEFKNEILLLAKLQHRNLVRLIG 418
Query: 371 FCVDGEEKLLVYEYLPNGSLDVVLF--EQHAQLDWTKRLDIINGIARGILYLHEDSRLQI 428
FC+ GEE+LLVYE++ N SLD +F E+ LDW R +I GIARG+LYLHEDSR +I
Sbjct: 419 FCIQGEERLLVYEFIKNASLDQFIFDTEKRQLLDWVVRYKMIGGIARGLLYLHEDSRFRI 478
Query: 429 IHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEAN--TTTIVGTYGYMAPEYAMEGLY 486
IHRDLKASN+LLD +MNPKI+DFG+A++F + + T+ I GTYGYMAPEYAM G +
Sbjct: 479 IHRDLKASNILLDQEMNPKIADFGLAKLFDSGQTMTHRFTSRIAGTYGYMAPEYAMHGQF 538
Query: 487 SIKSDVFGFGVLLLEIITGIR--NAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLLLCP 544
S+K+DVF FGVL++EIITG R N G + LL++ W W + L + DP L
Sbjct: 539 SVKTDVFSFGVLVIEIITGKRNNNGGSNGDEDAEDLLSWVWRSWREDTILSVIDPSLTAG 598
Query: 545 G-DQFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNESVMLGQPGKPPFSVGRLNFIDQNE 603
++ LR ++IGLLCVQE A RPTM++V LML + S L P +P F + + I N
Sbjct: 599 SRNEILRCIHIGLLCVQESAATRPTMATVSLMLNSYSFTLPTPLRPAFVLESV-VIPSNV 657
Query: 604 LDLEE--EYSVNFLTVSDILP 622
E + S N +TVS+ P
Sbjct: 658 SSSTEGLQMSSNDVTVSEFSP 678
>At4g23260 unknown protein
Length = 659
Score = 396 bits (1018), Expect = e-110
Identities = 242/625 (38%), Positives = 349/625 (55%), Gaps = 44/625 (7%)
Query: 9 KFIQFFLVLYTLDFLSYLALADPPYEICSTRNIYANGSSFDNNLSNLLLSLPFNDSNSIS 68
K + L + +S+ A++ +T + S +D N +L +L N +
Sbjct: 4 KSCELVLCFFVFFVISFSAISVSAQTCDNTTGTFIPNSPYDKNRRLILSTLASNVTAQEG 63
Query: 69 KFGNTSSGIGLDRVYGLYMCLDFVSNETCLKCVTNAIADTVKLCPQSKEAVVY---EEFC 125
F S GI D+V+ MC + C C+ + ++ C +A + E C
Sbjct: 64 YFIG-SIGIAPDQVFATGMCAPGSERDVCSLCIRSTSESLLQSCLDQADAFFWSGEETLC 122
Query: 126 QVRYSNKNFIGSLNVNG-----NIGKDNVQNISEPVKFETSVNKLLNGLTKIASFNVSAN 180
VRY+N+ F G L ++ N G+ N +++ + ++ G+T +S +++
Sbjct: 123 LVRYANRPFSGLLVMDPLGAIFNTGELNTNQTVFDIEWNNLTSSMIAGITSSSSGGNNSS 182
Query: 181 MYATGEVPF--EDKTIYALVQCTRDLAANDCSRCLLSAIGDIPGCCYASIGARVMSRSCY 238
Y + ++ + K I AL+QCT D+++ DC+ CL + D CC G + +C+
Sbjct: 183 KYYSDDIALVPDFKNISALMQCTPDVSSEDCNTCLRQNVVDYDNCCRGHQGGVMSRPNCF 242
Query: 239 LRYEFYPFYLGEKEQ-----------TKSSTNLGGKNNSSKIWMITVIAVGVGLVIIIFI 287
R+E YPF G +Q T S N S+I + A+ V V+ I +
Sbjct: 243 FRWEVYPFS-GAIDQINLPKSPPPSVTSPSPIANITKNDSRISGGKIAAIVVVTVVTIIL 301
Query: 288 CYLCFL---RNRQSKLFG-------------DCISVNFTDSNKLGEGGFGPVYKGILSDG 331
L F+ R +Q + + + NF++ NKLG+GGFG VYKG+L +G
Sbjct: 302 VVLGFVISNRRKQKQEMDLPTESVQFDLKTIESATSNFSERNKLGKGGFGEVYKGMLMNG 361
Query: 332 QEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVKLLGFCVDGEEKLLVYEYLPNGSLD 391
E+A+KRLS S QG EF NEV+++ KLQH NLV+LLGF + GEEKLLVYE++ N SLD
Sbjct: 362 TEIAVKRLSKTSGQGEVEFKNEVVVVAKLQHINLVRLLGFSLQGEEKLLVYEFVSNKSLD 421
Query: 392 VVLFE--QHAQLDWTKRLDIINGIARGILYLHEDSRLQIIHRDLKASNVLLDNDMNPKIS 449
LF+ + QLDWT R +II GI RGILYLH+DSRL+IIHRDLKASN+LLD DMNPKI+
Sbjct: 422 YFLFDPTKRNQLDWTMRRNIIGGITRGILYLHQDSRLKIIHRDLKASNILLDADMNPKIA 481
Query: 450 DFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEGLYSIKSDVFGFGVLLLEIITGIRNA 509
DFGMARIF + ANT +VGT+GYM+PEY G +S+KSDV+ FGVL+LEII+G +N+
Sbjct: 482 DFGMARIFGVDQTVANTGRVVGTFGYMSPEYVTHGQFSMKSDVYSFGVLILEIISGKKNS 541
Query: 510 GFC-YSKTTPSLLAYAWHLWNDGKGLELRDPLLL--CPGDQFLRYMNIGLLCVQEDAFDR 566
F +L+ Y W LW + EL DP + ++ +RY++IGLLCVQE+ DR
Sbjct: 542 SFYQMDGLVNNLVTYVWKLWENKSLHELLDPFINQDFTSEEVIRYIHIGLLCVQENPADR 601
Query: 567 PTMSSVVLMLMNESVMLGQPGKPPF 591
PTMS++ ML N S+ L P P F
Sbjct: 602 PTMSTIHQMLTNSSITLPVPLPPGF 626
>At4g23250 protein kinase - like protein
Length = 998
Score = 395 bits (1015), Expect = e-110
