

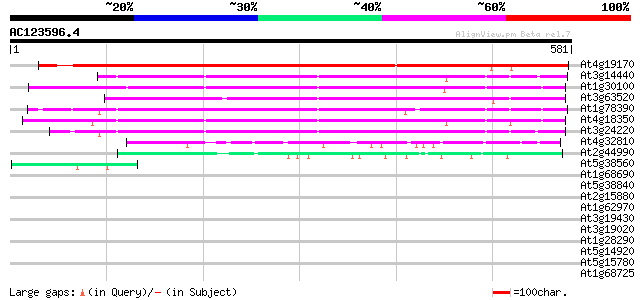
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123596.4 - phase: 0
(581 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g19170 neoxanthin cleavage enzyme-like protein 788 0.0
At3g14440 9-cis-epoxycarotenoid dioxygenase, putative 377 e-104
At1g30100 hypothetical protein 371 e-103
At3g63520 neoxanthin cleavage enzyme nc1 370 e-103
At1g78390 similar to 9-cis-epoxycarotenoid dioxygenase gb|AAF263... 361 e-100
At4g18350 neoxanthin cleavage enzyme - like protein 357 1e-98
At3g24220 9-cis-epoxycarotenoid dioxygenase, putative 327 1e-89
At4g32810 unknown protein 102 4e-22
At2g44990 hypothetical protein 96 4e-20
At5g38560 putative protein 46 7e-05
At1g68690 protein kinase, putative 41 0.002
At5g38840 kanadaptin - like protein 40 0.005
At2g15880 unknown protein 39 0.006
At1g62970 unknown protein 39 0.006
At3g19430 putative late embryogenesis abundant protein 37 0.023
At3g19020 hypothetical protein 37 0.023
At1g28290 proline-rich protein, putative 37 0.023
At5g14920 unknown protein 37 0.030
At5g15780 proline-rich protein 36 0.052
At1g68725 putative arabinogalactan protein AGP19 36 0.052
>At4g19170 neoxanthin cleavage enzyme-like protein
Length = 595
Score = 788 bits (2035), Expect = 0.0
Identities = 384/559 (68%), Positives = 456/559 (80%), Gaps = 27/559 (4%)
Query: 30 KEKPQTSQSQTLKTTTTTPPTKTLPTPPTITTTHNLKKPQTSSLPALLFNTFDDIINTFI 89
+ KP+T ++T T ++PP K +L LF T +D+INTFI
Sbjct: 54 RNKPKTLHNRTNHTLVSSPP----------------KLRPEMTLATALFTTVEDVINTFI 97
Query: 90 DPPIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPY 149
DPP +PSVDP+HVLS NFAPVLDELPPT C++I GTLP SLNG YIRNGPNPQFLPRGPY
Sbjct: 98 DPPSRPSVDPKHVLSDNFAPVLDELPPTDCEIIHGTLPLSLNGAYIRNGPNPQFLPRGPY 157
Query: 150 HLFDGDGMLHAITISNGKATLCSRYVQTYKYKIENEAGYQLIPNVFSGFNSLIASAARGS 209
HLFDGDGMLHAI I NGKATLCSRYV+TYKY +E + G ++PNVFSGFN + AS ARG+
Sbjct: 158 HLFDGDGMLHAIKIHNGKATLCSRYVKTYKYNVEKQTGAPVMPNVFSGFNGVTASVARGA 217
Query: 210 VTAARVISGQYNPSNGIGLANTSLALFGNKLFALGESDLPYEINLTPNGDIQTIGRYDFN 269
+TAARV++GQYNP NGIGLANTSLA F N+LFALGESDLPY + LT +GDI+TIGRYDF+
Sbjct: 218 LTAARVLTGQYNPVNGIGLANTSLAFFSNRLFALGESDLPYAVRLTESGDIETIGRYDFD 277
Query: 270 GKLSMSMTAHPKIDGDTGETFAFRYGPMPPFLTYFRFDANGVKHNDVPVFSMTRPSFLHD 329
GKL+MSMTAHPK D TGETFAFRYGP+PPFLTYFRFD+ G K DVP+FSMT PSFLHD
Sbjct: 278 GKLAMSMTAHPKTDPITGETFAFRYGPVPPFLTYFRFDSAGKKQRDVPIFSMTSPSFLHD 337
Query: 330 FAITKKYAVFTDIQLG--MNPLDMI-SGGSPVGSDPSKVSRIGILPRYASDESKMKWFDV 386
FAITK++A+F +IQLG MN LD++ GGSPVG+D K R+G++P+YA DES+MKWF+V
Sbjct: 338 FAITKRHAIFAEIQLGMRMNMLDLVLEGGSPVGTDNGKTPRLGVIPKYAGDESEMKWFEV 397
Query: 387 PGFNIMHAINSWDGEDDEETVTLIAPNVLSIEHTMERLELVHAMIEKVKINIKTGIVSRQ 446
PGFNI+HAIN+WD EDD +V LIAPN++SIEHT+ER++LVHA++EKVKI++ TGIV R
Sbjct: 398 PGFNIIHAINAWD-EDDGNSVVLIAPNIMSIEHTLERMDLVHALVEKVKIDLVTGIVRRH 456
Query: 447 PLSARNLDFAVINGDYMGKKNRFVYAAIGNPMPKISGVVKIDVLKGEEVGC----RLYGE 502
P+SARNLDFAVIN ++G+ +R+VYAAIG+PMPKISGVVK+DV KG+ C R+YG
Sbjct: 457 PISARNLDFAVINPAFLGRCSRYVYAAIGDPMPKISGVVKLDVSKGDRDDCTVARRMYGS 516
Query: 503 GCYGGEPFFVAREDGE---EEDDGYLVSYVHDEKKGESKFLVMDAKSPEFEIVAEVKLPR 559
GCYGGEPFFVAR+ G EEDDGY+V+YVHDE GESKFLVMDAKSPE EIVA V+LPR
Sbjct: 517 GCYGGEPFFVARDPGNPEAEEDDGYVVTYVHDEVTGESKFLVMDAKSPELEIVAAVRLPR 576
Query: 560 RVPYGFHGLFVKESDITKL 578
RVPYGFHGLFVKESD+ KL
Sbjct: 577 RVPYGFHGLFVKESDLNKL 595
