

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123975.11 - phase: 0
(206 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
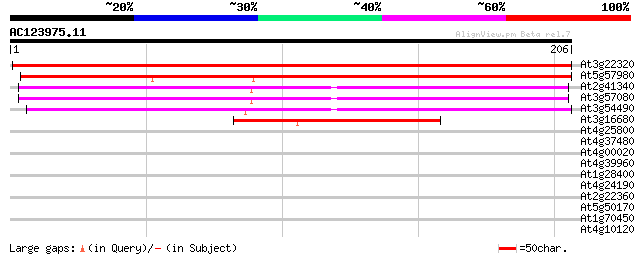
Score E
Sequences producing significant alignments: (bits) Value
At3g22320 RNA polymerase I, II and III 24.3 kDa subunit 323 4e-89
At5g57980 RNA polymerase I, II and III 24.3 kDa subunit-like pro... 189 7e-49
At2g41340 putative DNA-directed RNA polymerase 23kD subunit 140 6e-34
At3g57080 unknown protein 139 1e-33
At3g54490 RNA polymerase 24kDa subunit -like protein 123 6e-29
At3g16680 putative RNA polymerase 74 5e-14
At4g25800 putative calmodulin-binding protein 33 0.10
At4g37480 putative protein 32 0.18
At4g00020 putative BRCA2 homolog 30 0.89
At4g39960 DnaJ like protein 29 2.0
At1g28400 unknown protein 28 2.6
At4g24190 HSP90-like protein 28 3.4
At2g22360 DnaJ like protein 28 3.4
At5g50170 putative protein 27 5.8
At1g70450 putative protein kinase 27 5.8
At4g10120 sucrose-phosphate synthase - like protein 27 9.8
>At3g22320 RNA polymerase I, II and III 24.3 kDa subunit
Length = 205
Score = 323 bits (828), Expect = 4e-89
Identities = 157/205 (76%), Positives = 182/205 (88%)
Query: 2 VFSEEEITRLYRIRKTVMQMLKDRNYLVGDFELNMSKHDFKDKYGENMKREDLVINKTKK 61
+ +EEE+ RLYRI+KT+MQML+DR Y + D EL M+K F K+G+NMKREDLV K K+
Sbjct: 1 MLTEEELKRLYRIQKTLMQMLRDRGYFIADSELTMTKQQFIRKHGDNMKREDLVTLKAKR 60
Query: 62 DKPSDQIYVFFPEEAKVGVKTMKTYTNRMNSENVYRAILVCQTSLTPFAKTCVSEIASKF 121
+ SDQ+Y+FFP+EAKVGVKTMK YTNRM SENV+RAILV Q +LTPFA+TC+SEI+SKF
Sbjct: 61 NDNSDQLYIFFPDEAKVGVKTMKMYTNRMKSENVFRAILVVQQNLTPFARTCISEISSKF 120
Query: 122 HLEVFQEAELLVNIKEHVLVPEHQILNDTEKKTLLERYTVKETQLPRIQVTDPVARYYGL 181
HLEVFQEAE+LVNIKEHVLVPEHQ+L EKKTLLERYTVKETQLPRIQVTDP+ARY+GL
Sbjct: 121 HLEVFQEAEMLVNIKEHVLVPEHQVLTTEEKKTLLERYTVKETQLPRIQVTDPIARYFGL 180
Query: 182 KRGQVVKIIRPSETAGRYVTYRFVV 206
KRGQVVKIIRPSETAGRYVTYR+VV
Sbjct: 181 KRGQVVKIIRPSETAGRYVTYRYVV 205
>At5g57980 RNA polymerase I, II and III 24.3 kDa subunit-like
protein
Length = 210
Score = 189 bits (481), Expect = 7e-49
Identities = 97/206 (47%), Positives = 141/206 (68%), Gaps = 4/206 (1%)
Query: 5 EEEITRLYRIRKTVMQMLKDRNYLVGDFELNMSKHDFKDKYGENMKR---EDLVINKTKK 61
