

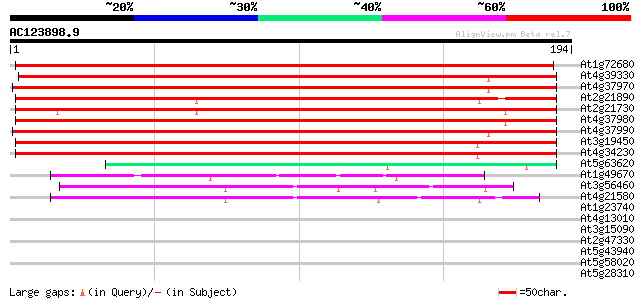
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123898.9 + phase: 0 /pseudo
(194 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g72680 putative cinnamyl-alcohol dehydrogenase 297 3e-81
At4g39330 cinnamyl-alcohol dehydrogenase CAD1 186 9e-48
At4g37970 cinnamyl alcohol dehydrogenase -like protein, LCADa 183 6e-47
At2g21890 putative cinnamyl-alcohol dehydrogenase 178 2e-45
At2g21730 putative cinnamyl-alcohol dehydrogenase 177 3e-45
At4g37980 cinnamyl-alcohol dehydrogenase ELI3-1 176 5e-45
At4g37990 cinnamyl-alcohol dehydrogenase ELI3-2 171 3e-43
At3g19450 putative cinnamyl alcohol dehydrogenase 2 166 5e-42
At4g34230 cinnamyl alcohol dehydrogenase like protein 166 9e-42
At5g63620 alcohol dehydrogenase-like protein 49 1e-06
At1g49670 ARP protein (At1g49670; F14J22.10) 47 6e-06
At3g56460 quinone reductase-like protein 45 3e-05
At4g21580 putative NADPH quinone oxidoreductase 41 5e-04
At1g23740 auxin-induced protein like 39 0.002
At4g13010 unknown protein 37 0.007
At3g15090 zinc-binding dehydrogenase, putative 36 0.015
At2g47330 putative ATP-dependent RNA helicase 32 0.16
At5g43940 alcohol dehydrogenase (EC 1.1.1.1) class III (pir||S71... 31 0.36
At5g58020 unknown protein 31 0.47
At5g28310 putative protein 30 0.80
>At1g72680 putative cinnamyl-alcohol dehydrogenase
Length = 355
Score = 297 bits (760), Expect = 3e-81
Identities = 140/186 (75%), Positives = 165/186 (88%)
Query: 3 VYTFNCVDYDGTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNM 62
V+TFN +D+DG++TKGGYS+ IVV+ERYC+ IP YPL SA PLLCAGITVY+PMMRHNM
Sbjct: 119 VFTFNGIDHDGSVTKGGYSSHIVVHERYCYKIPVDYPLESAAPLLCAGITVYAPMMRHNM 178
Query: 63 NQPGKSLGVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEE 122
NQPGKSLGV+GLGGLGHMAVKFGKAFGL VTVFSTS+SKKEEAL+ LGA+ FV+SS+ ++
Sbjct: 179 NQPGKSLGVIGLGGLGHMAVKFGKAFGLSVTVFSTSISKKEEALNLLGAENFVISSDHDQ 238
Query: 123 MRALAKTLDFIIDTASGDHPFDPYMSLLKISGVLALVGFPSEVKFSPASLNLGSRTVAGS 182
M+AL K+LDF++DTASGDH FDPYMSLLKI+G LVGFPSE+K SPA+LNLG R +AGS
Sbjct: 239 MKALEKSLDFLVDTASGDHAFDPYMSLLKIAGTYVLVGFPSEIKISPANLNLGMRMLAGS 298
Query: 183 AAGGTK 188
GGTK
Sbjct: 299 VTGGTK 304
>At4g39330 cinnamyl-alcohol dehydrogenase CAD1
Length = 360
Score = 186 bits (471), Expect = 9e-48
Identities = 94/187 (50%), Positives = 128/187 (68%), Gaps = 1/187 (0%)