>At3g14440 9-cis-epoxycarotenoid dioxygenase, putative
Length = 599
Score = 377 bits (967), Expect = e-104
Identities = 196/492 (39%), Positives = 297/492 (59%), Gaps = 13/492 (2%)
Query: 92 PIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHL 151
P+ + DP ++ NFAPV +E P + + G LP S+ GVY+RNG NP P +H
Sbjct: 112 PLPKTADPSVQIAGNFAPV-NEQPVRRNLPVVGKLPDSIKGVYVRNGANPLHEPVTGHHF 170
Query: 152 FDGDGMLHAITISNGKATLCSRYVQTYKYKIENEAGYQLIPNVFSGFNSLIASAARGSVT 211
FDGDGM+HA+ +G A+ R+ QT ++ E + G + P + AR +
Sbjct: 171 FDGDGMVHAVKFEHGSASYACRFTQTNRFVQERQLGRPVFPKAIGELHGH-TGIARLMLF 229
Query: 212 AARVISGQYNPSNGIGLANTSLALFGNKLFALGESDLPYEINLTPNGDIQTIGRYDFNGK 271
AR +G +P++G G+AN L F +L A+ E DLPY++ +TPNGD++T+GR+DF+G+
Sbjct: 230 YARAAAGIVDPAHGTGVANAGLVYFNGRLLAMSEDDLPYQVQITPNGDLKTVGRFDFDGQ 289
Query: 272 LSMSMTAHPKIDGDTGETFAFRYGPM-PPFLTYFRFDANGVKHNDVPVFSMTRPSFLHDF 330
L +M AHPK+D ++GE FA Y + P+L YFRF +G K DV + + +P+ +HDF
Sbjct: 290 LESTMIAHPKVDPESGELFALSYDVVSKPYLKYFRFSPDGTKSPDVEI-QLDQPTMMHDF 348
Query: 331 AITKKYAVFTDIQLGMNPLDMISGGSPVGSDPSKVSRIGILPRYASDESKMKWFDVPGFN 390
AIT+ + V D Q+ +MI GGSPV D +KV+R GIL +YA D S +KW D P
Sbjct: 349 AITENFVVVPDQQVVFKLPEMIRGGSPVVYDKNKVARFGILDKYAEDSSNIKWIDAPDCF 408
Query: 391 IMHAINSWDGEDDEETVTLIAPNVLSIEHTMERLELVHAMIEKVKINIKTGIVSRQPLSA 450
H N+W+ + +E V + + E E + +++ ++++N+KTG +R+P+ +
Sbjct: 409 CFHLWNAWEEPETDEVVVIGSCMTPPDSIFNESDENLKSVLSEIRLNLKTGESTRRPIIS 468
Query: 451 R-----NLDFAVINGDYMGKKNRFVYAAIGNPMPKISGVVKIDVLKGEEVGCRLYGEGCY 505
NL+ ++N + +G+K +F Y A+ P PK+SG K+D+ G EV LYG+ Y
Sbjct: 469 NEDQQVNLEAGMVNRNMLGRKTKFAYLALAEPWPKVSGFAKVDLTTG-EVKKHLYGDNRY 527
Query: 506 GGEPFFVAREDGEEEDDGYLVSYVHDEKKGESKFLVMDAKSPEFEIVAEVKLPRRVPYGF 565
GGEP F+ E G EED+GY++ +VHDEK +S+ +++A S E+ A VKLP RVPYGF
Sbjct: 528 GGEPLFLPGEGG-EEDEGYILCFVHDEKTWKSELQIVNAVS--LEVEATVKLPSRVPYGF 584
Query: 566 HGLFVKESDITK 577
HG F+ D+ K
Sbjct: 585 HGTFIGADDLAK 596
>At1g30100 hypothetical protein
Length = 589
Score = 371 bits (952), Expect = e-103
Identities = 211/564 (37%), Positives = 319/564 (56%), Gaps = 14/564 (2%)
Query: 20 PPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTITTTHNLKK--PQTSSLPALL 77
P ++S TK + + + ++ T P+P I + + P + A L
Sbjct: 28 PSSSVSFTNTKPRRRKLSANSVSDTPNLLNFPNYPSPNPIIPEKDTSRWNPLQRAASAAL 87
Query: 78 FNTFDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRN 137
++ P+ +VDPRH +S N+APV ++ + V G +P ++GVY+RN
Sbjct: 88 DFAETALLRRERSKPLPKTVDPRHQISGNYAPVPEQSVKSSLSV-DGKIPDCIDGVYLRN 146
Query: 138 GPNPQFLPRGPYHLFDGDGMLHAITISNGKATLCSRYVQTYKYKIENEAGYQLIPNVFSG 197
G NP F P +HLFDGDGM+HA+ I+NG A+ R+ +T + E + G + P
Sbjct: 147 GANPLFEPVSGHHLFDGDGMVHAVKITNGDASYSCRFTETERLVQEKQLGSPIFPKAIGE 206
Query: 198 FNSLIASAARGSVTAARVISGQYNPSNGIGLANTSLALFGNKLFALGESDLPYEINLTPN 257
+ + AR + AR + G N NG G+AN L F ++L A+ E DLPY++ +T N
Sbjct: 207 LHGH-SGIARLMLFYARGLFGLLNHKNGTGVANAGLVYFHDRLLAMSEDDLPYQVRVTDN 265
Query: 258 GDIQTIGRYDFNGKLSMSMTAHPKIDGDTGETFAFRYGPM-PPFLTYFRFDANGVKHNDV 316
GD++TIGR+DF+G+LS +M AHPKID T E FA Y + P+L YF+F G K DV
Sbjct: 266 GDLETIGRFDFDGQLSSAMIAHPKIDPVTKELFALSYDVVKKPYLKYFKFSPEGEKSPDV 325
Query: 317 PVFSMTRPSFLHDFAITKKYAVFTDIQLGMNPLDMISGGSPVGSDPSKVSRIGILPRYAS 376
+ + P+ +HDFAIT+ + V D Q+ DM G SPV D K+SR GILPR A
Sbjct: 326 EI-PLASPTMMHDFAITENFVVIPDQQVVFKLSDMFLGKSPVKYDGEKISRFGILPRNAK 384