++EITR++++R+TV+QML+DR Y + + +LN+ + +F ++ + M + E L ++ K
Sbjct: 5 DDEITRIFKVRRTVLQMLRDRGYTIEESDLNLKREEFVQRFCKTMNKVNKEALFVSANKG 64
Query: 62 DKPSDQIYVFFPEEAKVGVKTMKTYTN-RMNSENVYRAILVCQTSLTPFAKTCVSEIASK 120
P+D+IYVF+PE KVGV +K +M + V+R I+V ++T A+ VSE+
Sbjct: 65 PNPADKIYVFYPEGPKVGVPVIKKEVAIKMRDDKVHRGIVVVPMAITAPARMAVSELNKM 124
Query: 121 FHLEVFQEAELLVNIKEHVLVPEHQILNDTEKKTLLERYTVKETQLPRIQVTDPVARYYG 180
+EVF+EAEL+ NI EH LV ++ +L+D KK LL YTV++TQLPRI VTDP+ARYYG
Sbjct: 125 LTIEVFEEAELVTNITEHKLVNKYYVLDDQAKKKLLNTYTVQDTQLPRILVTDPLARYYG 184
Query: 181 LKRGQVVKIIRPSETAGRYVTYRFVV 206
LKRGQVVKI R T+ Y TYRF V
Sbjct: 185 LKRGQVVKIRRSDATSLDYYTYRFAV 210
>At2g41340 putative DNA-directed RNA polymerase 23kD subunit
Length = 218
Score = 140 bits (352), Expect = 6e-34
Identities = 79/203 (38%), Positives = 123/203 (59%), Gaps = 3/203 (1%)
Query: 4 SEEEITRLYRIRKTVMQMLKDRNYLVGDFELNMSKHDFKDKYGENMKREDLVINKTKKDK 63
S E + Y R+T M+ML+DR Y V D ++N+S F+ YGE+ + L I+ +
Sbjct: 17 SSVECHKYYLARRTTMEMLRDRGYDVSDEDINLSLQQFRALYGEHPDVDLLRISAKHRFD 76
Query: 64 PSDQIYVFFPEEAKVGVKTMKTYT-NRMNSENVYRAILVCQTSLTPFAKTCVSEIASKFH 122
S +I V F V V M+ + ++ EN+ ILV Q+ +T A V + F
Sbjct: 77 SSKKISVVFCGTGIVKVNAMRVIAADVLSRENITGLILVLQSHITNQALKAVELFS--FK 134
Query: 123 LEVFQEAELLVNIKEHVLVPEHQILNDTEKKTLLERYTVKETQLPRIQVTDPVARYYGLK 182
+E+F+ +LLVN+ +HVL P+HQ+LND EK++LL++++++E QLPR+ DP+ RYYGL+
Sbjct: 135 VELFEITDLLVNVSKHVLRPKHQVLNDKEKESLLKKFSIEEKQLPRLSSKDPIVRYYGLE 194
Query: 183 RGQVVKIIRPSETAGRYVTYRFV 205
GQV+K+ E + +VTYR V
Sbjct: 195 TGQVMKVTYKDELSESHVTYRCV 217
>At3g57080 unknown protein
Length = 222
Score = 139 bits (349), Expect = 1e-33
Identities = 77/203 (37%), Positives = 120/203 (58%), Gaps = 3/203 (1%)
Query: 4 SEEEITRLYRIRKTVMQMLKDRNYLVGDFELNMSKHDFKDKYGENMKREDLVINKTKKDK 63
S EE R Y R+ +QML+DR Y V D ++N+S HDF+ YGE + L I+ +
Sbjct: 21 SSEESHRYYLARRNGLQMLRDRGYEVSDEDINLSLHDFRTVYGERPDVDRLRISALHRSD 80
Query: 64 PSDQIYVFFPEEAKVGVKTMKTYT-NRMNSENVYRAILVCQTSLTPFAKTCVSEIASKFH 122
+ ++ + F + V V +++ + ++ E + ILV Q +T A + + F
Sbjct: 81 STKKVKIVFFGTSMVKVNAIRSVVADILSQETITGLILVLQNHVTNQALKAIELFS--FK 138
Query: 123 LEVFQEAELLVNIKEHVLVPEHQILNDTEKKTLLERYTVKETQLPRIQVTDPVARYYGLK 182