Query: 4 YTFNCVDYDGTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNMN 63
+T+N + DGT GGYS +IVV++R+ P++ P S PLLCAGITVYSPM + M
Sbjct: 122 FTYNAIGSDGTKNYGGYSENIVVDQRFVLRFPENLPSDSGAPLLCAGITVYSPMKYYGMT 181
Query: 64 QPGKSLGVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEEM 123
+ GK LGV GLGGLGH+AVK GKAFGL+VTV S+S +K EEA++ LGAD F+V+++ ++M
Sbjct: 182 EAGKHLGVAGLGGLGHVAVKIGKAFGLKVTVISSSSTKAEEAINHLGADSFLVTTDPQKM 241
Query: 124 RALAKTLDFIIDTASGDHPFDPYMSLLKISGVLALVGFPSE-VKFSPASLNLGSRTVAGS 182
+A T+D+IIDT S H P + LLK++G L +G P + ++ L LG + V GS
Sbjct: 242 KAAIGTMDYIIDTISAVHALYPLLGLLKVNGKLIALGLPEKPLELPMFPLVLGRKMVGGS 301
Query: 183 AAGGTKE 189
GG KE
Sbjct: 302 DVGGMKE 308
>At4g37970 cinnamyl alcohol dehydrogenase -like protein, LCADa
Length = 363
Score = 183 bits (464), Expect = 6e-47
Identities = 94/189 (49%), Positives = 128/189 (66%), Gaps = 1/189 (0%)
Query: 2 SVYTFNCVDYDGTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHN 61
++ T+N V +DGTI GGYS IVV+ERY IP + PL SA PLLCAGI++YSPM
Sbjct: 121 AIATYNGVHHDGTINYGGYSDHIVVDERYAVKIPHTLPLVSAAPLLCAGISMYSPMKYFG 180
Query: 62 MNQPGKSLGVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQE 121
+ P K +G+VGLGGLGH+ V+F KAFG +VTV S++ K ++AL LGAD F+VS++++
Sbjct: 181 LTGPDKHVGIVGLGGLGHIGVRFAKAFGTKVTVVSSTTGKSKDALDTLGADGFLVSTDED 240
Query: 122 EMRALAKTLDFIIDTASGDHPFDPYMSLLKISGVLALVGFPSE-VKFSPASLNLGSRTVA 180
+M+A T+D IIDT S H P + LLK +G L L+G + S SL LG +++A
Sbjct: 241 QMKAAMGTMDGIIDTVSASHSISPLIGLLKSNGKLVLLGATEKPFDISAFSLILGRKSIA 300
Query: 181 GSAAGGTKE 189
GS GG +E
Sbjct: 301 GSGIGGMQE 309
>At2g21890 putative cinnamyl-alcohol dehydrogenase
Length = 375
Score = 178 bits (451), Expect = 2e-45
Identities = 99/191 (51%), Positives = 123/191 (63%), Gaps = 6/191 (3%)
Query: 3 VYTFNCVDYDGTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNM 62
V+T+N DGT +GGYS IVV+ R+ IP P S PLLCAGITVYSPM + M
Sbjct: 115 VFTYNSRSSDGTRNQGGYSDVIVVDHRFVLSIPDGLPSDSGAPLLCAGITVYSPMKYYGM 174
Query: 63 N-QPGKSLGVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQE 121
+ GK LGV GLGGLGH+AVK GKAFGLRVTV S S K+ EA+ LGAD F+V+++ +
Sbjct: 175 TKESGKRLGVNGLGGLGHIAVKIGKAFGLRVTVISRSSEKEREAIDRLGADSFLVTTDSQ 234
Query: 122 EMRALAKTLDFIIDTASGDHPFDPYMSLLKISGVLALVGF---PSEVKFSPASLNLGSRT 178
+M+ T+DFIIDT S +H P SLLK+SG L +G P ++ P L LG +
Sbjct: 235 KMKEAVGTMDFIIDTVSAEHALLPLFSLLKVSGKLVALGLLEKPLDLPIFP--LVLGRKM 292