Query: 377 DESKMKWFDVPGFNIMHAINSWDGEDDEETVTLIAPNVLSIEHTMERLELVHAMIEKVKI 436
D S+M W + P H N+W+ + +E V + + + E E +++++ ++++
Sbjct: 385 DASEMVWVESPETFCFHLWNAWESPETDEVVVIGSCMTPADSIFNECDEQLNSVLSEIRL 444
Query: 437 NIKTGIVSRQPL----SARNLDFAVINGDYMGKKNRFVYAAIGNPMPKISGVVKIDVLKG 492
N+KTG +R+ + NL+ ++N + +G+K R+ Y AI P PK+SG K+D+ G
Sbjct: 445 NLKTGKSTRRTIIPGSVQMNLEAGMVNRNLLGRKTRYAYLAIAEPWPKVSGFAKVDLSTG 504
Query: 493 EEVGCRLYGEGCYGGEPFFVARE-DGEEEDDGYLVSYVHDEKKGESKFLVMDAKSPEFEI 551
EV YG YGGEPFF+ R + + EDDGY++S+VHDE+ ES+ +++A + E E
Sbjct: 505 -EVKNHFYGGKKYGGEPFFLPRGLESDGEDDGYIMSFVHDEESWESELHIVNAVTLELE- 562
Query: 552 VAEVKLPRRVPYGFHGLFVKESDI 575
A VKLP RVPYGFHG FV +D+
Sbjct: 563 -ATVKLPSRVPYGFHGTFVNSADM 585
>At3g63520 neoxanthin cleavage enzyme nc1
Length = 538
Score = 370 bits (951), Expect = e-103
Identities = 206/495 (41%), Positives = 290/495 (57%), Gaps = 24/495 (4%)
Query: 99 PRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHLFDGDGML 158
P H LS NFAP+ DE PP + + G LP LNG ++R GPNP+F YH FDGDGM+
Sbjct: 44 PLHYLSGNFAPIRDETPPVKDLPVHGFLPECLNGEFVRVGPNPKFDAVAGYHWFDGDGMI 103
Query: 159 HAITISNGKATLCSRYVQTYKYKIENEAGYQLIPNV--FSGFNSLIASAARGSVTAARVI 216
H + I +GKAT SRYV+T + K E G + GF L+ + T +++
Sbjct: 104 HGVRIKDGKATYVSRYVKTSRLKQEEFFGAAKFMKIGDLKGFFGLLMVNVQQLRTKLKIL 163
Query: 217 SGQYNPSNGIGLANTSLALFGNKLFALGESDLPYEINLTPNGDIQTIGRYDFNGKLSMSM 276
Y G G ANT+L KL AL E+D PY I + +GD+QT+G D++ +L+ S
Sbjct: 164 DNTY----GNGTANTALVYHHGKLLALQEADKPYVIKVLEDGDLQTLGIIDYDKRLTHSF 219
Query: 277 TAHPKIDGDTGETFAFRYGPMPPFLTYFRFDANGVKHNDVPVFSMTRPSFLHDFAITKKY 336
TAHPK+D TGE F F Y PP+LTY +G+ H+ VP+ +++ P +HDFAIT+ Y
Sbjct: 220 TAHPKVDPVTGEMFTFGYSHTPPYLTYRVISKDGIMHDPVPI-TISEPIMMHDFAITETY 278
Query: 337 AVFTDIQLGMNPLDMISGGSPVGS-DPSKVSRIGILPRYASDESKMKWFDVPGFNIMHAI 395
A+F D+ + P +M+ + S DP+K +R G+LPRYA DE ++WF++P I H
Sbjct: 279 AIFMDLPMHFRPKEMVKEKKMIYSFDPTKKARFGVLPRYAKDELMIRWFELPNCFIFHNA 338
Query: 396 NSWDGEDDEETVT--LIAPNVLSIE-HTMERLELVHAMIEKVKINIKTGIVSRQPLSARN 452
N+W+ ED+ +T L P++ + E+LE + +++ N+KTG S++ LSA
Sbjct: 339 NAWEEEDEVVLITCRLENPDLDMVSGKVKEKLENFGNELYEMRFNMKTGSASQKKLSASA 398
Query: 453 LDFAVINGDYMGKKNRFVYAAIGNPMPKISGVVKIDVLKGEEVGCRL------------Y 500
+DF IN Y GKK R+VY I + + K++G++K D+ E G R+
Sbjct: 399 VDFPRINECYTGKKQRYVYGTILDSIAKVTGIIKFDLHAEAETGKRMLEVGGNIKGIYDL 458
Query: 501 GEGCYGGEPFFVAREDGEEEDDGYLVSYVHDEKKGESKFLVMDAKSPEFEIVAEVKLPRR 560
GEG YG E +V RE EEDDGYL+ +VHDE G+S V+DAK+ E VA V+LP R
Sbjct: 459 GEGRYGSEAIYVPRETA-EEDDGYLIFFVHDENTGKSCVTVIDAKTMSAEPVAVVELPHR 517
Query: 561 VPYGFHGLFVKESDI 575
VPYGFH LFV E +
Sbjct: 518 VPYGFHALFVTEEQL 532
>At1g78390 similar to 9-cis-epoxycarotenoid dioxygenase
gb|AAF26356.1
Length = 657
Score = 361 bits (927), Expect = e-100
Identities = 212/574 (36%), Positives = 323/574 (55%), Gaps = 31/574 (5%)
Query: 19 IPPFNISSIKTKEKPQTSQSQTLKTTTTTPPT-KTLPTPPT--ITTTHNLKKPQTSSLPA 75
+P + SS+ K +S S +L++ T PP+ K L T I T N + P+ + +
Sbjct: 96 VPCYVSSSVNKK----SSVSSSLQSPTFKPPSWKKLCNDVTNLIPKTTN-QNPKLNPVQR 150
Query: 76 LLFNTFDDIINTFIDP-----PIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSL 130
D + N I P + DP ++ NF PV E P + GT+P +
Sbjct: 151 TAAMVLDAVENAMISHERRRHPHPKTADPAVQIAGNFFPV-PEKPVVHNLPVTGTVPECI 209
Query: 131 NGVYIRNGPNPQFLPRGPYHLFDGDGMLHAITISNGKATLCSRYVQTYKYKIENEAGYQL 190
GVY+RNG NP P +HLFDGDGM+HA+ NG + R+ +T + E E G +