Identities = 241/619 (38%), Positives = 347/619 (55%), Gaps = 58/619 (9%)
Query: 23 LSYLALADPPYEIC-STRNIYANGSSFDNNLSNLLLSLPFNDSNSISKFGNTSSGIGLDR 81
LS+ ++ + C +T + S++DNN LLLS ++ + + + N S G+G DR
Sbjct: 17 LSFRVISVSAQQTCDNTAGSFKPNSTYDNN-RRLLLSTFASNVTAQNGYFNGSFGLGTDR 75
Query: 82 VYGLYMCLDFVSNETCLKCVTNAIADTVKLCPQSKEAVVY---EEFCQVRYSNKNFIGSL 138
VY + MC + C C+ N +++C + + E C VRYSNK+F G L
Sbjct: 76 VYAMGMCAPGAEPDVCSNCIKNTAEGLLQICLNQTDGFSWSGEETLCLVRYSNKSFSGLL 135
Query: 139 NVNGNIGKDNVQNI--SEPVKFETSVNKLLNGLTKIASFNVSANMYATGEVPFEDKTIYA 196
+ + NV I + +F++ ++L+ + AS + A G +
Sbjct: 136 GLEPSNDFFNVNEIRKEDQKEFDSVFDELMFRTIQGASSSDVAPEPVYGNIS-------V 188
Query: 197 LVQCTRDLAANDCSRCLLSAIGDIPGCCYASIGARVMSRSCYLRYEFYPFYLG------- 249
++QCT D+++ DC+ CL ++ G ++ SC+ R+E Y F+
Sbjct: 189 VMQCTPDVSSKDCNLCLERSLDFYKKWYNGKRGTIILRPSCFFRWELYTFFGAFDSINAR 248
Query: 250 --------------EKEQTKSSTNLGGKNNSSKIWM------ITVIAVGVGLVIIIFICY 289
+ + TN+ KN S + V+ V V +++I+
Sbjct: 249 HPPPPPRPLSPPPLKTPSVTNQTNITKKNGKSFFGISGGTIAAIVVVVVVTIILIVVGLV 308
Query: 290 LCFLRNRQSKLFGDCISV------------NFTDSNKLGEGGFGPVYKGILSDGQEVAIK 337
+C R ++ ++ SV NF++ NKLG GGFG VYKG+L +G E+A+K
Sbjct: 309 ICKRRKQKQEIELPTESVQFDLKTIEAATGNFSEHNKLGAGGFGEVYKGMLLNGTEIAVK 368
Query: 338 RLSICSEQGSEEFINEVMLILKLQHKNLVKLLGFCVDGEEKLLVYEYLPNGSLDVVLFEQ 397
RLS S QG EF NEV+++ KLQH NLV+LLGF + GEEKLLVYE++PN SLD LF+
Sbjct: 369 RLSKTSGQGEIEFKNEVVVVAKLQHINLVRLLGFSLQGEEKLLVYEFVPNKSLDYFLFDP 428
Query: 398 HA--QLDWTKRLDIINGIARGILYLHEDSRLQIIHRDLKASNVLLDNDMNPKISDFGMAR 455
+ QLDWT R +II GI RGILYLH+DSRL+IIHRDLKASN+LLD DMNPKI+DFGMAR
Sbjct: 429 NKRNQLDWTVRRNIIGGITRGILYLHQDSRLKIIHRDLKASNILLDADMNPKIADFGMAR 488
Query: 456 IFAGSEGEANTTTIVGTYGYMAPEYAMEGLYSIKSDVFGFGVLLLEIITGIRNAGFC-YS 514
IF + ANT +VGT+GYM+PEY G +S+KSDV+ FGVL+LEII+G +N+ F
Sbjct: 489 IFGVDQTVANTARVVGTFGYMSPEYVTHGQFSMKSDVYSFGVLILEIISGKKNSSFYQMD 548
Query: 515 KTTPSLLAYAWHLWNDGKGLELRDPLLL--CPGDQFLRYMNIGLLCVQEDAFDRPTMSSV 572
+L+ Y W LW + EL DP + C D+ +RY++IGLLCVQE+ DRPTMS++
Sbjct: 549 GLVNNLVTYVWKLWENKTMHELIDPFIKEDCKSDEVIRYVHIGLLCVQENPADRPTMSTI 608
Query: 573 VLMLMNESVMLGQPGKPPF 591
+L S+ L P P F
Sbjct: 609 HQVLTTSSITLPVPQPPGF 627
Score = 100 bits (248), Expect = 3e-21
Identities = 75/283 (26%), Positives = 122/283 (42%), Gaps = 19/283 (6%)
Query: 36 CSTRNIYANGSSFDNNLSNLLLSLPFNDSNSISKFGNTSSGIGLDRVYGLYMCLDFVSNE 95
C ++ +D N LL SL N S + F N S G G DR+Y C+ E
Sbjct: 692 CGKTGLFKPNDKYDINRHLLLSSLASNVS-ARGGFYNASIGQGPDRLYASGTCIQGSEPE 750
Query: 96 TCLKCVTNAIADTVKLCPQSKEAVVYEEF-----CQVRYSNKNFIGSLNVNGNIGKDNVQ 150
C C+ +A +K C EA+ + F C +RYSN++F G L + N
Sbjct: 751 LCSACIDSAFIRVIKKCHNQTEALDWSSFNEEYPCMIRYSNRSFFGLLEMTPFFKNYNAT 810
Query: 151 NISEPV-KFETSVNKLLNGLTKIASFNVSANMYATGEVPFEDKTIYALVQCTRDLAANDC 209
+ + +F L+ G+ A + + Y G +T+YA V C++D++ +C
Sbjct: 811 DFQVNLTEFYQKWEALMLGVIADAISSPNPKFYGAGTGKIGIQTVYAFVLCSKDISPWNC 870
Query: 210 SRCLLSAIGDIPGCCYASIGARVMSRSCYLRYEFYPFY-----------LGEKEQTKSST 258
SRCL + + C S SCY+R++ Y FY EK + +
Sbjct: 871 SRCLRGNVDNYKLSCSGKPRGHSFSPSCYMRWDLYQFYGFIEYRASPTLPREKGRISELS 930
Query: 259 NLGGKNNSSKIWMITVIAVGVGLVIIIFICYLCFLRNRQSKLF 301
+ GGK ++ I ITV A+ + +++ + Y R + + F
Sbjct: 931 DDGGKISTRNILGITV-ALAFFITVLLVLGYALSRRRKAYQEF 972
>At4g11460 serine/threonine kinase-like protein
Length = 700
Score = 395 bits (1014), Expect = e-110
Identities = 235/613 (38%), Positives = 347/613 (56%), Gaps = 56/613 (9%)
Query: 34 EICSTR-NIYANGSSFDNNLSNLLLSLPFNDSNSISKFGNTSSGIGLDRVYGLYMCLDFV 92
++CS + + G +FD N +L SLP ++ + F N S G D++Y + MC+