Query: 179 VAGSAAGGTKE 189
V GS GG KE
Sbjct: 293 VGGSQIGGMKE 303
>At2g21730 putative cinnamyl-alcohol dehydrogenase
Length = 376
Score = 177 bits (449), Expect = 3e-45
Identities = 99/190 (52%), Positives = 123/190 (64%), Gaps = 3/190 (1%)
Query: 3 VYTFNCVDYDGTI-TKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHN 61
V+T+N DGT +GGYS IVV+ R+ IP P S PLLCAGITVYSPM +
Sbjct: 115 VFTYNSRSSDGTSRNQGGYSDVIVVDHRFVLSIPDGLPSDSGAPLLCAGITVYSPMKYYG 174
Query: 62 MN-QPGKSLGVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQ 120
M + GK LGV GLGGLGH+AVK GKAFGLRVTV S S K+ EA+ LGAD F+V+++
Sbjct: 175 MTKESGKRLGVNGLGGLGHIAVKIGKAFGLRVTVISRSSEKEREAIDRLGADSFLVTTDS 234
Query: 121 EEMRALAKTLDFIIDTASGDHPFDPYMSLLKISGVLALVGFPSEVKFSPA-SLNLGSRTV 179
++M+ T+DFIIDT S +H P SLLK++G L +G P + P SL LG + V
Sbjct: 235 QKMKEAVGTMDFIIDTVSAEHALLPLFSLLKVNGKLVALGLPEKPLDLPIFSLVLGRKMV 294
Query: 180 AGSAAGGTKE 189
GS GG KE
Sbjct: 295 GGSQIGGMKE 304
>At4g37980 cinnamyl-alcohol dehydrogenase ELI3-1
Length = 357
Score = 176 bits (447), Expect = 5e-45
Identities = 93/188 (49%), Positives = 121/188 (63%), Gaps = 1/188 (0%)
Query: 3 VYTFNCVDYDGTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNM 62
+ T ++D T+T GGYS +V E + IP + PL A PLLCAG+TVYSPM H +
Sbjct: 117 ILTSGAKNFDDTMTHGGYSDHMVCAEDFIIRIPDNLPLDGAAPLLCAGVTVYSPMKYHGL 176
Query: 63 NQPGKSLGVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEE 122
++PG +GVVGLGGLGH+AVKF KA G +VTV STS K++EA++ LGAD F+VS + ++
Sbjct: 177 DKPGMHIGVVGLGGLGHVAVKFAKAMGTKVTVISTSERKRDEAVTRLGADAFLVSRDPKQ 236
Query: 123 MRALAKTLDFIIDTASGDHPFDPYMSLLKISGVLALVGFPSEVKFSPA-SLNLGSRTVAG 181
M+ T+D IIDT S HP P + LLK G L +VG P+E P L G + V G
Sbjct: 237 MKDAMGTMDGIIDTVSATHPLLPLLGLLKNKGKLVMVGAPAEPLELPVFPLIFGRKMVVG 296
Query: 182 SAAGGTKE 189
S GG KE
Sbjct: 297 SMVGGIKE 304
>At4g37990 cinnamyl-alcohol dehydrogenase ELI3-2
Length = 359
Score = 171 bits (432), Expect = 3e-43
Identities = 91/189 (48%), Positives = 120/189 (63%), Gaps = 1/189 (0%)
Query: 2 SVYTFNCVDYDGTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHN 61
S+ T+ YD TIT GGYS +V E + IP + PL +A PLLCAGITVYSPM H
Sbjct: 116 SIQTYGFPYYDNTITYGGYSDHMVCEEGFVIRIPDNLPLDAAAPLLCAGITVYSPMKYHG 175
Query: 62 MNQPGKSLGVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQE 121
+++PG +GVVGLGGLGH+ VKF KA G +VTV STS K++EA++ LGAD F+VS + +
Sbjct: 176 LDKPGMHIGVVGLGGLGHVGVKFAKAMGTKVTVISTSEKKRDEAINRLGADAFLVSRDPK 235