Sbjct: 210 QGVYVRNGANPLHKPVSGHHLFDGDGMVHAVRFDNGSVSYACRFTETNRLVQERECGRPV 269
Query: 191 IPNVFSGFNSLIASAARGSVTAARVISGQYNPSNGIGLANTSLALFGNKLFALGESDLPY 250
P + + A+ + R + G +P+ G+G+AN L F L A+ E DLPY
Sbjct: 270 FPKAIGELHGHLG-IAKLMLFNTRGLFGLVDPTGGLGVANAGLVYFNGHLLAMSEDDLPY 328
Query: 251 EINLTPNGDIQTIGRYDFNGKLSMSMTAHPKIDGDTGETFAFRYGPM-PPFLTYFRFDAN 309
+ +T GD++T GRYDF+G+L +M AHPKID +T E FA Y + P+L YFRF ++
Sbjct: 329 HVKVTQTGDLETSGRYDFDGQLKSTMIAHPKIDPETRELFALSYDVVSKPYLKYFRFTSD 388
Query: 310 GVKHNDVPVFSMTRPSFLHDFAITKKYAVFTDIQLGMNPLDMISGGSPVGSDPSKVSRIG 369
G K DV + + +P+ +HDFAIT+ + V D Q+ +MI GGSPV D K SR G
Sbjct: 389 GEKSPDVEI-PLDQPTMIHDFAITENFVVIPDQQVVFRLPEMIRGGSPVVYDEKKKSRFG 447
Query: 370 ILPRYASDESKMKWFDVPGFNIMHAINSWDGEDDEETVT----LIAPNVLSIEHTMERLE 425
IL + A D S ++W +VP H NSW+ + +E V + P+ + EH E
Sbjct: 448 ILNKNAKDASSIQWIEVPDCFCFHLWNSWEEPETDEVVVIGSCMTPPDSIFNEHD----E 503
Query: 426 LVHAMIEKVKINIKTGIVSRQPLSAR--NLDFAVINGDYMGKKNRFVYAAIGNPMPKISG 483
+ +++ ++++N+KTG +R+P+ + NL+ ++N + +G+K R+ Y A+ P PK+SG
Sbjct: 504 TLQSVLSEIRLNLKTGESTRRPVISEQVNLEAGMVNRNLLGRKTRYAYLALTEPWPKVSG 563
Query: 484 VVKIDVLKGEEVGCRLYGEGCYGGEPFFVAREDGEEEDDGYLVSYVHDEKKGESKFLVMD 543
K+D+ G E+ +YGEG YGGEP F+ DG EED GY++ +VHDE+K +S+ +++
Sbjct: 564 FAKVDLSTG-EIRKYIYGEGKYGGEPLFLPSGDG-EEDGGYIMVFVHDEEKVKSELQLIN 621
Query: 544 AKSPEFEIVAEVKLPRRVPYGFHGLFVKESDITK 577
A + + E A V LP RVPYGFHG F+ + D++K
Sbjct: 622 AVNMKLE--ATVTLPSRVPYGFHGTFISKEDLSK 653
Score = 28.9 bits (63), Expect = 8.2
Identities = 18/54 (33%), Positives = 27/54 (49%), Gaps = 3/54 (5%)
Query: 23 NISSIKTKEKPQTS---QSQTLKTTTTTPPTKTLPTPPTITTTHNLKKPQTSSL 73
N+ K KPQT+ Q T+ +TT P T T T +T+ + +P+ SL
Sbjct: 30 NLVLKKPIPKPQTAAFNQESTMASTTLLPSTSTQFLDRTFSTSSSSSRPKLQSL 83
>At4g18350 neoxanthin cleavage enzyme - like protein
Length = 583
Score = 357 bits (915), Expect = 1e-98
Identities = 207/578 (35%), Positives = 319/578 (54%), Gaps = 22/578 (3%)
Query: 14 PTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTITTTHNLKKPQTSSL 73
P + I + + I +P Q + +K T T+ T PQ + L
Sbjct: 8 PMSGGIKTWPQAQIDLGFRPIKRQPKVIKCTVQIDVTELTKKRQLFTPRTTATPPQHNPL 67
Query: 74 PALLFNTFDDI---------INTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKG 124
+F I I+ D P+ + DPR ++ N++PV E + ++G
Sbjct: 68 RLNIFQKAAAIAIDAAERALISHEQDSPLPKTADPRVQIAGNYSPV-PESSVRRNLTVEG 126
Query: 125 TLPPSLNGVYIRNGPNPQFLPRGPYHLFDGDGMLHAITISNGKATLCSRYVQTYKYKIEN 184
T+P ++GVYIRNG NP F P +HLFDGDGM+HA+ I+NG A+ R+ +T + E
Sbjct: 127 TIPDCIDGVYIRNGANPMFEPTAGHHLFDGDGMVHAVKITNGSASYACRFTKTERLVQEK 186
Query: 185 EAGYQLIPNVFSGFNSLIASAARGSVTAARVISGQYNPSNGIGLANTSLALFGNKLFALG 244
G + P + + AR + AR + G N NG+G+AN L F N+L A+
Sbjct: 187 RLGRPVFPKAIGELHGH-SGIARLMLFYARGLCGLINNQNGVGVANAGLVYFNNRLLAMS 245
Query: 245 ESDLPYEINLTPNGDIQTIGRYDFNGKLSMSMTAHPKIDGDTGETFAFRYGPM-PPFLTY 303
E DLPY++ +T GD+QT+GRYDF+G+L +M AHPK+D T E A Y + P+L Y
Sbjct: 246 EDDLPYQLKITQTGDLQTVGRYDFDGQLKSAMIAHPKLDPVTKELHALSYDVVKKPYLKY 305
Query: 304 FRFDANGVKHNDVPVFSMTRPSFLHDFAITKKYAVFTDIQLGMNPLDMISGGSPVGSDPS 363
FRF +GVK ++ + + P+ +HDFAIT+ + V D Q+ +MISG SPV D
Sbjct: 306 FRFSPDGVKSPELEI-PLETPTMIHDFAITENFVVIPDQQVVFKLGEMISGKSPVVFDGE 364
Query: 364 KVSRIGILPRYASDESKMKWFDVPGFNIMHAINSWDGEDDEETVTLIAPNVLSIEHTMER 423
KVSR+GI+P+ A++ S++ W + P H N+W+ + EE V + + + ER
Sbjct: 365 KVSRLGIMPKDATEASQIIWVNSPETFCFHLWNAWESPETEEIVVIGSCMSPADSIFNER 424