Sbjct: 25 QLCSEKFGTFTPGGTFDKNRRIILSSLP-SEVTAQDGFYNASIGTDPDQLYAMGMCIPGA 83
Query: 93 SNETCLKCVTNAIADTVKLCPQSKEAVVY----EEFCQVRYSNKNFIGSLNVNGNIGKDN 148
+ C C+ + ++ CP A+ + + C RY N+ L++ N
Sbjct: 84 KQKLCRDCIMDVTRQLIQTCPNQTAAIHWSGGGKTVCMARYYNQPSSRPLDLESVSIGYN 143
Query: 149 VQNISEPVK-FETSVNKLL-NGLTKIASFNV------SANMYATGEVPFED-KTIYALVQ 199
V N+S + F+ +L+ + +TK +S ++ ++ YA E + + +YAL+Q
Sbjct: 144 VGNLSTNLTDFDRLWERLIAHMVTKASSASIKYLSFDNSRFYAADETNLTNSQMVYALMQ 203
Query: 200 CTRDLAANDCSRCLLSAIGDIPGCCYASIGARVMSRSCYLRYEFYPFY------------ 247
CT D++ ++C+ CL ++ D GCC+ G V SC R++ YPF
Sbjct: 204 CTPDVSPSNCNTCLKQSVDDYVGCCHGKQGGYVYRPSCIFRWDLYPFNGAFDLLTLAPPP 263
Query: 248 LGEKEQTKSSTNLGGK--NNSSKIWMITVIAVGVGLVIIIFICYLCFLRNR----QSKLF 301
+ + TN K + + I ++ V+A+ + + ++ +C R + S+
Sbjct: 264 SSQLQSPPPVTNKDEKTIHTGTIIGIVIVVAMVIIMALLALGVSVCRSRKKYQAFASETA 323
Query: 302 GDCISV---------------NFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQG 346
D +V NF SNK+G+GGFG VYKG LS+G EVA+KRLS S+QG
Sbjct: 324 DDITTVGYLQFDIKDIEAATSNFLASNKIGQGGFGEVYKGTLSNGTEVAVKRLSRTSDQG 383
Query: 347 SEEFINEVMLILKLQHKNLVKLLGFCVDGEEKLLVYEYLPNGSLDVVLF-----EQHAQL 401
EF NEV+L+ KLQH+NLV+LLGF + GEEK+LV+E++PN SLD LF + QL
Sbjct: 384 ELEFKNEVLLVAKLQHRNLVRLLGFALQGEEKILVFEFVPNKSLDYFLFGSTNPTKKGQL 443
Query: 402 DWTKRLDIINGIARGILYLHEDSRLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSE 461
DWT+R +II GI RG+LYLH+DSRL IIHRD+KASN+LLD DMNPKI+DFGMAR F +
Sbjct: 444 DWTRRYNIIGGITRGLLYLHQDSRLTIIHRDIKASNILLDADMNPKIADFGMARNFRDHQ 503
Query: 462 GEANTTTIVGTYGYMAPEYAMEGLYSIKSDVFGFGVLLLEIITGIRNAGFC-YSKTTPSL 520
E +T +VGT+GYM PEY G +S KSDV+ FGVL+LEI++G +N+ F + +L
Sbjct: 504 TEDSTGRVVGTFGYMPPEYVAHGQFSTKSDVYSFGVLILEIVSGRKNSSFYQMDGSVCNL 563
Query: 521 LAYAWHLWNDGKGLELRDPLL--LCPGDQFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMN 578
+ Y W LWN LEL DP + D+ R ++IGLLCVQE+ +RP +S++ ML N
Sbjct: 564 VTYVWRLWNTDSSLELVDPAISGSYEKDEVTRCIHIGLLCVQENPVNRPALSTIFQMLTN 623
Query: 579 ESVMLGQPGKPPF 591
S+ L P P F
Sbjct: 624 SSITLNVPQPPGF 636
>At4g11470 serine/threonine kinase-like protein
Length = 666
Score = 380 bits (977), Expect = e-106
Identities = 224/603 (37%), Positives = 329/603 (54%), Gaps = 49/603 (8%)
Query: 36 CSTRNIYANGSSFDNNLSNLLLSLPFNDSNSISKFGNTSSGIGLDRVYGLYMCLDFVSNE 95
C + ++D N +L +L N S+ ++F N S G G R+Y L +C+
Sbjct: 26 CGESVFFRPNGNYDTNRRLVLSTLASNVSSQNNRFYNVSVGEGAGRIYALGLCIPGSDPR 85
Query: 96 TCLKCVTNAIADTVKLCPQSKEAVVY------EEFCQVRYSNKNFIGSLNVNGNIGKDNV 149
C C+ A ++ CP ++ + + C VRYSN +F + + N
Sbjct: 86 VCSDCIQLASQGLLQTCPNQTDSFYWTGDNADKTLCFVRYSNNSFFNKMALEPTHAVYNT 145
Query: 150 QNISEPVKFETSV-NKLLNGL-TKIASFNVSANMYAT-GEVPFEDKTIYALVQCTRDLAA 206
+ T + +N + T++ A++ + P IYAL+QC +++
Sbjct: 146 MRFQGNLTAYTRTWDAFMNFMFTRVGQTRYLADISPRINQEPLSPDLIYALMQCIPGISS 205
Query: 207 NDCSRCLLSAIGDIPGCCYASIGARVMSRSCYLRYEFYPFY--LGEKEQTKSSTNL---- 260
DC CL + D CC IG V CY R++ Y +Y G++ ++ T L
Sbjct: 206 EDCETCLGKCVDDYQSCCNGFIGGVVNKPVCYFRWDGYKYYGAFGDEAPSQPPTPLPLPP 265
Query: 261 ------GGKNNSSKIWMITVIAVGVGLVIIIFICYLCFLRNRQS---------------- 298
GK S+ + I I V + +++ L + RQS
Sbjct: 266 PPPRDPDGKKISTGV--IVAIVVSAVIFVVLVALGLVIWKRRQSYKTLKYHTDDDMTSPQ 323
Query: 299 KLFGDCISV-----NFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINE 353
L D ++ NF+ +NKLG+GGFG VYKG+L + E+A+KRLS S QG++EF NE
Sbjct: 324 SLQFDFTTIEVATDNFSRNNKLGQGGFGEVYKGMLPNETEIAVKRLSSNSGQGTQEFKNE 383
Query: 354 VMLILKLQHKNLVKLLGFCVDGEEKLLVYEYLPNGSLDVVLFE--QHAQLDWTKRLDIIN 411
