

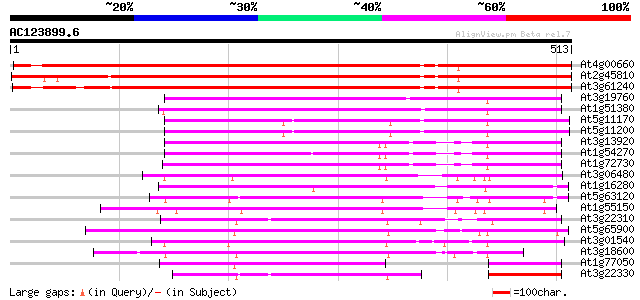
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123899.6 + phase: 0 /pseudo
(513 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g00660 RNA helicase like protein 706 0.0
At2g45810 putative ATP-dependent RNA helicase 666 0.0
At3g61240 DEAD box RNA helicase RH12 647 0.0
At3g19760 RNA helicase, putative 223 2e-58
At1g51380 putative RNA helicase 212 3e-55
At5g11170 DEAD BOX RNA helicase RH15 - like protein 196 3e-50
At5g11200 DEAD BOX RNA helicase RH15 195 6e-50
At3g13920 eukaryotic protein synthesis initiation factor 4A 192 4e-49
At1g54270 Eukaryotic Initiation Factor 4A-2 188 7e-48
At1g72730 Eukaryotic initiation factor 4A like protein 185 5e-47
At3g06480 putative RNA helicase 169 4e-42
At1g16280 putative ATP-dependent RNA helicase 169 4e-42
At5g63120 ATP-dependent RNA helicase-like protein 164 1e-40
At1g55150 unknown protein 159 3e-39
At3g22310 putative RNA helicase 159 5e-39
At5g65900 ATP-dependent RNA helicase-like 156 2e-38
At3g01540 AT3g01540/F4P13_9 156 2e-38
At3g18600 DEAD box helicase protein, putative 155 4e-38
At1g77050 140 2e-33
At3g22330 DEAD-Box RNA helicase like protein 132 5e-31
>At4g00660 RNA helicase like protein
Length = 505
Score = 706 bits (1823), Expect = 0.0
Identities = 381/517 (73%), Positives = 406/517 (77%), Gaps = 21/517 (4%)
Query: 4 NNRGRYPPGIGLGRGGSGGGGGGLTSNSNTGFQQRPHHQYQQQQQYVQR-HMMQNQHQQH 62
NNRGRYPPGIG GRG N N +Q R +Q QYVQR + QN QQ
Sbjct: 2 NNRGRYPPGIGAGRGAF---------NPNPNYQSRSGYQQHPPPQYVQRGNYAQNHQQQF 52
Query: 63 YQNQQQNQQQNQQQQQQQQWLRRNQL-GGGTDTNVVEEVEKTVQSEANDSSSQDWKARLK 121
Q Q Q QQQQQQQQWLRR Q+ GG ++ + V EVEKTVQSE D +S+DWKARLK
Sbjct: 53 QQAPSQPHQYQQQQQQQQQWLRRGQIPGGNSNGDAVVEVEKTVQSEVIDPNSEDWKARLK 112
Query: 122 LPPADTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDIL 181
LP DTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTG DIL
Sbjct: 113 LPAPDTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGRDIL 172
Query: 182 ARAKNGTGKTAAFSIPALEKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMV 241
ARAKNGTGKTAAF IP LEKIDQDNN+IQ VI+VPTRELALQTSQVCKELGKHL+IQVMV
Sbjct: 173 ARAKNGTGKTAAFCIPVLEKIDQDNNVIQAVIIVPTRELALQTSQVCKELGKHLKIQVMV 232
Query: 242 TTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQP 301
TTGGTSLKDDIMRLYQPVHLLVGTPGRILDL KKGVCVLKDCS+LVMDEADKLLS EFQP
Sbjct: 233 TTGGTSLKDDIMRLYQPVHLLVGTPGRILDLTKKGVCVLKDCSVLVMDEADKLLSQEFQP 292
Query: 302 SIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPYIINLMDELTLKGITQFYAFVEE 361
S+E LI FLP +RQILMFSATFPVTVKDFKDR+L PY+INLMDELTLKGITQFYAFVEE
Sbjct: 293 SVEHLISFLPESRQILMFSATFPVTVKDFKDRFLTNPYVINLMDELTLKGITQFYAFVEE 352
Query: 362 RQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSC-----KDVARPP 416
RQK+HCLNTLFSK I+ + F S E ++ YSC K +
Sbjct: 353 RQKIHCLNTLFSKL---QINQSIIFCNS--VNRVELLAKKITELGYSCFYIHAKMLQDHR 407
Query: 417 **SFPRLPQWCMQESRLYCLFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGL 476
F + LFTRGIDIQAVNVVINFDFPKN+ETYLHRVGRSGRFGHLGL
Sbjct: 408 NRVFHDFRNGACRNLVCTDLFTRGIDIQAVNVVINFDFPKNAETYLHRVGRSGRFGHLGL 467
Query: 477 AVNLITYEDRFNLYRIEQELGTEIKQIPPFIDQAIYC 513
AVNLITYEDRFNLYRIEQELGTEIKQIPP IDQAIYC
Sbjct: 468 AVNLITYEDRFNLYRIEQELGTEIKQIPPHIDQAIYC 504
>At2g45810 putative ATP-dependent RNA helicase
Length = 528
Score = 666 bits (1718), Expect = 0.0
Identities = 365/531 (68%), Positives = 406/531 (75%), Gaps = 26/531 (4%)
Query: 2 NNNNRGRYPPGIGLGRGGSGGGGGGLTSN---------SNTGFQQRPHHQ---YQQQQQY 49
NNNNRGR+PPGIG G N S T F Q+P Q Y Q Q
Sbjct: 4 NNNNRGRFPPGIGAAGPGPDPNFQSRNPNPPQPQQYLQSRTPFPQQPQPQPPQYLQSQSD 63
Query: 50 VQRHMMQNQHQQHYQNQQ-QNQQQNQQQQQQQQWLRRNQLGGGTDTNVVEEVEKTVQSEA 108
Q+++ + QQ Q QQ Q QQQ QQQQQ+QQW RR QL G D + ++EVEKTVQSEA
Sbjct: 64 AQQYVQRGYPQQIQQQQQLQQQQQQQQQQQEQQWSRRAQLPG--DPSYIDEVEKTVQSEA 121
Query: 109 -NDSSSQDWKARLKLPPADTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQ 167
+DS+++DWKA LKLPP D RY+TEDVTATKGNEFEDYFLKR+LL GIYEKGFE+PSPIQ
Sbjct: 122 ISDSNNEDWKATLKLPPRDNRYQTEDVTATKGNEFEDYFLKRDLLRGIYEKGFEKPSPIQ 181
Query: 168 EESIPIALTGSDILARAKNGTGKTAAFSIPALEKIDQDNNIIQVVILVPTRELALQTSQV 227
EESIPIALTGSDILARAKNGTGKT AF IP LEKID +NN+IQ VILVPTRELALQTSQV
Sbjct: 182 EESIPIALTGSDILARAKNGTGKTGAFCIPTLEKIDPENNVIQAVILVPTRELALQTSQV 241
Query: 228 CKELGKHLQIQVMVTTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLV 287