Query: 122 EMRALAKTLDFIIDTASGDHPFDPYMSLLKISGVLALVGFPSE-VKFSPASLNLGSRTVA 180
+++ T+D IIDT S H P + LLK G L +VG P + ++ L + V
Sbjct: 236 QIKDAMGTMDGIIDTVSATHSLLPLLGLLKHKGKLVMVGAPEKPLELPVMPLIFERKMVM 295
Query: 181 GSAAGGTKE 189
GS GG KE
Sbjct: 296 GSMIGGIKE 304
>At3g19450 putative cinnamyl alcohol dehydrogenase 2
Length = 365
Score = 166 bits (421), Expect = 5e-42
Identities = 84/188 (44%), Positives = 123/188 (64%), Gaps = 1/188 (0%)
Query: 3 VYTFNCVDYDGTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNM 62
++++N V DG T+GG++ +++VN+++ IP+ + A PLLCAG+TVYSP+ +
Sbjct: 119 IWSYNDVYTDGKPTQGGFADTMIVNQKFVVKIPEGMAVEQAAPLLCAGVTVYSPLSHFGL 178
Query: 63 NQPGKSLGVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEE 122
G G++GLGG+GHM VK KA G VTV S+S KKEEA+ LGAD +VVSS+ E
Sbjct: 179 MASGLKGGILGLGGVGHMGVKIAKAMGHHVTVISSSDKKKEEAIEHLGADDYVVSSDPAE 238
Query: 123 MRALAKTLDFIIDTASGDHPFDPYMSLLKISGVLALVG-FPSEVKFSPASLNLGSRTVAG 181
M+ LA +LD+IIDT HP DPY++ LK+ G L L+G + ++F + LG + ++G
Sbjct: 239 MQRLADSLDYIIDTVPVFHPLDPYLACLKLDGKLILMGVINTPLQFVTPLVILGRKVISG 298
Query: 182 SAAGGTKE 189
S G KE
Sbjct: 299 SFIGSIKE 306
>At4g34230 cinnamyl alcohol dehydrogenase like protein
Length = 357
Score = 166 bits (419), Expect = 9e-42
Identities = 83/188 (44%), Positives = 122/188 (64%), Gaps = 1/188 (0%)
Query: 3 VYTFNCVDYDGTITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNM 62
++++N V +G T+GG++ + VV++++ IP+ + A PLLCAG+TVYSP+ +
Sbjct: 118 IWSYNDVYINGQPTQGGFAKATVVHQKFVVKIPEGMAVEQAAPLLCAGVTVYSPLSHFGL 177
Query: 63 NQPGKSLGVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEE 122
QPG G++GLGG+GHM VK KA G VTV S+S K+EEAL LGAD +V+ S+Q +
Sbjct: 178 KQPGLRGGILGLGGVGHMGVKIAKAMGHHVTVISSSNKKREEALQDLGADDYVIGSDQAK 237
Query: 123 MRALAKTLDFIIDTASGDHPFDPYMSLLKISGVLALVG-FPSEVKFSPASLNLGSRTVAG 181
M LA +LD++IDT H +PY+SLLK+ G L L+G + ++F L LG + + G
Sbjct: 238 MSELADSLDYVIDTVPVHHALEPYLSLLKLDGKLILMGVINNPLQFLTPLLMLGRKVITG 297
Query: 182 SAAGGTKE 189
S G KE
Sbjct: 298 SFIGSMKE 305
>At5g63620 alcohol dehydrogenase-like protein
Length = 427
Score = 49.3 bits (116), Expect = 1e-06
Identities = 36/164 (21%), Positives = 66/164 (39%), Gaps = 8/164 (4%)
Query: 34 IPKSYPLASAGPLLCAGITVYSPMMRHNMNQPGKSLGVVGLGGLGHMAVKFGKAFGLRVT 93
+P+S P + + L CA T Y M +PG S+ V+G+GG+G ++ +AFG
Sbjct: 214 LPESLPYSESAILGCAVFTAYGAMAHAAEIRPGDSIAVIGIGGVGSSCLQIARAFGASDI 273