V+++ KLQHKNLV+LLGFC++ +E++LVYE++ N SLD LF+ +QLDW +R +II
Sbjct: 384 VVIVAKLQHKNLVRLLGFCIERDEQILVYEFVSNKSLDYFLFDPKMKSQLDWKRRYNIIG 443
Query: 412 GIARGILYLHEDSRLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVG 471
G+ RG+LYLH+DSRL IIHRD+KASN+LLD DMNPKI+DFGMAR F + E T +VG
Sbjct: 444 GVTRGLLYLHQDSRLTIIHRDIKASNILLDADMNPKIADFGMARNFRVDQTEDQTGRVVG 503
Query: 472 TYGYMAPEYAMEGLYSIKSDVFGFGVLLLEIITGIRNAGFC-YSKTTPSLLAYAWHLWND 530
T+GYM PEY G +S KSDV+ FGVL+LEI+ G +N+ F + +L+ + W LWN+
Sbjct: 504 TFGYMPPEYVTHGQFSTKSDVYSFGVLILEIVCGKKNSSFFQMDDSGGNLVTHVWRLWNN 563
Query: 531 GKGLELRDPLL--LCPGDQFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNESVMLGQPGK 588
L+L DP + D+ +R ++IG+LCVQE DRP MS++ ML N S+ L P
Sbjct: 564 DSPLDLIDPAIKESYDNDEVIRCIHIGILCVQETPADRPEMSTIFQMLTNSSITLPVPRP 623
Query: 589 PPF 591
P F
Sbjct: 624 PGF 626
>At4g23220 serine/threonine kinase - like protein
Length = 542
Score = 379 bits (972), Expect = e-105
Identities = 232/542 (42%), Positives = 313/542 (56%), Gaps = 48/542 (8%)
Query: 127 VRYSNKNFIGSLNVNGNIGKDNVQNI-SEPVKFETSVNKLLNGLTKIASFNVSANMYATG 185
VRYSN +F GSL + NV +I S +F+ +L + + S Y
Sbjct: 2 VRYSNSSFFGSLKAEPHFYIHNVDDITSNLTEFDQVWEELARRMIASTTSPSSKRKYYAA 61
Query: 186 EVPFED--KTIYALVQCTRDLAANDCSRCLLSAIGDIPGCCYASIGARVMSRSCYLRYEF 243
+V + IYAL+QCT DL+ DC CL ++GD CC G V SC R+E
Sbjct: 62 DVAALTAFQIIYALMQCTPDLSLEDCHICLRQSVGDYETCCNGKQGGIVYRASCVFRWEL 121
Query: 244 YPFY-------LGEKEQTKSS------TNLGGKNNSSKIWMITVIAVGVGLVIIIFICYL 290
+PF L Q+ + TN K S I + V A+ + VI++F+ L
Sbjct: 122 FPFSEAFSRISLAPPPQSPAFPTLPAVTNTATKKGSITISIGIVWAIIIPTVIVVFLVLL 181
Query: 291 CF----LRNRQS------------------KLFGDCISVNFTDSNKLGEGGFGPVYKGIL 328
R R+S K D + F++SN +G GGFG V+ G+L
Sbjct: 182 ALGFVVYRRRKSYQGSSTDITITHSLQFDFKAIEDATN-KFSESNIIGRGGFGEVFMGVL 240
Query: 329 SDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVKLLGFCVDGEEKLLVYEYLPNG 388
+ G EVAIKRLS S QG+ EF NEV+++ KL H+NLVKLLGFC++GEEK+LVYE++PN
Sbjct: 241 N-GTEVAIKRLSKASRQGAREFKNEVVVVAKLHHRNLVKLLGFCLEGEEKILVYEFVPNK 299
Query: 389 SLDVVLFE--QHAQLDWTKRLDIINGIARGILYLHEDSRLQIIHRDLKASNVLLDNDMNP 446
SLD LF+ + QLDWTKR +II GI RGILYLH+DSRL IIHRDLKASN+LLD DMNP
Sbjct: 300 SLDYFLFDPTKQGQLDWTKRYNIIRGITRGILYLHQDSRLTIIHRDLKASNILLDADMNP 359
Query: 447 KISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEGLYSIKSDVFGFGVLLLEIITGI 506
KI+DFGMARIF + ANT I GT GYM PEY +G +S +SDV+ FGVL+LEII G
Sbjct: 360 KIADFGMARIFGIDQSGANTKKIAGTRGYMPPEYVRQGQFSTRSDVYSFGVLVLEIICGR 419
Query: 507 RNAGFCYSKTT-PSLLAYAWHLWNDGKGLELRDPLLL--CPGDQFLRYMNIGLLCVQEDA 563
N S TT +L+ YAW LW + LEL DP + C ++ R ++I LLCVQ +
Sbjct: 420 NNRFIHQSDTTVENLVTYAWRLWRNDSPLELVDPTISENCETEEVTRCIHIALLCVQHNP 479
Query: 564 FDRPTMSSVVLMLMNESVMLGQPGKPPFSVGRLNFIDQNELD---LEEEYSVNFLTVSDI 620
DRP++S++ +ML+N S +L P +P F ++ +++ LD ++N +T++D
Sbjct: 480 TDRPSLSTINMMLINNSYVLPDPQQPGFFFPIISNQERDGLDSMNRSNPQTINDVTITDF 539
Query: 621 LP 622
P
Sbjct: 540 EP 541
Score = 33.9 bits (76), Expect = 0.28
Identities = 23/105 (21%), Positives = 44/105 (41%), Gaps = 4/105 (3%)
Query: 29 ADPPYEICSTRNIYANGSSFDNNLSNLLLSLPFNDSNSISK----FGNTSSGIGLDRVYG 84
A+P + I + +I +N + FD L + + ++ SK + ++ +Y
Sbjct: 15 AEPHFYIHNVDDITSNLTEFDQVWEELARRMIASTTSPSSKRKYYAADVAALTAFQIIYA 74
Query: 85 LYMCLDFVSNETCLKCVTNAIADTVKLCPQSKEAVVYEEFCQVRY 129
L C +S E C C+ ++ D C + +VY C R+
Sbjct: 75 LMQCTPDLSLEDCHICLRQSVGDYETCCNGKQGGIVYRASCVFRW 119
>At4g21400 serine/threonine protein kinase - like protein
Length = 711
Score = 374 bits (960), Expect = e-104