CKEL K+L+I+VMVTTGGTSL+DDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDC+MLV
Sbjct: 242 CKELSKYLKIEVMVTTGGTSLRDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCAMLV 301
Query: 288 MDEADKLLSPEFQPSIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPYIINLMDEL 347
MDEADKLLS EFQPSIE+LIQFLP +RQILMFSATFPVTVK FKDRYL+KPYIINLMD+L
Sbjct: 302 MDEADKLLSVEFQPSIEELIQFLPESRQILMFSATFPVTVKSFKDRYLKKPYIINLMDQL 361
Query: 348 TLKGITQFYAFVEERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLY 407
TL G+TQ+YAFVEERQKVHCLNTLFSK I+ + F S E ++ Y
Sbjct: 362 TLMGVTQYYAFVEERQKVHCLNTLFSKL---QINQSIIFCNS--VNRVELLAKKITELGY 416
Query: 408 SC-----KDVARPP**SFPRLPQWCMQESRLYCLFTRGIDIQAVNVVINFDFPKNSETYL 462
SC K V F + LFTRGIDIQAVNVVINFDFP+ SE+YL
Sbjct: 417 SCFYIHAKMVQDHRNRVFHDFRNGACRNLVCTDLFTRGIDIQAVNVVINFDFPRTSESYL 476
Query: 463 HRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIKQIPPFIDQAIYC 513
HRVGRSGRFGHLGLAVNL+TYEDRF +Y+ EQELGTEIK IP ID+AIYC
Sbjct: 477 HRVGRSGRFGHLGLAVNLVTYEDRFKMYQTEQELGTEIKPIPSLIDKAIYC 527
>At3g61240 DEAD box RNA helicase RH12
Length = 498
Score = 647 bits (1668), Expect = 0.0
Identities = 354/518 (68%), Positives = 391/518 (75%), Gaps = 29/518 (5%)
Query: 3 NNNRGRYPPGIGLGRGGSGGGGGGLTSNSNTGFQQRPHHQYQ-QQQQYVQRHMMQNQHQQ 61
N NRGRYPPG+G GRG N + Q Q Q QQYVQR QN Q
Sbjct: 2 NTNRGRYPPGVGTGRGAP----------PNPDYHQSYRQQQPPQDQQYVQRGYSQNPQQM 51
Query: 62 HYQNQQQNQQQNQQQQQQQQWLRRNQLGGGTDTNVVEEVEKTVQSEAN-DSSSQDWKARL 120
Q QQQ+QQQQQQQQW RR QL G +N E V++T Q EA+ D++ QDWKA L
Sbjct: 52 ------QLQQQHQQQQQQQQWSRRPQLPGNA-SNANEVVQQTTQPEASSDANGQDWKATL 104
Query: 121 KLPPADTRYRTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDI 180
+LPP DTRY+T DVTATKGNEFEDYFLKR+LL GIYEKGFE+PSPIQEESIPIALTGSDI
Sbjct: 105 RLPPPDTRYQTADVTATKGNEFEDYFLKRDLLKGIYEKGFEKPSPIQEESIPIALTGSDI 164
Query: 181 LARAKNGTGKTAAFSIPALEKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVM 240
LARAKNGTGKT AF IP LEKID +NN+IQ +ILVPTRELALQTSQVCKEL K+L IQVM
Sbjct: 165 LARAKNGTGKTGAFCIPVLEKIDPNNNVIQAMILVPTRELALQTSQVCKELSKYLNIQVM 224
Query: 241 VTTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQ 300
VTTGGTSL+DDIMRL+QPVHLLVGTPGRILDL KKGVCVLKDC+MLVMDEADKLLS EFQ
Sbjct: 225 VTTGGTSLRDDIMRLHQPVHLLVGTPGRILDLTKKGVCVLKDCAMLVMDEADKLLSAEFQ 284
Query: 301 PSIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPYIINLMDELTLKGITQFYAFVE 360
PS+E+LIQFLP RQ LMFSATFPVTVK FKDR+LRKPY+INLMD+LTL G+TQ+YAFVE
Sbjct: 285 PSLEELIQFLPQNRQFLMFSATFPVTVKAFKDRHLRKPYVINLMDQLTLMGVTQYYAFVE 344
Query: 361 ERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSC-----KDVARP 415
ERQKVHCLNTLFSK I+ + F S E ++ YSC K V
Sbjct: 345 ERQKVHCLNTLFSKL---QINQSIIFCNS--VNRVELLAKKITELGYSCFYIHAKMVQDH 399
Query: 416 P**SFPRLPQWCMQESRLYCLFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLG 475
F + LFTRGIDIQAVNVVINFDFP+ SE+YLHRVGRSGRFGHLG
Sbjct: 400 RNRVFHEFRNGACRNLVCTDLFTRGIDIQAVNVVINFDFPRTSESYLHRVGRSGRFGHLG 459
Query: 476 LAVNLITYEDRFNLYRIEQELGTEIKQIPPFIDQAIYC 513
LAVNL+TYEDRF +Y+ EQELGTEIK IP IDQAIYC
Sbjct: 460 LAVNLVTYEDRFKMYQTEQELGTEIKPIPSNIDQAIYC 497
>At3g19760 RNA helicase, putative
Length = 408
Score = 223 bits (568), Expect = 2e-58
Identities = 128/367 (34%), Positives = 208/367 (55%), Gaps = 6/367 (1%)
Query: 142 FEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALEK 201
F D +K ++L G+YE GFE+PS IQ+ ++ L G D++A+A++GTGKT+ ++ +
Sbjct: 37 FNDMGIKEDVLRGVYEYGFEKPSAIQQRAVMPILQGRDVIAQAQSGTGKTSMIALSVCQV 96
Query: 202 IDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPVHL 261
+D + +Q +IL PTRELA QT + + +G H IQ GG S+ +DI +L VH+
Sbjct: 97 VDTSSREVQALILSPTRELATQTEKTIQAIGLHANIQAHACIGGNSVGEDIRKLEHGVHV 156
Query: 262 LVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQILMFSA 321
+ GTPGR+ D+ K+ + +L++DE+D++LS F+ I + ++LPP Q+ + SA
Sbjct: 157 VSGTPGRVCDMIKRRSLRTRAIKLLILDESDEMLSRGFKDQIYDVYRYLPPDLQVCLVSA 216
Query: 322 TFPVTVKDFKDRYLRKPY-IINLMDELTLKGITQFYAFVEERQKVHCLNTLFSKAANKPI 380
T P + + +++ +P I+ DELTL+GI QF+ VE+ + +TL I
Sbjct: 217 TLPHEILEMTSKFMTEPVKILVKRDELTLEGIKQFFVAVEKEE--WKFDTLCDLYDTLTI 274
Query: 381 HHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDVARPP**SFPRLPQWCMQESRLYC---LF 437
+ F + + R F S P + ++ +SR+ ++
Sbjct: 275 TQAVIFCNTKRKVDYLSEKMRSHNFTVSSMHGDMPQKERDAIMNEFRSGDSRVLITTDVW 334
Query: 438 TRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQELG 497
RGID+Q V++VIN+D P N E Y+HR+GRSGRFG G+A+N + +D L IEQ