Query: 94 VFSTSMSKKEEALSFLGADQFVVSSNQEEMRALAKT-----LDFIIDTASGDHPFDPYMS 148
+ K + LGA V ++ ++ + + + +D ++ F
Sbjct: 274 IAVDVQDDKLQKAKTLGATHIVNAAKEDAVERIREITGGMGVDVAVEALGKPQTFMQCTL 333
Query: 149 LLKISGVLALVGFPSEVKFSPASLNLGSR---TVAGSAAGGTKE 189
+K G ++G +N R V GS G ++
Sbjct: 334 SVKDGGKAVMIGLSQAGSVGEIDINRLVRRKIKVIGSYGGRARQ 377
>At1g49670 ARP protein (At1g49670; F14J22.10)
Length = 629
Score = 47.0 bits (110), Expect = 6e-06
Identities = 41/156 (26%), Positives = 76/156 (48%), Gaps = 11/156 (7%)
Query: 15 ITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNMNQPGKS-LGVVG 73
+T G YS ++V+ ++ +P+ P A +L +G+T + + + G++ L
Sbjct: 379 MTFGAYSEYMIVSSKHVLPVPRPDPEVVA--MLTSGLTALIALEKAGQMKSGETVLVTAA 436
Query: 74 LGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEEMRALAKTLDF- 132
GG G AV+ K G +V + + S+K + L LG D+ V+ E ++ + K +F
Sbjct: 437 AGGTGQFAVQLAKLSGNKV-IATCGGSEKAKLLKELGVDR-VIDYKSENIKTVLKK-EFP 493
Query: 133 ----IIDTASGDHPFDPYMSLLKISGVLALVGFPSE 164
II + G FD ++ L + G L ++G S+
Sbjct: 494 KGVNIIYESVGGQMFDMCLNALAVYGRLIVIGMISQ 529
>At3g56460 quinone reductase-like protein
Length = 348
Score = 44.7 bits (104), Expect = 3e-05
Identities = 45/167 (26%), Positives = 80/167 (46%), Gaps = 12/167 (7%)
Query: 18 GGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNMNQPGKSLGVVG-LGG 76
G ++ IV ++ FL+P+ + +A L A T + ++ G+ L V+G GG
Sbjct: 103 GSFAQFIVADQSRLFLVPERCDMVAAAALPVAFGTSHVALVHRARLTSGQVLLVLGAAGG 162
Query: 77 LGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGAD-------QFVVSSNQEEMRA-LAK 128
+G AV+ GK G V + ++K + L +G D + V+SS +E ++ K
Sbjct: 163 VGLAAVQIGKVCGAIVIAVARG-TEKIQLLKSMGVDHVVDLGTENVISSVKEFIKTRKLK 221
Query: 129 TLDFIIDTASGDHPFDPYMSLLKISGVLALVGFPS-EVKFSPASLNL 174
+D + D G + M +LK + ++GF S E+ PA++ L
Sbjct: 222 GVDVLYDPVGGKLTKES-MKVLKWGAQILVIGFASGEIPVIPANIAL 267
>At4g21580 putative NADPH quinone oxidoreductase
Length = 325
Score = 40.8 bits (94), Expect = 5e-04
Identities = 47/179 (26%), Positives = 75/179 (41%), Gaps = 14/179 (7%)
Query: 15 ITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNMNQPGKSLGVVG- 73
++ GGY+ + V F IP L A TV+S + G+S + G
Sbjct: 89 LSGGGYAEKVSVPAGQIFPIPAGISLKDAAAFPEVACTVWSTVFMMGRLSVGESFLIHGG 148
Query: 74 LGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEEMRAL-----AK 128
G+G A++ K G+RV V + S +K A LGAD + ++ + + K