+E+FQ +LLVNI +H L P+HQ+LND EK TLL++++++E QLPRI D + RYYGL+
Sbjct: 139 VEIFQITDLLVNITKHSLKPQHQVLNDEEKTTLLKKFSIEEKQLPRISKKDAIVRYYGLE 198
Query: 183 RGQVVKIIRPSETAGRYVTYRFV 205
+GQVVK+ E +V +R V
Sbjct: 199 KGQVVKVNYRGELTESHVAFRCV 221
>At3g54490 RNA polymerase 24kDa subunit -like protein
Length = 233
Score = 123 bits (309), Expect = 6e-29
Identities = 71/201 (35%), Positives = 113/201 (55%), Gaps = 3/201 (1%)
Query: 7 EITRLYRIRKTVMQMLKDRNYLVGDFELNMSKHDFKDKYGENMKREDLVINKTKKDKPSD 66
E R Y R T +ML+DR Y V + EL+++ +F+ +GE + E L I + P
Sbjct: 35 ESKRFYLARTTAFEMLRDRGYEVNEAELSLTLSEFRSVFGEKPELERLRICVPLRSDPKK 94
Query: 67 QIYVFFPEEAKVGVKTMKT-YTNRMNSENVYRAILVCQTSLTPFAKTCVSEIASKFHLEV 125
+I V F + VK+++ + N+ ++ ILV Q+ + FA+ ++ F +E
Sbjct: 95 KILVVFMGTEPITVKSVRALHIQISNNVGLHAMILVLQSKMNHFAQKALTTFP--FTVET 152
Query: 126 FQEAELLVNIKEHVLVPEHQILNDTEKKTLLERYTVKETQLPRIQVTDPVARYYGLKRGQ 185
F +LLVNI +H+ P+ +ILN EK+ LL ++ +++ QLP +Q D RYYGLK+ Q
Sbjct: 153 FPIEDLLVNITKHIQQPKIEILNKEEKEQLLRKHALEDKQLPYLQEKDSFVRYYGLKKKQ 212
Query: 186 VVKIIRPSETAGRYVTYRFVV 206
VVKI E G +VTYR ++
Sbjct: 213 VVKITYSKEPVGDFVTYRCII 233
>At3g16680 putative RNA polymerase
Length = 87
Score = 73.9 bits (180), Expect = 5e-14
Identities = 39/78 (50%), Positives = 51/78 (65%), Gaps = 2/78 (2%)
Query: 83 MKTYTNRMNSENVYRAILVCQT--SLTPFAKTCVSEIASKFHLEVFQEAELLVNIKEHVL 140
MK Y +++ S NV+RAILV Q + + A + + FH+EVFQE EL+VN+KEHV
Sbjct: 1 MKKYIDQLKSANVFRAILVVQDIKAFSRQALVFLGAVYPIFHIEVFQEKELIVNVKEHVF 60
Query: 141 VPEHQILNDTEKKTLLER 158
VPEHQ L EK+ LER
Sbjct: 61 VPEHQALTTEEKQKFLER 78
>At4g25800 putative calmodulin-binding protein
Length = 536
Score = 33.1 bits (74), Expect = 0.10
Identities = 31/129 (24%), Positives = 59/129 (45%), Gaps = 7/129 (5%)
Query: 46 GENMKREDLVINKTKKDKPSDQIYVFFPEEAK-VGVKTMKTY-TNRMNSENVYRAILVCQ 103
G + K DL++ K S ++Y+++ E+++ VGV Y + + +E+ Y +
Sbjct: 211 GMSNKMWDLLVEHAKTCVLSGKLYIYYTEDSRSVGVVFNNIYELSGLITEDQYLSADSLS 270
Query: 104 TSLTPFAKTCVSEIASKFHLEVFQEAELLVNIKEHVLVPEHQILNDTEKKTLLERY-TVK 162
S + V + ++ V E E L+N+ + PE ++ T+ T L Y TV
Sbjct: 271 ESQKVYVDGLVKKAYENWNQVVEYEGESLLNLNQ----PERLDISQTDPVTALASYSTVP 326
Query: 163 ETQLPRIQV 171
+Q P +
Sbjct: 327 LSQFPEFAI 335
>At4g37480 putative protein