Sbjct: 335 ARGIDVQQVSLVINYDLPNNRELYIHRIGRSGRFGRKGVAINFVKSDDIKILRDIEQYYS 394
Query: 498 TEIKQIP 504
T+I ++P
Sbjct: 395 TQIDEMP 401
>At1g51380 putative RNA helicase
Length = 392
Score = 212 bits (540), Expect = 3e-55
Identities = 127/377 (33%), Positives = 213/377 (55%), Gaps = 12/377 (3%)
Query: 137 TKG----NEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTA 192
TKG F+D + ++L G+Y+ G+++PS IQ+ ++ L G D++A+A++GTGKT+
Sbjct: 15 TKGIKPIKSFDDMGMNDKVLRGVYDYGYKKPSEIQQRALVPILKGRDVIAQAQSGTGKTS 74
Query: 193 AFSIPALEKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDI 252
+I + ++ + +QV++L P+RELA QT + + +G H IQ GG S+ +DI
Sbjct: 75 MIAISVCQIVNISSRKVQVLVLSPSRELASQTEKTIQAIGAHTNIQAHACIGGKSIGEDI 134
Query: 253 MRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPP 312
+L + VH + GTPGR+ D+ K+G K +LV+DE+D++LS + I + + LP
Sbjct: 135 KKLERGVHAVSGTPGRVYDMIKRGSLQTKAVKLLVLDESDEMLSKGLKDQIYDVYRALPH 194
Query: 313 TRQILMFSATFPVTVKDFKDRYLRKPY-IINLMDELTLKGITQFYAFVE-ERQKVHCLNT 370
Q+ + SAT P + + ++++ P I+ DELTL+GI Q+Y V+ E K L
Sbjct: 195 DIQVCLISATLPQEILEMTEKFMTDPVRILVKPDELTLEGIKQYYVDVDKEEWKFDTLCD 254
Query: 371 LFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDVARPP**SFPRLPQWCMQE 430
L+ + I+ + F + R F+ S + + Q+ +
Sbjct: 255 LYGRLT---INQAIIFCNTRQKVDWLTEKMRSSNFIVSSMHGDKRQKERDDIMNQFRSFK 311
Query: 431 SRLYC---LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYEDRF 487
SR+ ++ RGID+Q V+ VIN+D P N E Y+HR+GR+GRFG G+A+N + D
Sbjct: 312 SRVLIASDVWARGIDVQTVSHVINYDIPNNPELYIHRIGRAGRFGREGVAINFVKSSDMK 371
Query: 488 NLYRIEQELGTEIKQIP 504
+L IE+ GT+I+++P
Sbjct: 372 DLKDIERHYGTKIREMP 388
>At5g11170 DEAD BOX RNA helicase RH15 - like protein
Length = 427
Score = 196 bits (498), Expect = 3e-50
Identities = 129/381 (33%), Positives = 203/381 (52%), Gaps = 14/381 (3%)
Query: 142 FEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALEK 201
F D+ LK ELL I + GFE PS +Q E IP A+ G D++ +AK+G GKTA F + L++
Sbjct: 48 FRDFLLKPELLRAIVDSGFEHPSEVQHECIPQAILGMDVICQAKSGMGKTAVFVLSTLQQ 107
Query: 202 IDQDNNIIQVVILVPTRELALQTSQVCKELGKHL-QIQVMVTTGGTSLK--DDIMRLYQP 258
I+ + ++L TRELA Q +L +V V GG ++K D+++ P
Sbjct: 108 IEPSPGQVSALVLCHTRELAYQICNEFVRFSTYLPDTKVSVFYGGVNIKIHKDLLKNECP 167
Query: 259 VHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLL-SPEFQPSIEQLIQFLPPTRQIL 317
H++VGTPGR+L LA++ LK+ ++DE DK+L S + + ++++ + P +Q++
Sbjct: 168 -HIVVGTPGRVLALAREKDLSLKNVRHFILDECDKMLESLDMRRDVQEIFKMTPHDKQVM 226
Query: 318 MFSATFPVTVKDFKDRYLRKPYIINLMDE--LTLKGITQFYAFVEERQKVHCLNTLFSKA 375
MFSAT ++ ++++ P I + DE LTL G+ Q Y + E +K LN L
Sbjct: 227 MFSATLSKEIRPVCKKFMQDPMEIYVDDEAKLTLHGLVQHYIKLSEMEKTRKLNDLLDAL 286
Query: 376 ANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDVARPP**SFPRLPQWCMQESRLYC 435
+ ++ F KS N F C R + R+
Sbjct: 287 ---DFNQVVIFVKSVSRAAELNKLLVECNFPSICIHSGMSQEERLTRYKSFKEGHKRILV 343
Query: 436 ---LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLI-TYEDRFNLYR 491
L RGIDI+ VN+VIN+D P +++TYLHRVGR+GRFG GLA+ + + D L +
Sbjct: 344 ATDLVGRGIDIERVNIVINYDMPDSADTYLHRVGRAGRFGTKGLAITFVASASDSEVLNQ 403
Query: 492 IEQELGTEIKQIPPFIDQAIY 512
+++ +IK++P ID + Y
Sbjct: 404 VQERFEVDIKELPEQIDTSTY 424
>At5g11200 DEAD BOX RNA helicase RH15
Length = 427
Score = 195 bits (495), Expect = 6e-50
Identities = 129/381 (33%), Positives = 203/381 (52%), Gaps = 14/381 (3%)
Query: 142 FEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALEK 201
F D+ LK ELL I + GFE PS +Q E IP A+ G D++ +AK+G GKTA F + L++
Sbjct: 48 FRDFLLKPELLRAIVDSGFEHPSEVQHECIPQAILGMDVICQAKSGMGKTAVFVLSTLQQ 107
Query: 202 IDQDNNIIQVVILVPTRELALQTSQVCKELGKHL-QIQVMVTTGGTSLK--DDIMRLYQP 258
I+ + ++L TRELA Q +L +V V GG ++K D+++ P
Sbjct: 108 IEPSPGQVSALVLCHTRELAYQICNEFVRFSTYLPDTKVSVFYGGVNIKIHKDLLKNECP 167
Query: 259 VHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLL-SPEFQPSIEQLIQFLPPTRQIL 317
H++VGTPGR+L LA++ LK+ ++DE DK+L S + + ++++ + P +Q++
Sbjct: 168 -HIVVGTPGRVLALAREKDLSLKNVRHFILDECDKMLESLDMRRDVQEIFKMTPHDKQVM 226
Query: 318 MFSATFPVTVKDFKDRYLRKPYIINLMDE--LTLKGITQFYAFVEERQKVHCLNTLFSKA 375
MFSAT ++ ++++ P I + DE LTL G+ Q Y + E +K LN L
Sbjct: 227 MFSATLSKEIRPVCKKFMQDPMEIYVDDEAKLTLHGLVQHYIKLSEMEKNRKLNDLLDAL 286
Query: 376 ANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDVARPP**SFPRLPQWCMQESRLYC 435
+ ++ F KS N F C R + R+
Sbjct: 287 ---DFNQVVIFVKSVSRAAELNKLLVECNFPSICIHSGMSQEERLTRYKSFKEGHKRILV 343