Identities = 250/706 (35%), Positives = 350/706 (49%), Gaps = 99/706 (14%)
Query: 13 FFLVLYTLDFLSYLALADPP-YEICSTRNIYANG------SSFDNNLSNLLLSLPFNDSN 65
FF L + ++ LA YE N A+G SSF NL+ L+ SL S
Sbjct: 8 FFFFACVLKIVPFICLAQKDKYEFPPGFNCVASGGNFTANSSFAGNLNGLVSSLSSLTSK 67
Query: 66 SISKFGNTSSGIGLDRVYGLYMCLDFVSNETCLKCVTNAIADTVKLCPQSKEAVVYEEFC 125
+ +S +R Y + +C V + CL C+ A + ++ CP + +AVV+ C
Sbjct: 68 PYGFYNLSSGDSSGERAYAIGLCRREVKRDDCLSCIQIAARNLIEQCPLTNQAVVWYTHC 127
Query: 126 QVRYSNKNFIGSLNVNGNIGKDNVQNIS-EPVKFETSVNKLLNGLTKIASFNVSANMYA- 183
RYSN G + +NIS +F+ +LL+ L IA+ YA
Sbjct: 128 MFRYSNMIIYGRKETTPTLSFQAGKNISANRDEFDRLQIELLDRLKGIAAAGGPNRKYAQ 187
Query: 184 -TGEVPFEDKTIYALVQCTRDLAANDCSRCLLSAIGDIPGCCYASIGARVMSRSCYLRYE 242
+G Y CT DL+ DC+ CL+ IPGCC +G R SC R+E
Sbjct: 188 GSGSGVAGYPQFYGSAHCTPDLSEQDCNDCLVFGFEKIPGCCAGQVGLRWFFPSCSYRFE 247
Query: 243 FYPFYLGEKE----------------QTKSSTNLGGKNNSSKIWMITVIAVGVGLVIIIF 286
+ FY + + ++ GK S I I + V V L I
Sbjct: 248 TWRFYEFDADLEPDPPAIQPADSPTSAARTERTGKGKGGSKVIVAIVIPIVFVALFAICL 307
Query: 287 ICYLCFLRNR---------------------QSKLFGDCISV----------NFTDSNKL 315
L + +N+ Q F D + V NF+ N+L
Sbjct: 308 CLLLKWKKNKSVGRVKGNKHNLLLLVIVILLQKDEFSDSLVVDFETLKAATDNFSPENEL 367
Query: 316 GEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVKLLGFCVDG 375
G GGFG VYKG+ S GQE+A+KRLS S QG EF NE++L+ KLQH+NLV+LLGFC++G
Sbjct: 368 GRGGFGSVYKGVFSGGQEIAVKRLSCTSGQGDSEFKNEILLLAKLQHRNLVRLLGFCIEG 427
Query: 376 EEKLLVYEYLPNGSLDVVLF------------------------------EQHAQLDWTK 405
+E++LVYE++ N SLD +F ++ LDW
Sbjct: 428 QERILVYEFIKNASLDNFIFGNCFPPFSPYDDPTVLFFLLCVDLYAVTDLKKRQLLDWGV 487
Query: 406 RLDIINGIARGILYLHEDSRLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEAN 465
R +I G+ARG+LYLHEDSR +IIHRDLKASN+LLD +MNPKI+DFG+A+++ + +
Sbjct: 488 RYKMIGGVARGLLYLHEDSRYRIIHRDLKASNILLDQEMNPKIADFGLAKLYDTDQTSTH 547
Query: 466 --TTTIVGTYGYMAPEYAMEGLYSIKSDVFGFGVLLLEIIT--GIRNAGFCYSKTTPSLL 521
T+ I GTYGYMAPEYA+ G +S+K+DVF FGVL++EIIT G N + +LL
Sbjct: 548 RFTSKIAGTYGYMAPEYAIYGQFSVKTDVFSFGVLVIEIITGKGNNNGRSNDDEEAENLL 607
Query: 522 AYAWHLWNDGKGLELRDPLLLCPG-DQFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNES 580
++ W W + L + DP L + LR ++IGLLCVQE RPTM SV LML + S
Sbjct: 608 SWVWRCWREDIILSVIDPSLTTGSRSEILRCIHIGLLCVQESPASRPTMDSVALMLNSYS 667
Query: 581 VMLGQPGKPPFS----VGRLNFIDQNELDLEEEYSVNFLTVSDILP 622
L P +P F+ + +N E L S+N +TVS++ P
Sbjct: 668 YTLPTPSRPAFALESVMPSMNVSSSTEPLL---MSLNDVTVSELSP 710
>At4g11480 serine/threonine kinase-like protein
Length = 656
Score = 369 bits (948), Expect = e-102
Identities = 224/599 (37%), Positives = 323/599 (53%), Gaps = 51/599 (8%)
Query: 36 CSTRNIYANGSSFDNNLSNLLLSLPFNDSNSISKFGNTSSGIGLDRVYGLYMCLDFVSNE 95
C + ++D N +L +L N S S + N S G G DR+Y L +C+ +
Sbjct: 26 CVDSMFFRPNGTYDTNRHLILSNLASNVS-SRDGYYNGSVGEGPDRIYALGLCIPGTDPK 84
Query: 96 TCLKCVTNAIADTVKLCPQSKEAVVYEE---FCQVRYSNKNFIGSLNVN-----GNIGKD 147
C C+ A ++ CP ++ + C VRYSN +F +++ G++
Sbjct: 85 VCDDCMQIASTGILQNCPNQTDSYDWRSQKTLCFVRYSNSSFFNKMDLEPTMVIGDLNSG 144
Query: 148 NVQNISEPVKFETSVNKLLNGLTKIASFNVSANMYATGEVP-FEDKTIYALVQCTRDLAA 206
Q + + + + +N + V Y P IYAL+QC R +++
Sbjct: 145 LFQG--DLAAYTRTWEEFMNSMIT----RVGRTRYLADISPRIGSARIYALMQCIRGISS 198
Query: 207 NDCSRCLLSAIGDIPGCCYASIGARVMSRSCYLRYEFYPFYLGEKEQTKSSTNLGGKNNS 266
+C C+ + CC IG + C+ R++ YLG T S +
Sbjct: 199 MECETCIRDNVRMYQSCCNGFIGGTIRKPVCFFRWDGSE-YLGAFGDTPSLPPPSPDGKT 257
Query: 267 SKIWMITVIAVGVGLVIIIFICYLCFLRNRQS------KLFGDCISVN------------ 308