Length = 523
Score = 32.3 bits (72), Expect = 0.18
Identities = 22/93 (23%), Positives = 46/93 (48%), Gaps = 3/93 (3%)
Query: 70 VFFPEEAKVGVKTMKTYTNRMNS--ENVYRAILVCQTSLTPFAKTCVSEIASKFHLEVFQ 127
+F P + + + + + +NR EN Y + V +TS K +A + H ++ +
Sbjct: 30 LFNPSQGRTRLLSGEAESNRNEFPVENAYDILNVSETSSIAEIKASFRRLAKETHPDLIE 89
Query: 128 EAELLVNIKEHV-LVPEHQILNDTEKKTLLERY 159
+ N + V ++ ++IL+D+EK+ +RY
Sbjct: 90 SKKDPSNSRRFVQILAAYEILSDSEKRAHYDRY 122
>At4g00020 putative BRCA2 homolog
Length = 765
Score = 30.0 bits (66), Expect = 0.89
Identities = 24/110 (21%), Positives = 52/110 (46%), Gaps = 7/110 (6%)
Query: 3 FSEEEITRLYRIRKTVMQMLKDRNYLVGDFELNMSKHDFKDKYGENMKREDLVINK---- 58
+++++ T+ R+ KTV + + + F+ KH + G ++ D + +K
Sbjct: 437 YNKQDSTQKKRLGKTV-SVSPFKRPRISSFKTPSKKHALQASSGLSVVSCDTLTSKKVLS 495
Query: 59 TKKDKPSDQIYV--FFPEEAKVGVKTMKTYTNRMNSENVYRAILVCQTSL 106
T+ + S ++Y+ FF +TM +N++ N+Y + + QT L
Sbjct: 496 TRYPEKSPRVYIKDFFGMHPTATTRTMSEESNQVMQINMYSVMSLLQTRL 545
>At4g39960 DnaJ like protein
Length = 447
Score = 28.9 bits (63), Expect = 2.0
Identities = 17/64 (26%), Positives = 34/64 (52%), Gaps = 2/64 (3%)
Query: 96 YRAILVCQTSLTPFAKTCVSEIASKFHLEVFQEAELLVNIKEHVLVPEHQILNDTEKKTL 155
Y + V + + K+ ++A +H +V ++A KE + ++IL+D EK++L
Sbjct: 87 YSVLGVSKNATKAEIKSAYRKLARSYHPDVNKDAGAEDKFKE--ISNAYEILSDDEKRSL 144
Query: 156 LERY 159
+RY
Sbjct: 145 YDRY 148
>At1g28400 unknown protein
Length = 335
Score = 28.5 bits (62), Expect = 2.6
Identities = 13/40 (32%), Positives = 19/40 (47%)
Query: 24 DRNYLVGDFELNMSKHDFKDKYGENMKREDLVINKTKKDK 63
D NY +F N + +++ KY EN+K E N K
Sbjct: 173 DENYAKEEFNNNNNNNNYNYKYDENVKEESFPENNEDNKK 212
>At4g24190 HSP90-like protein
Length = 823
Score = 28.1 bits (61), Expect = 3.4
Identities = 35/141 (24%), Positives = 61/141 (42%), Gaps = 32/141 (22%)
Query: 40 DFKDKYGENMKREDLVINKTKKDKPSDQIYVFFPEEAKVGVKTM-KTYTNRMNSENVYRA 98
D++DK +N+ +E L + K KDK E K K + K + + SENV
Sbjct: 612 DYEDKKFQNVSKEGLKVGKDSKDK-----------ELKEAFKELTKWWKGNLASENVDDV 660
Query: 99 ILVCQTSLTPFAKTCVSEIASKF----HLEVFQEAELLVNIKEHVLV---------PEHQ 145
+ + + TP CV + SKF ++E +++ L + + + P H
Sbjct: 661 KISNRLADTP----CV-VVTSKFGWSANMERIMQSQTLSDANKQAYMRGKRVLEINPRHP 715