Query: 436 ---LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLI-TYEDRFNLYR 491
L RGIDI+ VN+VIN+D P +++TYLHRVGR+GRFG GLA+ + + D L +
Sbjct: 344 ATDLVGRGIDIERVNIVINYDMPDSADTYLHRVGRAGRFGTKGLAITFVASASDSEVLNQ 403
Query: 492 IEQELGTEIKQIPPFIDQAIY 512
+++ +IK++P ID + Y
Sbjct: 404 VQERFEVDIKELPEQIDTSTY 424
>At3g13920 eukaryotic protein synthesis initiation factor 4A
Length = 412
Score = 192 bits (488), Expect = 4e-49
Identities = 128/393 (32%), Positives = 195/393 (49%), Gaps = 58/393 (14%)
Query: 142 FEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALEK 201
F+ L+ LL GIY GFE+PS IQ+ I G D++ +A++GTGKTA F L++
Sbjct: 41 FDAMGLQENLLRGIYAYGFEKPSAIQQRGIVPFCKGLDVIQQAQSGTGKTATFCSGVLQQ 100
Query: 202 IDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPVHL 261
+D Q ++L PTRELA Q +V + LG +L ++V GGTS+++D L VH+
Sbjct: 101 LDFSLIQCQALVLAPTRELAQQIEKVMRALGDYLGVKVHACVGGTSVREDQRILQAGVHV 160
Query: 262 LVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQILMFSA 321
+VGTPGR+ D+ K+ + M V+DEAD++LS F+ I + Q LPP Q+ +FSA
Sbjct: 161 VVGTPGRVFDMLKRQSLRADNIKMFVLDEADEMLSRGFKDQIYDIFQLLPPKIQVGVFSA 220
Query: 322 TFPVTVKDFKDRYLR-----------------KPYIIN----------LMDELTLKGITQ 354
T P + +++ K + +N L D ITQ
Sbjct: 221 TMPPEALEITRKFMSKPVRILVKRDELTLEGIKQFYVNVEKEEWKLETLCDLYETLAITQ 280
Query: 355 FYAFVEERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDVAR 414
FV R+KV + L K ++ G + +D+
Sbjct: 281 SVIFVNTRRKV---DWLTDKMRSRDHTVSATHGDMD----------------QNTRDII- 320
Query: 415 PP**SFPRLPQWCMQESRLYC---LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRF 471
+ ++ SR+ L RGID+Q V++VINFD P E YLHR+GRSGRF
Sbjct: 321 --------MREFRSGSSRVLITTDLLARGIDVQQVSLVINFDLPTQPENYLHRIGRSGRF 372
Query: 472 GHLGLAVNLITYEDRFNLYRIEQELGTEIKQIP 504
G G+A+N +T +D L+ I++ ++++P
Sbjct: 373 GRKGVAINFVTRDDERMLFDIQKFYNVVVEELP 405
>At1g54270 Eukaryotic Initiation Factor 4A-2
Length = 412
Score = 188 bits (477), Expect = 7e-48
Identities = 128/394 (32%), Positives = 197/394 (49%), Gaps = 60/394 (15%)
Query: 142 FEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALEK 201
F+ L+ LL GIY GFE+PS IQ+ I G D++ +A++GTGKTA F L++
Sbjct: 41 FDAMGLQENLLRGIYAYGFEKPSAIQQRGIVPFCKGLDVIQQAQSGTGKTATFCSGVLQQ 100
Query: 202 IDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPVHL 261
+D Q ++L PTRELA Q +V + LG + ++V GGTS+++D L VH+
Sbjct: 101 LDYALLQCQALVLAPTRELAQQIEKVMRALGDYQGVKVHACVGGTSVREDQRILQAGVHV 160
Query: 262 LVGTPGRILDLAKKGVCVLKDC-SMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQILMFS 320
+VGTPGR+ D+ ++ + DC M V+DEAD++LS F+ I + Q LPP Q+ +FS
Sbjct: 161 VVGTPGRVFDMLRRQ-SLRPDCIKMFVLDEADEMLSRGFKDQIYDIFQLLPPKIQVGVFS 219
Query: 321 ATFPVTVKDFKDRYLR-----------------KPYIIN----------LMDELTLKGIT 353
AT P + +++ K + +N L D IT
Sbjct: 220 ATMPPEALEITRKFMSKPVRILVKRDELTLEGIKQFYVNVEKEDWKLETLCDLYETLAIT 279
Query: 354 QFYAFVEERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDVA 413
Q FV R+KV + L K ++ G + +D+
Sbjct: 280 QSVIFVNTRRKV---DWLTDKMRSRDHTVSATHGDMD----------------QNTRDII 320
Query: 414 RPP**SFPRLPQWCMQESRLYC---LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGR 470
+ ++ SR+ L RGID+Q V++VINFD P E YLHR+GRSGR
Sbjct: 321 ---------MREFRSGSSRVLITTDLLARGIDVQQVSLVINFDLPTQPENYLHRIGRSGR 371
Query: 471 FGHLGLAVNLITYEDRFNLYRIEQELGTEIKQIP 504
FG G+A+N +T +D+ L+ I++ ++++P
Sbjct: 372 FGRKGVAINFVTLDDQRMLFDIQKFYNVVVEELP 405
>At1g72730 Eukaryotic initiation factor 4A like protein
Length = 414
Score = 185 bits (470), Expect = 5e-47
Identities = 126/395 (31%), Positives = 193/395 (47%), Gaps = 58/395 (14%)
Query: 140 NEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPAL 199
+ F+ L+ +LL GIY GFE+PS IQ+ I G D++ +A++GTGKTA F L
Sbjct: 41 DSFDAMELQPDLLRGIYAYGFEKPSAIQQRGIIPFCKGLDVIQQAQSGTGKTATFCSGVL 100
Query: 200 EKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPV 259
+++D Q ++L PTRELA Q +V + LG +L ++ GGTS+++D L V
Sbjct: 101 QQLDISLVQCQALVLAPTRELAQQIEKVMRALGDYLGVKAQACVGGTSVREDQRVLQSGV 160
Query: 260 HLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQILMF 319
H++VGTPGR+ DL ++ M V+DEAD++LS F+ I + Q LP Q+ +F
Sbjct: 161 HVVVGTPGRVFDLLRRQSLRADAIKMFVLDEADEMLSRGFKDQIYDIFQLLPSKVQVGVF 220
Query: 320 SATFPVTVKDFKDRYLR-----------------KPYIIN----------LMDELTLKGI 352
SAT P + +++ K + +N L D I
Sbjct: 221 SATMPPEALEITRKFMNKPVRILVKRDELTLEGIKQFYVNVDKEEWKLETLCDLYETLAI 280
Query: 353 TQFYAFVEERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDV 412