Query: 424 LELVHAMIEKVKINIKTGIVSRQPLSAR---NLDFAVINGDYMGKKNRFVYAAIGNPMPK 480
E + +++ +++IN++T +R+ L NL+ ++N + +G+K RF + AI P PK
Sbjct: 425 DESLRSVLSEIRINLRTRKTTRRSLLVNEDVNLEIGMVNRNRLGRKTRFAFLAIAYPWPK 484
Query: 481 ISGVVKIDVLKGEEVGCRLYGEGCYGGEPFFVAREDG---EEEDDGYLVSYVHDEKKGES 537
+SG K+D+ G E+ +YG YGGEPFF+ G E EDDGY+ +VHDE+ S
Sbjct: 485 VSGFAKVDLCTG-EMKKYIYGGEKYGGEPFFLPGNSGNGEENEDDGYIFCHVHDEETKTS 543
Query: 538 KFLVMDAKSPEFEIVAEVKLPRRVPYGFHGLFVKESDI 575
+ +++A + + E A +KLP RVPYGFHG FV +++
Sbjct: 544 ELQIINAVNLKLE--ATIKLPSRVPYGFHGTFVDSNEL 579
>At3g24220 9-cis-epoxycarotenoid dioxygenase, putative
Length = 577
Score = 327 bits (837), Expect = 1e-89
Identities = 195/545 (35%), Positives = 300/545 (54%), Gaps = 22/545 (4%)
Query: 42 KTTTTTPPTKTLPTPPTITTTHNLKKPQTSSLPALLFNTFDDIINTFIDP-----PIKPS 96
K T T +P+P + T+ P + L L D I ++ + P P+
Sbjct: 39 KIPTLPDLTSPVPSPVKLKPTY----PNLNLLQKLAATMLDKIESSIVIPMEQNRPLPKP 94
Query: 97 VDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHLFDGDG 156
DP LS NFAPV +E P + G +P L GVYIRNG NP F P +HLFDGDG
Sbjct: 95 TDPAVQLSGNFAPV-NECPVQNGLEVVGQIPSCLKGVYIRNGANPMFPPLAGHHLFDGDG 153
Query: 157 MLHAITIS-NGKATLCSRYVQTYKYKIENEAGYQLIPNVFSGFNSLIASAARGSVTAARV 215
M+HA++I + + + RY +T + E G + P + + AR ++ AR
Sbjct: 154 MIHAVSIGFDNQVSYSCRYTKTNRLVQETALGRSVFPKPIGELHGH-SGLARLALFTARA 212
Query: 216 ISGQYNPSNGIGLANTSLALFGNKLFALGESDLPYEINLTPNGDIQTIGRYDFNGKLSMS 275
G + + G+G+AN + F +L A+ E DLPY++ + GD++TIGR+ F+ ++ S
Sbjct: 213 GIGLVDGTRGMGVANAGVVFFNGRLLAMSEDDLPYQVKIDGQGDLETIGRFGFDDQIDSS 272
Query: 276 MTAHPKIDGDTGETFAFRYGPM-PPFLTYFRFDANGVKHNDVPVFSMTRPSFLHDFAITK 334
+ AHPK+D TG+ Y + P L Y +F+ G K DV + ++ P+ +HDFAIT+
Sbjct: 273 VIAHPKVDATTGDLHTLSYNVLKKPHLRYLKFNTCGKKTRDVEI-TLPEPTMIHDFAITE 331
Query: 335 KYAVFTDIQLGMNPLDMISGGSPVGSDPSKVSRIGILPRYASDESKMKWFDVPGFNIMHA 394
+ V D Q+ +MI GGSPV K++R G+L + S + W DVP H
Sbjct: 332 NFVVIPDQQMVFKLSEMIRGGSPVIYVKEKMARFGVLSKQDLTGSDINWVDVPDCFCFHL 391
Query: 395 INSWDGEDDE-ETVTLIAPNVLSIEHTM--ERLELVHAMIEKVKINIKTGIVSRQPL-SA 450
N+W+ +E + V ++ + +S T+ E E + ++++N++T +R+ + +
Sbjct: 392 WNAWEERTEEGDPVIVVIGSCMSPPDTIFSESGEPTRVELSEIRLNMRTKESNRKVIVTG 451
Query: 451 RNLDFAVINGDYMGKKNRFVYAAIGNPMPKISGVVKIDVLKGEEVGCRLYGEGCYGGEPF 510
NL+ IN Y+G+K++FVY AI +P PK SG+ K+D+ G V YG +GGEP
Sbjct: 452 VNLEAGHINRSYVGRKSQFVYIAIADPWPKCSGIAKVDIQNG-TVSEFNYGPSRFGGEPC 510
Query: 511 FVAREDGEEEDDGYLVSYVHDEKKGESKFLVMDAKSPEFEIVAEVKLPRRVPYGFHGLFV 570
FV +G EED GY++ +V DE+K ES+F+V+DA + + VA V+LP RVPYGFHG FV
Sbjct: 511 FVPEGEG-EEDKGYVMGFVRDEEKDESEFVVVDA--TDMKQVAAVRLPERVPYGFHGTFV 567
Query: 571 KESDI 575
E+ +
Sbjct: 568 SENQL 572
>At4g32810 unknown protein
Length = 570
Score = 102 bits (255), Expect = 4e-22
Identities = 125/508 (24%), Positives = 214/508 (41%), Gaps = 96/508 (18%)
Query: 122 IKGTLPPSLNGVYIRNGPNPQFLPRGPY-HLFDGDGMLHAITISNGKATLCSRYVQTYKY 180
++G +P LNG Y+RNGP + + HLFDG L + G+ R +++ Y
Sbjct: 97 VQGKIPTWLNGTYLRNGPGLWNIGDHDFRHLFDGYSTLVKLQFDGGRIFAAHRLLESDAY 156
Query: 181 KI-------------ENEAGYQLIPNVFSGFNSLIASAARGSVTAARVISGQYNPSNGIG 227
K E + N FSG ++ R+ SG+ N
Sbjct: 157 KAAKKHNRLCYREFSETPKSVIINKNPFSGIGEIV-----------RLFSGESLTDN--- 202
Query: 228 LANTSLALFGN-KLFALGESDLPYEINLTPNGDIQTIGRYDFNGKLS--MSMTAHPKIDG 284
ANT + G+ ++ L E+ L + ++TIG+++++ LS M +AHP +
Sbjct: 203 -ANTGVIKLGDGRVMCLTETQKGSI--LVDHETLETIGKFEYDDVLSDHMIQSAHPIVT- 258