I + V V + +++ L + RQS K D S
Sbjct: 258 ISTGAIVAVVVSVVIFVVLLALVLVIRKRRQSYKTLKPKTDDDMTSPQSLQFDFMTLEAA 317
Query: 309 ---FTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNL 365
F+ +NKLG+GGFG VYKG+L + EVA+KRLS S QG++EF NEV+++ KLQHKNL
Sbjct: 318 TDKFSRNNKLGKGGFGEVYKGMLPNETEVAVKRLSSNSGQGTQEFKNEVVIVAKLQHKNL 377
Query: 366 VKLLGFCVDGEEKLLVYEYLPNGSLDVVLF---EQH-------AQLDWTKRLDIINGIAR 415
V+LLGFC++ +E++LVYE++PN SL+ LF ++H +QLDW +R +II GI R
Sbjct: 378 VRLLGFCLERDEQILVYEFVPNKSLNYFLFGNKQKHLLDPTKKSQLDWKRRYNIIGGITR 437
Query: 416 GILYLHEDSRLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGY 475
G+LYLH+DSRL IIHRD+KASN+LLD DMNPKI+DFGMAR F + E NT +VGT+GY
Sbjct: 438 GLLYLHQDSRLTIIHRDIKASNILLDADMNPKIADFGMARNFRVDQTEDNTRRVVGTFGY 497
Query: 476 MAPEYAMEGLYSIKSDVFGFGVLLLEIITGIRNAGFC-YSKTTPSLLAYAWHLWNDGKGL 534
M PEY G +S KSDV+ FGVL+LEI+ G +N+ F + +L+ + W LWN+ L
Sbjct: 498 MPPEYVTHGQFSTKSDVYSFGVLILEIVCGKKNSSFYKIDDSGGNLVTHVWRLWNNDSPL 557
Query: 535 ELRDPLL--LCPGDQFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNESVMLGQPGKPPF 591
+L DP + C D+ +R ++IGLLCVQE DRP MS++ ML N S+ L P P F
Sbjct: 558 DLIDPAIEESCDNDKVIRCIHIGLLCVQETPVDRPEMSTIFQMLTNSSITLPVPRPPGF 616
>At4g11490 serine/threonine kinase-like protein
Length = 636
Score = 365 bits (937), Expect = e-101
Identities = 223/590 (37%), Positives = 330/590 (55%), Gaps = 43/590 (7%)
Query: 36 CSTRNIYANGSSFDNNLSNLLLSLPFNDSNSISKFGNTSSGIGL--DRVYGLYMCLDFVS 93
C+ + ++D N +L SL S + +G +S IG D V+ + MC+D
Sbjct: 28 CNETGYFEPWKTYDTNRRQILTSLA---SKVVDHYGFYNSSIGKVPDEVHVMGMCIDGTE 84
Query: 94 NETCLKCVTNAIADTVKLCPQSKEAVVY---EEFCQVRYSNKNFIGSLNVNGNIGKDNVQ 150
C C+ A + CP EA + + C RYSN +F + ++ + +
Sbjct: 85 PTVCSDCLKVAADQLQENCPNQTEAYTWTPHKTLCFARYSNSSFFKRVGLHPLYMEHSNV 144
Query: 151 NISEPVKF-----ETSVNKLLNGLTKIASFNVSANMYATGEVP--FEDKTIYALVQCTRD 203
+I + + E ++L++ + + ++S+ Y V + IYAL+ CT D
Sbjct: 145 DIKSNLTYLNTIWEALTDRLMSDASSDYNASLSSRRYYAANVTNLTNFQNIYALMLCTPD 204
Query: 204 LAANDCSRCLLSAIGDIPGCCYASIGAR---VMSRSCYLRYEFYPFYLGEKEQTKSSTNL 260
L C CL A+ + Y ++ + V SC R++ YPF +G T S
Sbjct: 205 LEKGACHNCLEKAVSE-----YGNLRMQRGIVAWPSCCFRWDLYPF-IGAFNLTLSPPPG 258
Query: 261 GGKNNSSKIWMITVIAVGVGLVIIIFICYLCFLRNR------QSKLFG--------DCIS 306
+N S ++ V+A GV + ++ + + R R +S + + +
Sbjct: 259 SKRNISVGFFVAIVVATGVVISVLSTLVVVLVCRKRKTDPPEESPKYSLQYDLKTIEAAT 318
Query: 307 VNFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLV 366
F+ N LG+GGFG V+KG+L DG E+A+KRLS S QG +EF NE L+ KLQH+NLV
Sbjct: 319 CTFSKCNMLGQGGFGEVFKGVLQDGSEIAVKRLSKESAQGVQEFQNETSLVAKLQHRNLV 378
Query: 367 KLLGFCVDGEEKLLVYEYLPNGSLDVVLFE--QHAQLDWTKRLDIINGIARGILYLHEDS 424
+LGFC++GEEK+LVYE++PN SLD LFE + QLDW KR II G ARGILYLH DS
Sbjct: 379 GVLGFCMEGEEKILVYEFVPNKSLDQFLFEPTKKGQLDWAKRYKIIVGTARGILYLHHDS 438
Query: 425 RLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEG 484
L+IIHRDLKASN+LLD +M PK++DFGMARIF + A+T +VGT+GY++PEY M G
Sbjct: 439 PLKIIHRDLKASNILLDAEMEPKVADFGMARIFRVDQSRADTRRVVGTHGYISPEYLMHG 498
Query: 485 LYSIKSDVFGFGVLLLEIITGIRNAGFCYS-KTTPSLLAYAWHLWNDGKGLELRDPLL-- 541
+S+KSDV+ FGVL+LEII+G RN+ F + ++ +L+ YAW W +G LEL D L
Sbjct: 499 QFSVKSDVYSFGVLVLEIISGKRNSNFHETDESGKNLVTYAWRHWRNGSPLELVDSELEK 558
Query: 542 LCPGDQFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNESVMLGQPGKPPF 591
++ R ++I LLCVQ D RP +S++++ML + S+ L P P +
Sbjct: 559 NYQSNEVFRCIHIALLCVQNDPEQRPNLSTIIMMLTSNSITLPVPQSPVY 608
>At1g65800 receptor kinase, putative
Length = 847
Score = 363 bits (933), Expect = e-100