TQ FV R+KV + L K ++ G + +D+
Sbjct: 281 TQSVIFVNTRRKV---DWLTDKMRSRDHTVSATHGDMD----------------QNTRDI 321
Query: 413 ARPP**SFPRLPQWCMQESRLYC---LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSG 469
+ ++ SR+ L RGID+Q V++VINFD P E YLHR+GRSG
Sbjct: 322 I---------MREFRSGSSRVLITTDLLARGIDVQQVSLVINFDLPTQPENYLHRIGRSG 372
Query: 470 RFGHLGLAVNLITYEDRFNLYRIEQELGTEIKQIP 504
RFG G+A+N +T ED + I++ ++++P
Sbjct: 373 RFGRKGVAINFMTSEDERMMADIQRFYNVVVEELP 407
>At3g06480 putative RNA helicase
Length = 1088
Score = 169 bits (427), Expect = 4e-42
Identities = 126/418 (30%), Positives = 194/418 (46%), Gaps = 55/418 (13%)
Query: 122 LPPADTRYRTEDVTATKGN------EFEDYFLKRELLMGIYEKGFERPSPIQEESIPIAL 175
L P + + +VT T N FE L E+L + GF P+PIQ ++ PIAL
Sbjct: 411 LSPVEIYRKQHEVTTTGENIPAPYITFESSGLPPEILRELLSAGFPSPTPIQAQTWPIAL 470
Query: 176 TGSDILARAKNGTGKTAAFSIPALEKI----DQDNNIIQVVILVPTRELALQTSQVCKEL 231
DI+A AK G+GKT + IPA + + N V+IL PTRELA Q
Sbjct: 471 QSRDIVAIAKTGSGKTLGYLIPAFILLRHCRNDSRNGPTVLILAPTRELATQIQDEALRF 530
Query: 232 GKHLQIQVMVTTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEA 291
G+ +I GG + L + ++V TPGR+ D+ + + + S+LV+DEA
Sbjct: 531 GRSSRISCTCLYGGAPKGPQLKELERGADIVVATPGRLNDILEMKMIDFQQVSLLVLDEA 590
Query: 292 DKLLSPEFQPSIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPYIINL--MDELTL 349
D++L F+P I +++ +PP RQ LM++AT+P V+ L P +N+ +DEL
Sbjct: 591 DRMLDMGFEPQIRKIVNEIPPRRQTLMYTATWPKEVRKIASDLLVNPVQVNIGRVDELAA 650
Query: 350 -KGITQFYAFVEERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYS 408
K ITQ+ V + +K L + + ++V +F +
Sbjct: 651 NKAITQYVEVVPQMEKERRLEQILRS---------------------QERGSKVIIFCST 689
Query: 409 ---CKDVARPP**SFPRL----------PQWCMQESR--LYCLF------TRGIDIQAVN 447
C +AR F + W + + R C+ RG+DI+ +
Sbjct: 690 KRLCDHLARSVGRHFGAVVIHGDKTQGERDWVLNQFRSGKSCVLIATDVAARGLDIKDIR 749
Query: 448 VVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIKQIPP 505
VVIN+DFP E Y+HR+GR+GR G G+A T +D + + L +Q+PP
Sbjct: 750 VVINYDFPTGVEDYVHRIGRTGRAGATGVAFTFFTEQDWKYAPDLIKVLEGANQQVPP 807
Score = 32.7 bits (73), Expect = 0.49
Identities = 19/47 (40%), Positives = 23/47 (48%), Gaps = 1/47 (2%)
Query: 34 GFQQRPHHQYQQQQQYVQRHMMQNQHQQHYQNQQQNQQQNQQQQQQQ 80
G + H QQ QQ+ +H+ QQH QQ QQ QQ QQQ
Sbjct: 88 GHVSQQHGFQQQPQQFPSQHVRPQMMQQH-PAQQMPQQSGQQFPQQQ 133
Score = 29.6 bits (65), Expect = 4.2
Identities = 23/74 (31%), Positives = 31/74 (41%), Gaps = 19/74 (25%)
Query: 34 GFQQRP------HHQYQQQQQYVQRHMMQNQHQQHYQNQQQNQ-------------QQNQ 74
GFQQ+P H + Q QQ+ + M Q QQ Q Q Q+ Q
Sbjct: 95 GFQQQPQQFPSQHVRPQMMQQHPAQQMPQQSGQQFPQQQSQSMVPHPHGHPSVQTYQPTT 154
Query: 75 QQQQQQQWLRRNQL 88
QQQQQ + +Q+
Sbjct: 155 QQQQQGMQNQHSQM 168
>At1g16280 putative ATP-dependent RNA helicase
Length = 491
Score = 169 bits (427), Expect = 4e-42
Identities = 120/392 (30%), Positives = 190/392 (47%), Gaps = 30/392 (7%)
Query: 137 TKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSI 196
T FE L + E G +P+P+Q +P L G D+L A+ G+GKTAAF++
Sbjct: 55 TSATNFEGLGLAEWAVETCKELGMRKPTPVQTHCVPKILAGRDVLGLAQTGSGKTAAFAL 114
Query: 197 PALEKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLY 256
P L ++ +D + +++ PTRELA Q ++ K LG L ++ V GG + M L
Sbjct: 115 PILHRLAEDPYGVFALVVTPTRELAFQLAEQFKALGSCLNLRCSVIVGGMDMLTQTMSLV 174
Query: 257 QPVHLLVGTPGRILDLAKKG---VCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPT 313
H+++ TPGRI L + V LV+DEAD++L FQ + + Q LP +
Sbjct: 175 SRPHIVITTPGRIKVLLENNPDVPPVFSRTKFLVLDEADRVLDVGFQDELRTIFQCLPKS 234
Query: 314 RQILMFSATFPVTVKDFKDRYLRKPYIINLMDEL-TLKGITQFYAFVEERQKVHCLNTLF 372
RQ L+FSAT ++ + K Y + L T+ +TQ + F ++ K L +
Sbjct: 235 RQTLLFSATMTSNLQALLEHSSNKAYFYEAYEGLKTVDTLTQQFIFEDKDAKELYLVHIL 294
Query: 373 SKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDVARPP**SFPRLPQWCMQESR 432
S+ +K I + F + R L D + L M+ S
Sbjct: 295 SQMEDKGIRSAMIFVST----------CRTCQRLSLMLDELEVENIAMHSLNSQSMRLSA 344
Query: 433 L-------------YCLFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVN 479
L + +RG+DI V++VIN+D P++ Y+HRVGR+ R G GLAV+
Sbjct: 345 LSKFKSGKVPILLATDVASRGLDIPTVDLVINYDIPRDPRDYVHRVGRTARAGRGGLAVS 404
Query: 480 LITYEDRFNLYRIEQELGTEIKQIPPFIDQAI 511
+IT D +++IE+E+G K++ P+ + I
Sbjct: 405 IITETDVKLIHKIEEEVG---KKMEPYNKKVI 433
>At5g63120 ATP-dependent RNA helicase-like protein
Length = 591
Score = 164 bits (415), Expect = 1e-40
Identities = 124/428 (28%), Positives = 205/428 (46%), Gaps = 75/428 (17%)