Query: 285 DTGETFAFRYGP--MPPFLTYFRFDANGVKHNDVPVFSMTR----PSFLHDFAITKKYAV 338
ET + P + P R +A K V P ++H FA+T+ Y V
Sbjct: 259 ---ETEMWTLIPDLVKPGYRVVRMEAGSNKREVVGRVRCRSGSWGPGWVHSFAVTENYVV 315
Query: 339 FTDIQLGMNPLDMISGGSPVGSDPSKVSRIGILPR---YASDESKMKW-----FDVPGFN 390
++ L + +++ ++P+ + + P+ + SK+ +VP +
Sbjct: 316 IPEMPLRYSVKNLLR------AEPTPLYKFEWCPQDGAFIHVMSKLTGEVVASVEVPAYV 369
Query: 391 IMHAINSW--DGEDDEETVTLIAPNVLSIEHT-------MERLELV-----HAMIEKVKI 436
H IN++ D D + +IA EH M RL+ + H ++ +I
Sbjct: 370 TFHFINAYEEDKNGDGKATVIIAD---CCEHNADTRILDMLRLDTLRSSHGHDVLPDARI 426
Query: 437 N-------------IKTGIVSRQPLSARNLDFAVINGDYMGKKNRFVYAA-IGNPMPKIS 482
++T + + + R +D IN Y+G+K R+VYA P +
Sbjct: 427 GRFRIPLDGSKYGKLETAVEAEK--HGRAMDMCSINPLYLGQKYRYVYACGAQRPCNFPN 484
Query: 483 GVVKIDVLKGEEVGCRLYGEGCYGGEPFFVAREDGEEEDDGYLVSYVHDEKKGESKFLVM 542
+ K+D++ E+ + G EPFFV R EDDG ++S V E+ G S +++
Sbjct: 485 ALSKVDIV--EKKVKNWHEHGMIPSEPFFVPRPGATHEDDGVVISIV-SEENGGSFAILL 541
Query: 543 DAKSPEFEIVAEVKLPRRVPYGFHGLFV 570
D S FE +A K P +PYG HG ++
Sbjct: 542 DGSS--FEEIARAKFPYGLPYGLHGCWI 567
>At2g44990 hypothetical protein
Length = 618
Score = 96.3 bits (238), Expect = 4e-20
Identities = 141/566 (24%), Positives = 231/566 (39%), Gaps = 123/566 (21%)
Query: 112 DELPPTQCKVIKGTLPPSL-NGVYIRNGPNPQFLPRGP-YHLFDGDGMLHAITISNGK-- 167
+ + P K I+G++P + +G Y GP G H DG G L A I K
Sbjct: 71 ETIEPVVIKPIEGSIPVNFPSGTYYLAGPGLFTDDHGSTVHPLDGHGYLRAFHIDGNKRK 130
Query: 168 ATLCSRYVQTYKYKIENEAGYQLIPNVFSGFNSLIASAARGSVTAARVISGQYNPSNGIG 227
AT ++YV+T K E++ G S++ R T
Sbjct: 131 ATFTAKYVKTEAKKEEHDPVTDTWRFTHRGPFSVLKGGKRFGNTKVMK-----------N 179
Query: 228 LANTSLALFGNKLFALGESDLPYEINLTPNGDIQTIGRYDFNGKLSMSMTAHPKIDGDTG 287
+ANTS+ + +L L E PYEI +G + T+GR++ S D D
Sbjct: 180 VANTSVLKWAGRLLCLWEGGEPYEIE---SGSLDTVGRFNVENNGCESCDDDDSSDRDLS 236
Query: 288 -----ETFAFRYGP-------MPP--FLTYFRFDA------------------------- 308
+T A P MPP FL++++ D
Sbjct: 237 GHDIWDTAADLLKPILQGVFKMPPKRFLSHYKVDGRRKRLLTVTCNAEDMLLPRSNFTFC 296
Query: 309 ---NGVKHNDVPVFSMTRPSFLHDFAITKKYAVFTDIQLGMNPLDMIS---GGSPVGS-- 360
+ K F + +HD+A T + + ++ +NP+ I+ G SP+ S
Sbjct: 297 EYDSEFKLIQTKEFKIDDHMMIHDWAFTDTHYILFANRVKLNPIGSIAAMCGMSPMVSAL 356
Query: 361 --DPSKVSR-IGILPRYASDESKM-KWFDVP-----GFNIMHAINSWDGEDDEETVTL-- 409
+PS S I ILPR++ S+ + + VP ++H+ N+++ +D + +
Sbjct: 357 SLNPSNESSPIYILPRFSDKYSRGGRDWRVPVEVSSQLWLIHSGNAYETREDNGDLKIQI 416
Query: 410 ------------------------IAPNVLSIEHTMERLELVHAMIEKVKINI-KTGIVS 444
+ P+V+++ ++L L H + KV + + TG +
Sbjct: 417 QASACSYRWFDFQKMFGYDWQSNKLDPSVMNLNRGDDKL-LPHLV--KVSMTLDSTGNCN 473
Query: 445 R---QPLSARNL--DFAVINGDYMGKKNRFVYAAIGN------PMPKISGVVKIDVLKGE 493
+PL+ N DF VIN + GKKN+++Y+A + P VVK D L
Sbjct: 474 SCDVEPLNGWNKPSDFPVINSSWSGKKNKYMYSAASSGTRSELPHFPFDMVVKFD-LDSN 532
Query: 494 EVGCRLYGEGCYGGEPFFVAR---EDGEEEDDGYLVSYVHDEKKGESKFLVMDAK--SPE 548
V G + GEP FV + E+GEEEDDGY+V + +++DAK
Sbjct: 533 LVRTWSTGARRFVGEPMFVPKNSVEEGEEEDDGYIVVVEYAVSVERCYLVILDAKKIGES 592
Query: 549 FEIVAEVKLPRRV--PYGFHGLFVKE 572
+V+ +++PR + P GFHGL+ +
Sbjct: 593 DAVVSRLEVPRNLTFPMGFHGLWASD 618
>At5g38560 putative protein
Length = 681
Score = 45.8 bits (107), Expect = 7e-05
Identities = 43/148 (29%), Positives = 55/148 (37%), Gaps = 18/148 (12%)
Query: 3 PKPIIITSKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTITTT 62