Sbjct: 149 SSGIGTFAIQIAKHLGVRVFVTAGS-DEKLAACKELGADVCINYKTEDFVAKVKAETDGK 207
Query: 129 TLDFIIDTASGDHPFDPYMSLLKISGVLALVGF----PSEVKFSPASLNLGSRTVAGSA 183
+D I+D + + L G L ++G +E+K S SL TV G+A
Sbjct: 208 GVDVILDCIGAPY-LQKNLDSLNFDGRLCIIGLMGGANAEIKLS--SLLPKRLTVLGAA 263
>At1g23740 auxin-induced protein like
Length = 386
Score = 38.9 bits (89), Expect = 0.002
Identities = 40/145 (27%), Positives = 65/145 (44%), Gaps = 7/145 (4%)
Query: 18 GGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNMNQPGKSLGVV-GLGG 76
G + V E+ L PK+ A A L A T ++R + GKS+ V+ G GG
Sbjct: 180 GSLAEYTAVEEKLLALKPKNIDFAQAAGLPLAIETADEGLVRTEFSA-GKSILVLNGAGG 238
Query: 77 LGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEEMRALAKTLDFIIDT 136
+G + ++ K V +T+ ++K E + LGAD + +E + L D + D
Sbjct: 239 VGSLVIQLAKHVYGASKVAATASTEKLELVRSLGAD-LAIDYTKENIEDLPDKYDVVFDA 297
Query: 137 ASGDHPFDPYMSLLKISG-VLALVG 160
D + ++K G V+AL G
Sbjct: 298 IG---MCDKAVKVIKEGGKVVALTG 319
>At4g13010 unknown protein
Length = 329
Score = 37.0 bits (84), Expect = 0.007
Identities = 40/147 (27%), Positives = 62/147 (41%), Gaps = 14/147 (9%)
Query: 18 GGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRH---NMNQPGKSLGVV-- 72
GG + V E+ P+ A A L AG+T + ++ GK ++
Sbjct: 103 GGLAEFAVATEKLTVKRPQEVGAAEAAALPVAGLTALQALTNPAGLKLDGTGKKANILVT 162
Query: 73 -GLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEEMRAL----A 127
GG+GH AV+ K VT +T ++ E + LGAD+ V+ E AL
Sbjct: 163 AASGGVGHYAVQLAKLANAHVT--ATCGARNIEFVKSLGADE-VLDYKTPEGAALKSPSG 219
Query: 128 KTLDFIIDTASGDHPFDPYMSLLKISG 154
K D ++ A+G PF + L +G
Sbjct: 220 KKYDAVVHCANG-IPFSVFEPNLSENG 245
>At3g15090 zinc-binding dehydrogenase, putative
Length = 366
Score = 35.8 bits (81), Expect = 0.015
Identities = 31/127 (24%), Positives = 55/127 (42%), Gaps = 3/127 (2%)
Query: 14 TITKGGYSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNMNQPGKSLGVVG 73
T +G Y+ +++E P S A + A +T + + + G+ L V G
Sbjct: 126 TALRGTYTDYGILSEDELTEKPSSISHVEASAIPFAALTAWRALKSNARITEGQRLLVFG 185
Query: 74 LGG-LGHMAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEEMRALAKTLDF 132
GG +G A++ A G VT +K + + GA+Q V + ++ A+ D
Sbjct: 186 GGGAVGFSAIQLAVASGCHVTASCVGQTK--DRILAAGAEQAVDYTTEDIELAVKGKFDA 243
Query: 133 IIDTASG 139
++DT G
Sbjct: 244 VLDTIGG 250
>At2g47330 putative ATP-dependent RNA helicase
Length = 760
Score = 32.3 bits (72), Expect = 0.16