Query: 129 YRTEDVTATKGNE-------FEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDIL 181
YRTE + +G + F+D +L I + GF P+PIQ + P+AL G D++
Sbjct: 147 YRTERDISVEGRDVPKPMKMFQDANFPDNILEAIAKLGFTEPTPIQAQGWPMALKGRDLI 206
Query: 182 ARAKNGTGKTAAFSIPAL------EKIDQDNNIIQVVILVPTRELALQTSQVCKELGKHL 235
A+ G+GKT A+ +PAL ++ QD+ I V+IL PTRELA+Q + ++ G
Sbjct: 207 GIAETGSGKTLAYLLPALVHVSAQPRLGQDDGPI-VLILAPTRELAVQIQEESRKFGLRS 265
Query: 236 QIQVMVTTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLL 295
++ GG I L + V +++ TPGR++D+ + LK + LV+DEAD++L
Sbjct: 266 GVRSTCIYGGAPKGPQIRDLRRGVEIVIATPGRLIDMLECQHTNLKRVTYLVLDEADRML 325
Query: 296 SPEFQPSIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPY--IINLMDELTLKGIT 353
F+P I +++ + P RQ L++SAT+P V+ ++LR PY II D + I
Sbjct: 326 DMGFEPQIRKIVSQIRPDRQTLLWSATWPREVETLARQFLRDPYKAIIGSTDLKANQSIN 385
Query: 354 QFYAFVEERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYS---CK 410
Q V +K + L TL + + +++ +F+ + C
Sbjct: 386 QVIEIVPTPEKYNRLLTLLKQLMDG---------------------SKILIFVETKRGCD 424
Query: 411 DVARPP**SFPRLPQW----------CMQESRLYCLF--------------TRGIDIQAV 446
V R R+ W + R+ F RG+D++ +
Sbjct: 425 QVTRQ-----LRMDGWPALAIHGDKTQSERDRVLAEFKSGRSPIMTATDVAARGLDVKDI 479
Query: 447 NVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYED-RF--NLYRIEQELGTEIKQI 503
V+N+DFP E Y+HR+GR+GR G G+A T+++ +F L +I QE G + +
Sbjct: 480 KCVVNYDFPNTLEDYIHRIGRTGRAGAKGMAFTFFTHDNAKFARELVKILQEAG---QVV 536
Query: 504 PPFIDQAI 511
PP + +
Sbjct: 537 PPTLSALV 544
>At1g55150 unknown protein
Length = 501
Score = 159 bits (402), Expect = 3e-39
Identities = 130/475 (27%), Positives = 207/475 (43%), Gaps = 79/475 (16%)
Query: 84 RRNQLGGGTDTNVVEEVEKTVQSEANDSSSQDWKARLK-LPPADTRYRTED--VTATKGN 140
RR+ G G ++ T + ND + K L L P + + E V A
Sbjct: 17 RRSDSGFGGTSSYGSSGSHTSSKKDNDGNESPRKLDLDGLTPFEKNFYVESPAVAAMTDT 76
Query: 141 EFEDYFLKREL-----------------------LMGIYEKGFERPSPIQEESIPIALTG 177
E E+Y RE+ L + + GF P+PIQ + P+A+ G
Sbjct: 77 EVEEYRKLREITVEGKDIPKPVKSFRDVGFPDYVLEEVKKAGFTEPTPIQSQGWPMAMKG 136
Query: 178 SDILARAKNGTGKTAAFSIPALEKIDQDNNIIQ-----VVILVPTRELALQTSQVCKELG 232
D++ A+ G+GKT ++ +PA+ ++ + V++L PTRELA+Q Q + G
Sbjct: 137 RDLIGIAETGSGKTLSYLLPAIVHVNAQPMLAHGDGPIVLVLAPTRELAVQIQQEASKFG 196
Query: 233 KHLQIQVMVTTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEAD 292
+I+ GG + L + V +++ TPGR++D+ + L+ + LV+DEAD
Sbjct: 197 SSSKIKTTCIYGGVPKGPQVRDLQKGVEIVIATPGRLIDMMESNNTNLRRVTYLVLDEAD 256
Query: 293 KLLSPEFQPSIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPY--IINLMDELTLK 350
++L F P I +++ + P RQ L +SAT+P V+ ++L PY II D +
Sbjct: 257 RMLDMGFDPQIRKIVSHIRPDRQTLYWSATWPKEVEQLSKKFLYNPYKVIIGSSDLKANR 316
Query: 351 GITQFYAFVEERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMFL---Y 407
I Q + E QK + L L + +R+ +FL
Sbjct: 317 AIRQIVDVISESQKYNKLVKLLEDIMDG---------------------SRILVFLDTKK 355
Query: 408 SCKDVARP-P**SFPRLP----------QWCMQESR--------LYCLFTRGIDIQAVNV 448
C + R +P L W + E R + RG+D++ V
Sbjct: 356 GCDQITRQLRMDGWPALSIHGDKSQAERDWVLSEFRSGKSPIMTATDVAARGLDVKDVKY 415
Query: 449 VINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYED-RF--NLYRIEQELGTEI 500
VIN+DFP + E Y+HR+GR+GR G G A T + RF L I QE G ++
Sbjct: 416 VINYDFPGSLEDYVHRIGRTGRAGAKGTAYTFFTVANARFAKELTNILQEAGQKV 470
>At3g22310 putative RNA helicase
Length = 610
Score = 159 bits (401), Expect = 5e-39
Identities = 114/399 (28%), Positives = 184/399 (45%), Gaps = 59/399 (14%)
Query: 139 GNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPA 198
G D + E++ + +G E+ PIQ+ + A+ G D++ RA+ GTGKT AF IP
Sbjct: 115 GLAIADLGISPEIVKALKGRGIEKLFPIQKAVLEPAMEGRDMIGRARTGTGKTLAFGIPI 174
Query: 199 LEKIDQDN------NIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDI 252
++KI + N Q ++L PTRELA Q + +E L + GGT + +
Sbjct: 175 IDKIIKFNAKHGRGKNPQCLVLAPTRELARQVEKEFRESAPSLD--TICLYGGTPIGQQM 232
Query: 253 MRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPP 312
L + + VGTPGRI+DL K+G L + +V+DEAD++L F +E ++Q LP
Sbjct: 233 RELNYGIDVAVGTPGRIIDLMKRGALNLSEVQFVVLDEADQMLQVGFAEDVEIILQKLPA 292
Query: 313 TRQILMFSATFPVTVKDFKDRYLRKPYIINLM---DELTLKGITQFYAFVEERQKVHCLN 369
RQ +MFSAT P ++ +YL P I+L+ D+ GIT + + + +
Sbjct: 293 KRQSMMFSATMPSWIRSLTKKYLNNPLTIDLVGDSDQKLADGITMYSIAADSYGRASIIG 352
Query: 370 TLFSK----------AANKPIHHLLQFGKSS*TPC*ENH*TRVFMFLYSCKDVARPP**S 419
L + K L FG + C H D+++