Identities = 192/323 (59%), Positives = 238/323 (73%), Gaps = 10/323 (3%)
Query: 308 NFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVK 367
NF+ NKLG+GGFG VYKG+L DG+E+A+KRLS S QG++EF+NEV LI KLQH NLV+
Sbjct: 522 NFSTDNKLGQGGFGIVYKGMLLDGKEIAVKRLSKMSSQGTDEFMNEVRLIAKLQHINLVR 581
Query: 368 LLGFCVDGEEKLLVYEYLPNGSLDVVLFEQ--HAQLDWTKRLDIINGIARGILYLHEDSR 425
LLG CVD EK+L+YEYL N SLD LF+Q + L+W KR DIINGIARG+LYLH+DSR
Sbjct: 582 LLGCCVDKGEKMLIYEYLENLSLDSHLFDQTRSSNLNWQKRFDIINGIARGLLYLHQDSR 641
Query: 426 LQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEGL 485
+IIHRDLKASNVLLD +M PKISDFGMARIF E EANT +VGTYGYM+PEYAM+G+
Sbjct: 642 CRIIHRDLKASNVLLDKNMTPKISDFGMARIFGREETEANTRRVVGTYGYMSPEYAMDGI 701
Query: 486 YSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLLL--- 542
+S+KSDVF FGVLLLEII+G RN GF S +LL + W W +GK LE+ DP+ +
Sbjct: 702 FSMKSDVFSFGVLLLEIISGKRNKGFYNSNRDLNLLGFVWRHWKEGKELEIVDPINIDAL 761
Query: 543 ---CPGDQFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNESVMLGQPGKPPFSVGR--LN 597
P + LR + IGLLCVQE A DRP MSSV++ML +E+ + QP +P F VGR L
Sbjct: 762 SSEFPTHEILRCIQIGLLCVQERAEDRPVMSSVMVMLGSETTAIPQPKRPGFCVGRSSLE 821
Query: 598 FIDQNELDLEEEYSVNFLTVSDI 620
+ ++E +VN +T+S I
Sbjct: 822 VDSSSSTQRDDECTVNQVTLSVI 844
>At4g04490 putative receptor-like protein kinase
Length = 658
Score = 360 bits (925), Expect = e-99
Identities = 231/640 (36%), Positives = 346/640 (53%), Gaps = 63/640 (9%)
Query: 35 ICSTRNIYANGSSFDNNLSNLLLSLPFNDSNSISKFGNTSSGIGLDRVYGLYMCLDFVSN 94
+C + N +S+ NL +LL SL SN I + G + + LD VY L +C
Sbjct: 29 VCGDEDFSPN-TSYVENLESLLPSLA---SNVIRERGFYN--VSLDGVYALALCRKHYEV 82
Query: 95 ETCLKCVTNAIADTVKLCPQSKEAVVYEE------FCQVRYSNKNFIGSLNVN--GNIGK 146
+ C +CV A + C EA ++ C VRYSN + G L + GN+
Sbjct: 83 QACRRCVDRASRTLLTQCRGKTEAYHWDSENDANVSCLVRYSNIHRFGKLKLEPIGNVPH 142
Query: 147 DNVQNISEPVKFETSVNKLLNGLTKIASFNVSANM---YATGEVPFEDKT-IYALVQCTR 202
++ S + N ++AS +++ Y F D + L+QCT
Sbjct: 143 SSLDPSSNLTRISQEFAARANRTVEVASTADESSVLKYYGVSSAEFTDTPEVNMLMQCTP 202
Query: 203 DLAANDCSRCLLSAIGDIPGCCYASIGARVMSRSCYLRYEFYPFYLG-----------EK 251
DL+++DC+ CL + + +G V SCY R++ Y F
Sbjct: 203 DLSSSDCNHCLRENVRYNQEHNWDRVGGTVARPSCYFRWDDYRFAGAFDNLERVPAPPRS 262
Query: 252 EQTKSSTNLGGKNNSSKIWMITVIAVGVGLVIIIFICYLCFLRNRQSKLFGDC------- 304
QT+ + K + W + V+ G+ + +F+ ++ R + +++ +
Sbjct: 263 PQTRQDYRVK-KGRMFQPWSVVVVVFPTGINLAVFVAFVLAYRRMRRRIYTEINKNSDSD 321
Query: 305 -------------ISVN-FTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEF 350
I+ N F+ NKLG+GGFG VYKGIL GQE+A+KRL+ S QG EF
Sbjct: 322 GQATLRFDLGMILIATNEFSLENKLGQGGFGSVYKGILPSGQEIAVKRLAGGSGQGELEF 381
Query: 351 INEVMLILKLQHKNLVKLLGFCVDGEEKLLVYEYLPNGSLDVVLFEQHAQ--LDWTKRLD 408
NEV+L+ +LQH+NLVKLLGFC +G E++LVYE++PN SLD +F++ + L W R
Sbjct: 382 KNEVLLLTRLQHRNLVKLLGFCNEGNEEILVYEHVPNSSLDHFIFDEDKRWLLTWDVRYR 441
Query: 409 IINGIARGILYLHEDSRLQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTT 468
II G+ARG+LYLHEDS+L+IIHRDLKASN+LLD +MNPK++DFGMAR+F E T+
Sbjct: 442 IIEGVARGLLYLHEDSQLRIIHRDLKASNILLDAEMNPKVADFGMARLFNMDETRGETSR 501
Query: 469 IVGTYGYMAPEYAMEGLYSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHLW 528
+VGTYGYMAPEY G +S KSDV+ FGV+LLE+I+G +N F T L A+AW W
Sbjct: 502 VVGTYGYMAPEYVRHGQFSAKSDVYSFGVMLLEMISGEKNKNF----ETEGLPAFAWKRW 557
Query: 529 NDGKGLELRDPLL-LCPGDQFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNESVM-LGQP 586
+G+ + DP L P ++ ++ + IGLLCVQE+A RPTM+SV+ L + + +P
Sbjct: 558 IEGELESIIDPYLNENPRNEIIKLIQIGLLCVQENAAKRPTMNSVITWLARDGTFTIPKP 617