Query: 146 ILNDTEKKTLL--ERYTVKET 164
I+ + + + E +VKET
Sbjct: 716 IIKELKDRIASDPEDESVKET 736
>At2g22360 DnaJ like protein
Length = 442
Score = 28.1 bits (61), Expect = 3.4
Identities = 16/64 (25%), Positives = 33/64 (51%), Gaps = 2/64 (3%)
Query: 96 YRAILVCQTSLTPFAKTCVSEIASKFHLEVFQEAELLVNIKEHVLVPEHQILNDTEKKTL 155
Y + V + + K+ ++A +H +V ++ KE + +++L+D EKK+L
Sbjct: 88 YSVLGVSKNATKAEIKSAYRKLARNYHPDVNKDPGAEEKFKE--ISNAYEVLSDDEKKSL 145
Query: 156 LERY 159
+RY
Sbjct: 146 YDRY 149
>At5g50170 putative protein
Length = 1027
Score = 27.3 bits (59), Expect = 5.8
Identities = 20/70 (28%), Positives = 34/70 (48%), Gaps = 5/70 (7%)
Query: 24 DRNYLVGDFELNMSKHDFKDKYGENMKREDLVINKTKKDKPSDQIYVFFPEEAKVGVKTM 83
D+ +G E+N KH + ++ LV N + + Q+ +F E K GV+TM
Sbjct: 614 DQGASLGHAEINFLKHTADELADLSVA---LVGNHAQASQSKLQLRIFL--ENKNGVETM 668
Query: 84 KTYTNRMNSE 93
K Y +++ E
Sbjct: 669 KDYLSKVEKE 678
>At1g70450 putative protein kinase
Length = 394
Score = 27.3 bits (59), Expect = 5.8
Identities = 13/42 (30%), Positives = 23/42 (53%)
Query: 156 LERYTVKETQLPRIQVTDPVARYYGLKRGQVVKIIRPSETAG 197
LE++ VK I+ RY G KR ++V+++R ++ G
Sbjct: 286 LEKHYVKNEVFRMIETAAACVRYSGPKRPRMVQVLRALDSEG 327
>At4g10120 sucrose-phosphate synthase - like protein
Length = 1083
Score = 26.6 bits (57), Expect = 9.8
Identities = 16/62 (25%), Positives = 31/62 (49%), Gaps = 2/62 (3%)
Query: 4 SEEEITRLYRIRKTVMQMLKDRNYLVGDFELNMSKHDFKDKYGENMKREDLV--INKTKK 61
+ E RL I + + + + +V D + +SK + + G N EDL+ +++ +K
Sbjct: 93 TRERSNRLENICWRIWHLARKKKQIVWDDGVRLSKRRIEREQGRNDAEEDLLSELSEGEK 152
Query: 62 DK 63
DK
Sbjct: 153 DK 154
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.135 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,338,141
Number of Sequences: 26719
Number of extensions: 175148
Number of successful extensions: 479
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 464
Number of HSP's gapped (non-prelim): 17
length of query: 206
length of database: 11,318,596
effective HSP length: 95
effective length of query: 111
effective length of database: 8,780,291
effective search space: 974612301
effective search space used: 974612301
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC123975.11