Sbjct: 353 PLVKEHGKGGKCIVFTQTKRDADRLAFGLAKSYKCEALH-----------GDISQ----- 396
Query: 420 FPRLPQWCMQESRLYCLFTRG--------------IDIQAVNVVINFDFPKNSETYLHRV 465
Q R F G +D+ V++VI+++ P N+ET++HR
Sbjct: 397 --------AQRERTLAGFRDGNFSILVATDVAARGLDVPNVDLVIHYELPNNTETFVHRT 448
Query: 466 GRSGRFGHLGLAVNLITYEDRFNLYRIEQELGTEIKQIP 504
GR+GR G G A+ + + + IE+E+G+ ++P
Sbjct: 449 GRTGRAGKKGSAILIHGQDQTRAVKMIEKEVGSRFNELP 487
>At5g65900 ATP-dependent RNA helicase-like
Length = 633
Score = 156 bits (395), Expect = 2e-38
Identities = 129/468 (27%), Positives = 220/468 (46%), Gaps = 33/468 (7%)
Query: 70 QQQNQQQQQQQQWLRRNQLGGGTDTNVVEEVEKTVQSEANDSSSQDWKARLKLPPADTRY 129
++ +++++ ++ +R D + EE E + + ++ ++ +
Sbjct: 84 EEPKKKKKKNKKLQQRGDTNDEEDEVIAEEEEPKKKKKKQRKDTEAKSEEEEVEDKEEEK 143
Query: 130 RTEDVTATKGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTG 189
+ E+ + FE L I E GF R + IQ ++IP + G D+L A+ G+G
Sbjct: 144 KLEETSIMTNKTFESLSLSDNTYKSIKEMGFARMTQIQAKAIPPLMMGEDVLGAARTGSG 203
Query: 190 KTAAFSIPALEKIDQ----DNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGG 245
KT AF IPA+E + + N V+++ PTRELA+Q+ V KEL K+ V GG
Sbjct: 204 KTLAFLIPAVELLYRVKFTPRNGTGVLVICPTRELAIQSYGVAKELLKYHSQTVGKVIGG 263
Query: 246 TSLKDDIMRLYQPVHLLVGTPGRILD-LAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIE 304
K + L + V+LLV TPGR+LD L + K+ LVMDEAD++L F+ ++
Sbjct: 264 EKRKTEAEILAKGVNLLVATPGRLLDHLENTNGFIFKNLKFLVMDEADRILEQNFEEDLK 323
Query: 305 QLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPYIINLMD---ELTLKGITQFYAFVEE 361
+++ LP TRQ +FSAT V+D L P I++ + E+T +G+ Q Y V
Sbjct: 324 KILNLLPKTRQTSLFSATQSAKVEDLARVSLTSPVYIDVDEGRKEVTNEGLEQGYCVVPS 383
Query: 362 RQKVHCLNTLFSK-AANKPIHHLLQFGKSS*TPC*ENH*TRVFMFL-YSCKDVARPP**S 419
++ L T + K I KS+ +F ++ + C ++ R
Sbjct: 384 AMRLLFLLTFLKRFQGKKKIMVFFSTCKST------KFHAELFRYIKFDCLEI-RGGIDQ 436
Query: 420 FPRLPQWCM----QESRLYC--LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGR-FG 472
R P + + L C + RG+D V+ ++ +D P N Y+HRVGR+ R G
Sbjct: 437 NKRTPTFLQFIKAETGILLCTNVAARGLDFPHVDWIVQYDPPDNPTDYIHRVGRTARGEG 496
Query: 473 HLGLAVNLITYED-RFNLYRIEQELGTE--------IKQIPPFIDQAI 511
G A+ ++T ++ +F Y ++ E + + PF++ I
Sbjct: 497 AKGKALLVLTPQELKFIQYLKAAKIPVEEHEFEEKKLLDVKPFVENLI 544
>At3g01540 AT3g01540/F4P13_9
Length = 619
Score = 156 bits (395), Expect = 2e-38
Identities = 121/396 (30%), Positives = 198/396 (49%), Gaps = 23/396 (5%)
Query: 130 RTEDVTATKGN------EFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILAR 183
R ++T + G FE ELL + GF P+PIQ +S PIA+ G DI+A
Sbjct: 142 RRHEITVSGGQVPPPLMSFEATGFPPELLREVLSAGFSAPTPIQAQSWPIAMQGRDIVAI 201
Query: 184 AKNGTGKTAAFSIPA---LEKIDQDNNI-IQVVILVPTRELALQTSQVCKELGKHLQIQV 239
AK G+GKT + IP L++I D+ + +++L PTRELA Q + + G+ +I
Sbjct: 202 AKTGSGKTLGYLIPGFLHLQRIRNDSRMGPTILVLSPTRELATQIQEEAVKFGRSSRISC 261
Query: 240 MVTTGGTSLKDDIMRLYQPVHLLVGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEF 299
GG + L + ++V TPGR+ D+ + L+ S LV+DEAD++L F
Sbjct: 262 TCLYGGAPKGPQLRDLERGADIVVATPGRLNDILEMRRISLRQISYLVLDEADRMLDMGF 321
Query: 300 QPSIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPYIINL--MDELTL-KGITQFY 356
+P I ++++ +P RQ LM++AT+P V+ L P +N+ +DEL K ITQ
Sbjct: 322 EPQIRKIVKEIPTKRQTLMYTATWPKGVRKIAADLLVNPAQVNIGNVDELVANKSITQHI 381
Query: 357 AFVEERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*E--NH*TRVFMFLYSCKDVAR 414
V +K L + + +P ++ F S+ C + + TR F D ++
Sbjct: 382 EVVAPMEKQRRLEQIL--RSQEPGSKVIIF-CSTKRMCDQLTRNLTRQFGAAAIHGDKSQ 438
Query: 415 PP**SFPRLPQWCMQESRLYC---LFTRGIDIQAVNVVINFDFPKNSETYLHRVGRSGRF 471
P + L Q+ + + + RG+D++ + V+N+DFP E Y+HR+GR+GR
Sbjct: 439 PERDNV--LNQFRSGRTPVLVATDVAARGLDVKDIRAVVNYDFPNGVEDYVHRIGRTGRA 496
Query: 472 GHLGLAVNLITYEDRFNLYRIEQELGTEIKQIPPFI 507
G G A +D + + + L +++PP I
Sbjct: 497 GATGQAFTFFGDQDSKHASDLIKILEGANQRVPPQI 532
>At3g18600 DEAD box helicase protein, putative
Length = 568
Score = 155 bits (393), Expect = 4e-38
Identities = 124/428 (28%), Positives = 202/428 (46%), Gaps = 44/428 (10%)
Query: 77 QQQQQWLRRNQLGGGTDTNVVEEVEKTVQSEANDSSSQDWKARLKLPPADTRYRTEDVTA 136
++ ++ +R+ G + EE TV+ A+++ + K ++K E V A
Sbjct: 10 EELKKRVRKRSRGKKNEQQKAEEKTHTVEENADETQKKSEK-KVKKVRGKIEEEEEKVEA 68
Query: 137 TKGNE-----------------FEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSD 179
+ E F+ L + + I E GF+ + IQ SI L G D
Sbjct: 69 MEDGEDEKNIVIVGKGIMTNVTFDSLDLSEQTSIAIKEMGFQYMTQIQAGSIQPLLEGKD 128