P P +ITS P SS PP + + P T+ +T + PP P+PP TTT
Sbjct: 72 PSPPVITSPPPTVASSPPPPVVIASPPPSTPATTPPAPPQTVSPPPPPDASPSPPAPTTT 131
Query: 63 HNLKKP-------------QTSSLPALLFNTFDDIINTFIDPPIKPSVDP---RHVLSQN 106
+ KP +T S P +T T PP S P
Sbjct: 132 NPPPKPSPSPPGETPSPPGETPSPPKPSPSTPTPTTTTSPPPPPATSASPPSSNPTDPST 191
Query: 107 FAPVLDELP--PTQCKVIKGTLPPSLNG 132
AP LP P + + K T P S NG
Sbjct: 192 LAPPPTPLPVVPREKPIAKPTGPASNNG 219
Score = 40.4 bits (93), Expect = 0.003
Identities = 29/97 (29%), Positives = 38/97 (38%), Gaps = 3/97 (3%)
Query: 3 PKPIIITSKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTITTT 62
P P++ +S PP SS PP SS P + ++ PP PP+ T
Sbjct: 48 PPPVVSSSPPPPVVSSPPP---SSSPPPSPPVITSPPPTVASSPPPPVVIASPPPSTPAT 104
Query: 63 HNLKKPQTSSLPALLFNTFDDIINTFIDPPIKPSVDP 99
PQT S P + T +PP KPS P
Sbjct: 105 TPPAPPQTVSPPPPPDASPSPPAPTTTNPPPKPSPSP 141
>At1g68690 protein kinase, putative
Length = 708
Score = 40.8 bits (94), Expect = 0.002
Identities = 41/149 (27%), Positives = 55/149 (36%), Gaps = 24/149 (16%)
Query: 10 SKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTITTT------- 62
S PP TS PP N ++ P TS + PP + P PP +TT+
Sbjct: 11 SNSPPVTSPPPPLNNATSPATPPPVTSP----LPPSAPPPNRAPPPPPPVTTSPPPVANG 66
Query: 63 ---HNLKKPQTSSLPALLFNTFDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQC 119
L KP SS P +I + PP S P+ V+ P PP
Sbjct: 67 APPPPLPKPPESSSPPP-----QPVIPS---PPPSTSPPPQPVIPS--PPPSASPPPALV 116
Query: 120 KVIKGTLPPSLNGVYIRNGPNPQFLPRGP 148
+ + PP + R P+P L R P
Sbjct: 117 PPLPSSPPPPASVPPPRPSPSPPILVRSP 145
Score = 38.1 bits (87), Expect = 0.014
Identities = 33/126 (26%), Positives = 46/126 (36%), Gaps = 18/126 (14%)
Query: 3 PKPIIITSKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTITTT 62
P P +T+ PP + PP + KP S S + +PP T P P + +
Sbjct: 51 PPPPPVTTSPPPVANGAPPPPLP------KPPESSSPPPQPVIPSPPPSTSPPPQPVIPS 104
Query: 63 HNLKKPQTSSLPALLFNTFDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQCKVI 122
P S PAL+ PP +PS P P+L PP + I
Sbjct: 105 ---PPPSASPPPALVPPLPSSPPPPASVPPPRPSPSP---------PILVRSPPPSVRPI 152
Query: 123 KGTLPP 128
+ PP
Sbjct: 153 QSPPPP 158
Score = 32.3 bits (72), Expect = 0.74
Identities = 31/136 (22%), Positives = 50/136 (35%), Gaps = 6/136 (4%)
Query: 12 QPPTTSSIPPFNI--SSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTITTTHNLKKPQ 69
+PP +SS PP + S + P + + + PP P P + ++ P+
Sbjct: 74 KPPESSSPPPQPVIPSPPPSTSPPPQPVIPSPPPSASPPPALVPPLPSSPPPPASVPPPR 133
Query: 70 TSSLPALLFNTFDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPS 129
S P +L + + PP PS P +Q+ P PP++ PPS
Sbjct: 134 PSPSPPILVRSPPPSVRPIQSPPPPPSDRP----TQSPPPPSPPSPPSERPTQSPPSPPS 189
Query: 130 LNGVYIRNGPNPQFLP 145
P+P P
Sbjct: 190 ERPTQSPPPPSPPSPP 205
Score = 31.2 bits (69), Expect = 1.7
Identities = 24/99 (24%), Positives = 40/99 (40%), Gaps = 6/99 (6%)
Query: 3 PKPIIITSKQPPT------TSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTP 56
P P + + PP T S PP + S ++ Q+ S + T +PP + P+P
Sbjct: 145 PPPSVRPIQSPPPPPSDRPTQSPPPPSPPSPPSERPTQSPPSPPSERPTQSPPPPSPPSP 204
Query: 57 PTITTTHNLKKPQTSSLPALLFNTFDDIINTFIDPPIKP 95
P+ + + P + P + + TF PP P
Sbjct: 205 PSDRPSQSPPPPPEDTKPQPPRRSPNSPPPTFSSPPRSP 243
>At5g38840 kanadaptin - like protein
Length = 730
Score = 39.7 bits (91), Expect = 0.005
Identities = 25/77 (32%), Positives = 38/77 (48%), Gaps = 7/77 (9%)
Query: 20 PPFNIS-SIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTITTTHNLKKPQTSSLPALLF 78