Query: 587 GKPPFSVGRLNFIDQN----ELDLEEEYSVNFLTVSDILP 622
+ F L+ +N E ++ +SV+ ++++ + P
Sbjct: 618 TEAAFVTLPLSVKPENRSMSERKDKDPFSVDEVSITVLYP 657
>At1g65790 receptor kinase, putative
Length = 843
Score = 360 bits (925), Expect = e-99
Identities = 190/323 (58%), Positives = 237/323 (72%), Gaps = 10/323 (3%)
Query: 308 NFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVK 367
NF++ NKLG+GGFG VYKG L DG+E+A+KRLS S QG++EF+NEV LI KLQH NLV+
Sbjct: 518 NFSNDNKLGQGGFGIVYKGRLLDGKEIAVKRLSKMSSQGTDEFMNEVRLIAKLQHINLVR 577
Query: 368 LLGFCVDGEEKLLVYEYLPNGSLDVVLFEQ--HAQLDWTKRLDIINGIARGILYLHEDSR 425
LLG CVD EK+L+YEYL N SLD LF+Q + L+W KR DIINGIARG+LYLH+DSR
Sbjct: 578 LLGCCVDKGEKMLIYEYLENLSLDSHLFDQTRSSNLNWQKRFDIINGIARGLLYLHQDSR 637
Query: 426 LQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEGL 485
+IIHRDLKASNVLLD +M PKISDFGMARIF E EANT +VGTYGYM+PEYAM+G+
Sbjct: 638 CRIIHRDLKASNVLLDKNMTPKISDFGMARIFGREETEANTRRVVGTYGYMSPEYAMDGI 697
Query: 486 YSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLLL--- 542
+S+KSDVF FGVLLLEII+G RN GF S +LL + W W +G LE+ DP+ +
Sbjct: 698 FSMKSDVFSFGVLLLEIISGKRNKGFYNSNRDLNLLGFVWRHWKEGNELEIVDPINIDSL 757
Query: 543 ---CPGDQFLRYMNIGLLCVQEDAFDRPTMSSVVLMLMNESVMLGQPGKPPFSVGR--LN 597
P + LR + IGLLCVQE A DRP MSSV++ML +E+ + QP +P F +GR L
Sbjct: 758 SSKFPTHEILRCIQIGLLCVQERAEDRPVMSSVMVMLGSETTAIPQPKRPGFCIGRSPLE 817
Query: 598 FIDQNELDLEEEYSVNFLTVSDI 620
+ ++E +VN +T+S I
Sbjct: 818 ADSSSSTQRDDECTVNQITLSVI 840
>At4g03230 putative receptor kinase
Length = 852
Score = 358 bits (919), Expect = 5e-99
Identities = 180/293 (61%), Positives = 227/293 (77%), Gaps = 5/293 (1%)
Query: 308 NFTDSNKLGEGGFGPVYKGILSDGQEVAIKRLSICSEQGSEEFINEVMLILKLQHKNLVK 367
NF+++NKLG+GGFGPVYKG+ QE+A+KRLS CS QG EEF NEV+LI KLQH+NLV+
Sbjct: 531 NFSNANKLGQGGFGPVYKGMFPGDQEIAVKRLSRCSGQGLEEFKNEVVLIAKLQHRNLVR 590
Query: 368 LLGFCVDGEEKLLVYEYLPNGSLDVVLFEQHA--QLDWTKRLDIINGIARGILYLHEDSR 425
LLG+CV GEEKLL+YEY+P+ SLD +F++ +LDW R +II GIARG+LYLH+DSR
Sbjct: 591 LLGYCVAGEEKLLLYEYMPHKSLDFFIFDRKLCQRLDWKMRCNIILGIARGLLYLHQDSR 650
Query: 426 LQIIHRDLKASNVLLDNDMNPKISDFGMARIFAGSEGEANTTTIVGTYGYMAPEYAMEGL 485
L+IIHRDLK SN+LLD +MNPKISDFG+ARIF GSE ANT +VGTYGYM+PEYA+EGL
Sbjct: 651 LRIIHRDLKTSNILLDEEMNPKISDFGLARIFGGSETSANTNRVVGTYGYMSPEYALEGL 710
Query: 486 YSIKSDVFGFGVLLLEIITGIRNAGFCYSKTTPSLLAYAWHLWNDGKGLELRDPLL--LC 543
+S KSDVF FGV+++E I+G RN GF + + SLL +AW LW +G+EL D L C
Sbjct: 711 FSFKSDVFSFGVVVIETISGKRNTGFHEPEKSLSLLGHAWDLWKAERGIELLDQALQESC 770
Query: 544 PGDQFLRYMNIGLLCVQEDAFDRPTMSSVVLML-MNESVMLGQPGKPPFSVGR 595
+ FL+ +N+GLLCVQED DRPTMS+VV ML +E+ L P +P F + R
Sbjct: 771 ETEGFLKCLNVGLLCVQEDPNDRPTMSNVVFMLGSSEAATLPTPKQPAFVLRR 823
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.141 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,234,101
Number of Sequences: 26719
Number of extensions: 631081
Number of successful extensions: 5122
Number of sequences better than 10.0: 1061
Number of HSP's better than 10.0 without gapping: 921
Number of HSP's successfully gapped in prelim test: 140
Number of HSP's that attempted gapping in prelim test: 1455
Number of HSP's gapped (non-prelim): 1299
length of query: 622
length of database: 11,318,596
effective HSP length: 105
effective length of query: 517
effective length of database: 8,513,101
effective search space: 4401273217
effective search space used: 4401273217
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC123975.9