Identities = 31/105 (29%), Positives = 46/105 (43%), Gaps = 16/105 (15%)
Query: 56 PMMRHNMNQPG--KSLGVVGL-----GGLGHM----AVKFGKAFGLRVTVFSTSMSKKEE 104
PM+ H M+QP + G +G+ L H A KF KA+GLRV+ MSK E+
Sbjct: 285 PMIVHIMDQPELQRDEGPIGVICAPTRELAHQIFLEAKKFSKAYGLRVSAVYGGMSKHEQ 344
Query: 105 ALSFLGADQFVVSS-----NQEEMRALAKTLDFIIDTASGDHPFD 144
+ VV++ + +M+AL + D FD
Sbjct: 345 FKELKAGCEIVVATPGRLIDMLKMKALTMMRASYLVLDEADRMFD 389
>At5g43940 alcohol dehydrogenase (EC 1.1.1.1) class III
(pir||S71244)
Length = 379
Score = 31.2 bits (69), Expect = 0.36
Identities = 43/184 (23%), Positives = 73/184 (39%), Gaps = 21/184 (11%)
Query: 20 YSTSIVVNERYCFLIPKSYPLASAGPLLCAGITVYSPMMRHNMNQPGKSLGVVGLGGLGH 79
+S VV++ I + PL L C T + +PG ++ + GLG +G
Sbjct: 149 FSQYTVVHDVSVAKIDPTAPLDKVCLLGCGVPTGLGAVWNTAKVEPGSNVAIFGLGTVGL 208
Query: 80 MAVKFGKAFGLRVTVFSTSMSKKEEALSFLGADQFVVSSNQEEMRALAKTLDFIIDTASG 139
+ K G + SKK E G ++FV + ++ + I+D G
Sbjct: 209 AVAEGAKTAGASRIIGIDIDSKKYETAKKFGVNEFVNPKDHDK-----PIQEVIVDLTDG 263
Query: 140 --DHPFD--PYMSLLKIS--------GVLALVGFPS---EVKFSPASLNLGSRTVAGSAA 184
D+ F+ +S+++ + G +VG + E+ P L G R G+A
Sbjct: 264 GVDYSFECIGNVSVMRAALECCHKGWGTSVIVGVAASGQEISTRPFQLVTG-RVWKGTAF 322
Query: 185 GGTK 188
GG K
Sbjct: 323 GGFK 326
>At5g58020 unknown protein
Length = 354
Score = 30.8 bits (68), Expect = 0.47
Identities = 14/39 (35%), Positives = 22/39 (55%)
Query: 144 DPYMSLLKISGVLALVGFPSEVKFSPASLNLGSRTVAGS 182
DP SLL +SG+ +L+ + FS S+ L + + GS
Sbjct: 23 DPAQSLLTLSGITSLLESSQRISFSACSITLDGKLLNGS 61
>At5g28310 putative protein
Length = 233
Score = 30.0 bits (66), Expect = 0.80
Identities = 16/48 (33%), Positives = 27/48 (55%), Gaps = 2/48 (4%)
Query: 61 NMNQPGKSLGVVGLGGLGHMAVKFGKAFGLRVTVFSTSMSKKEEALSF 108
N Q K +G+VGLG +G KAFG +++ +S ++K A+ +
Sbjct: 109 NQYQSKKRIGIVGLGSIGSKVATRLKAFGCQISY--SSRNRKPYAVPY 154
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,221,425
Number of Sequences: 26719
Number of extensions: 165358
Number of successful extensions: 423
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 400
Number of HSP's gapped (non-prelim): 30
length of query: 194
length of database: 11,318,596
effective HSP length: 94
effective length of query: 100
effective length of database: 8,807,010
effective search space: 880701000
effective search space used: 880701000
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC123898.9