Query: 180 ILARAKNGTGKTAAFSIPALEKIDQDN----NIIQVVILVPTRELALQTSQVCKELGKHL 235
+L A+ G+GKT AF IPA+E + ++ N V+++ PTRELA+QT V +EL KH
Sbjct: 129 VLGAARTGSGKTLAFLIPAVELLFKERFSPRNGTGVIVICPTRELAIQTKNVAEELLKHH 188
Query: 236 QIQVMVTTGGTSLKDDIMRLYQPVHLLVGTPGRILD-LAKKGVCVLKDCSMLVMDEADKL 294
V + GG + + + R+ +L++ TPGR+LD L + K LV+DEAD++
Sbjct: 189 SQTVSMVIGGNNRRSEAQRIASGSNLVIATPGRLLDHLQNTKAFIYKHLKCLVIDEADRI 248
Query: 295 LSPEFQPSIEQLIQFLPPTRQILMFSATFPVTVKDFKDRYLRKPYIINLMD---ELTLKG 351
L F+ + ++++ LP TRQ +FSAT VKD L P +++ D ++T +G
Sbjct: 249 LEENFEEDMNKILKILPKTRQTALFSATQTSKVKDLARVSLTSPVHVDVDDGRRKVTNEG 308
Query: 352 ITQFYAFVEERQKVHCLNTLFSKAANKPIHHLLQFGKSS*TPC*ENH*TRVFMF------ 405
+ Q Y V +Q++ L + K NK I KS + H T +
Sbjct: 309 LEQGYCVVPSKQRLILLISFLKKNLNKKIMVFFSTCKSV-----QFH-TEIMKISDVDVS 362
Query: 406 -LYSCKDVARPP**SFPRLPQWCMQESRLYC--LFTRGIDIQAVNVVINFDFPKNSETYL 462
++ D R F + ++ L C + RG+DI +V+ +I +D P Y+
Sbjct: 363 DIHGGMDQNRRTKTFFDFMK---AKKGILLCTDVAARGLDIPSVDWIIQYDPPDKPTEYI 419
Query: 463 HRVGRSGR 470
HRVGR+ R
Sbjct: 420 HRVGRTAR 427
>At1g77050
Length = 513
Score = 140 bits (352), Expect = 2e-33
Identities = 78/209 (37%), Positives = 122/209 (58%), Gaps = 3/209 (1%)
Query: 138 KGNEFEDYFLKRELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIP 197
K FE L + I +KG++ P+PIQ +++P+ L+G D++A A+ G+GKTAAF IP
Sbjct: 26 KSGGFESLNLGPNVFNAIKKKGYKVPTPIQRKTMPLILSGVDVVAMARTGSGKTAAFLIP 85
Query: 198 ALEKIDQ--DNNIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRL 255
LEK+ Q ++ +IL PTR+LA QT + KELGK ++V + GG S++D L
Sbjct: 86 MLEKLKQHVPQGGVRALILSPTRDLAEQTLKFTKELGKFTDLRVSLLVGGDSMEDQFEEL 145
Query: 256 YQPVHLLVGTPGRILD-LAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTR 314
+ +++ TPGR++ L++ L+ +V DEAD L F + Q++ L R
Sbjct: 146 TKGPDVIIATPGRLMHLLSEVDDMTLRTVEYVVFDEADSLFGMGFAEQLHQILTQLSENR 205
Query: 315 QILMFSATFPVTVKDFKDRYLRKPYIINL 343
Q L+FSAT P + +F LR+P ++ L
Sbjct: 206 QTLLFSATLPSALAEFAKAGLREPQLVRL 234
Score = 51.2 bits (121), Expect = 1e-06
Identities = 25/66 (37%), Positives = 38/66 (56%)
Query: 439 RGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQELGT 498
RGIDI ++ VIN+DFP + ++HRVGR+ R G G A + +T ED + + L
Sbjct: 333 RGIDIPLLDNVINWDFPPRPKIFVHRVGRAARAGRTGCAYSFVTPEDMPYMLDLHLFLSK 392
Query: 499 EIKQIP 504
++ P
Sbjct: 393 PVRPAP 398
>At3g22330 DEAD-Box RNA helicase like protein
Length = 616
Score = 132 bits (332), Expect = 5e-31
Identities = 79/237 (33%), Positives = 128/237 (53%), Gaps = 13/237 (5%)
Query: 150 ELLMGIYEKGFERPSPIQEESIPIALTGSDILARAKNGTGKTAAFSIPALEKIDQDN--- 206
E++ + KG E+ PIQ+ + A+ G D++ RA+ GTGKT AF IP ++KI + N
Sbjct: 114 EIVKALSSKGIEKLFPIQKAVLEPAMEGRDMIGRARTGTGKTLAFGIPIIDKIIKYNAKH 173
Query: 207 ----NIIQVVILVPTRELALQTSQVCKELGKHLQIQVMVTTGGTSLKDDIMRLYQPVHLL 262
N + ++L PTRELA Q + +E L + GGT + + +L V +
Sbjct: 174 GRGRNPL-CLVLAPTRELARQVEKEFRESAPSLD--TICLYGGTPIGQQMRQLDYGVDVA 230
Query: 263 VGTPGRILDLAKKGVCVLKDCSMLVMDEADKLLSPEFQPSIEQLIQFLPPTRQILMFSAT 322
VGTPGR++DL K+G L + +V+DEAD++L F +E +++ LP RQ +MFSAT
Sbjct: 231 VGTPGRVIDLMKRGALNLSEVQFVVLDEADQMLQVGFAEDVEIILEKLPEKRQSMMFSAT 290
Query: 323 FPVTVKDFKDRYLRKPYIINLM---DELTLKGITQFYAFVEERQKVHCLNTLFSKAA 376
P ++ +YL P ++L+ D+ GIT + + + + L ++ A
Sbjct: 291 MPSWIRSLTKKYLNNPLTVDLVGDSDQKLADGITTYSIIADSYGRASIIGPLVTEHA 347
Score = 57.4 bits (137), Expect = 2e-08
Identities = 23/66 (34%), Positives = 45/66 (67%)
Query: 439 RGIDIQAVNVVINFDFPKNSETYLHRVGRSGRFGHLGLAVNLITYEDRFNLYRIEQELGT 498
RG+D+ V+++I+++ P N+ET++HR GR+GR G G A+ + + + + IE+E+G+
Sbjct: 410 RGLDVPNVDLIIHYELPNNTETFVHRTGRTGRAGKKGSAILIYSQDQSRAVKIIEREVGS 469
Query: 499 EIKQIP 504
++P
Sbjct: 470 RFTELP 475
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.140 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,592,487
Number of Sequences: 26719
Number of extensions: 516677
Number of successful extensions: 10039
Number of sequences better than 10.0: 266
Number of HSP's better than 10.0 without gapping: 203
Number of HSP's successfully gapped in prelim test: 69
Number of HSP's that attempted gapping in prelim test: 5006
Number of HSP's gapped (non-prelim): 2044
length of query: 513
length of database: 11,318,596
effective HSP length: 104
effective length of query: 409
effective length of database: 8,539,820
effective search space: 3492786380
effective search space used: 3492786380
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC123899.6