PP N S I+ E TS SQ+ +T+T PP P PP + TT + +P+ +
Sbjct: 5 PPRNPSHDIEPPEPNSTSISQSDETSTMNPPPPRNPNPPDLKTTEVVVEPEP------IE 58
Query: 79 NTFDDIINTFIDPPIKP 95
+ DD + D P++P
Sbjct: 59 ESKDDSVTVDADKPVRP 75
>At2g15880 unknown protein
Length = 727
Score = 39.3 bits (90), Expect = 0.006
Identities = 40/146 (27%), Positives = 51/146 (34%), Gaps = 12/146 (8%)
Query: 12 QPPTTSSIP--PFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPT-PPTITTTHNLKKP 68
QPP S P P+N S +K + P Q + PP + PT PP +T + KP
Sbjct: 410 QPPKESPQPNDPYNQSPVKFRRSPPPPQQPHHHVVHSPPPASSPPTSPPVHSTPSPVHKP 469
Query: 69 QTSSLPALLFNTFDDIINTF----IDPPIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKG 124
Q + +D F PP+ P + S PV PP
Sbjct: 470 QPPKESPQPNDPYDQSPVKFRRSPPPPPVHSPPPPSPIHSPPPPPVYSPPPPPPV----- 524
Query: 125 TLPPSLNGVYIRNGPNPQFLPRGPYH 150
PP VY P P P P H
Sbjct: 525 YSPPPPPPVYSPPPPPPVHSPPPPVH 550
Score = 28.9 bits (63), Expect = 8.2
Identities = 30/119 (25%), Positives = 39/119 (32%), Gaps = 13/119 (10%)
Query: 3 PKPIIITSKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTITTT 62
P P + + PP S PP + P S +PP PP +
Sbjct: 580 PPPPVYSPPPPPVHSPPPPVHSPPPPVHSPPPPVYSPPPPPPVHSPPPPVFSPPPPV--- 636
Query: 63 HNLKKPQTSSLPALLFNTFDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQCKV 121
H+ P S P + + PP+K S P V S P PPTQ V
Sbjct: 637 HSPPPPVYSPPPP---------VYSPPPPPVK-SPPPPPVYSPPLLPPKMSSPPTQTPV 685
>At1g62970 unknown protein
Length = 825
Score = 39.3 bits (90), Expect = 0.006
Identities = 29/111 (26%), Positives = 45/111 (40%), Gaps = 13/111 (11%)
Query: 4 KPIIITSKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPP----TKTLPTPPTI 59
KP + QPP TS P + +K P + T ++T + P + P PP
Sbjct: 523 KPFV---SQPPNTSKPMPVSQPPTTSKPLPVSQPPPTFQSTCPSQPPAASSSLSPLPPVF 579
Query: 60 TTTHNLKKPQTSSLPALLFNTFDDIINTFIDPPI-KPSVDPRHVLSQNFAP 109
+T + + P S+ P+ + +T PP P P HV +Q P
Sbjct: 580 NSTQSFQSPPVSTTPSAVPEA-----STIPSPPAPAPVAQPTHVFNQTPPP 625
>At3g19430 putative late embryogenesis abundant protein
Length = 550
Score = 37.4 bits (85), Expect = 0.023
Identities = 32/108 (29%), Positives = 42/108 (38%), Gaps = 25/108 (23%)
Query: 2 VPKPIIITSKQPPTTSSIPPFNISSIKTKEKPQ------------TSQSQTLKTTTTTPP 49
VP P S PPT + P + T P T + T TPP
Sbjct: 144 VPSPTPPVSPPPPTPTPSVPSPTPPVPTDPMPSPPPPVSPPPPTPTPSVPSPPDVTPTPP 203
Query: 50 TKTLPTPPTITTTHNLKKPQTSSLPALLFNTFDDIINTFIDPPIKPSV 97
T ++P+PP +T T P T S+P+ D+ T PP PSV
Sbjct: 204 TPSVPSPPDVTPT-----PPTPSVPSP-----PDVTPT---PPTPPSV 238
>At3g19020 hypothetical protein
Length = 951
Score = 37.4 bits (85), Expect = 0.023
Identities = 38/140 (27%), Positives = 51/140 (36%), Gaps = 19/140 (13%)
Query: 13 PPTTSSIP--PFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTITTTHNLKKPQT 70
PP S +P P++ S IK K +PQ T +T TT+P + + +PP H+ P
Sbjct: 598 PPLESPVPNDPYDASPIK-KRRPQPPSPSTEETKTTSPQSPPVHSPPPPPPVHSPPPPVF 656
Query: 71 SSLPALLFNTFDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSL 130
S P + PP P V S PV PP PP
Sbjct: 657 SPPPPM------------HSPPPPVYSPPPPVHSPPPPPVHSPPPPVHSPPPPVHSPPP- 703
Query: 131 NGVYIRNGPNPQFLPRGPYH 150
+ + P P P P H
Sbjct: 704 ---PVHSPPPPVHSPPPPVH 720
>At1g28290 proline-rich protein, putative
Length = 359
Score = 37.4 bits (85), Expect = 0.023
Identities = 36/130 (27%), Positives = 45/130 (33%), Gaps = 14/130 (10%)
Query: 4 KPIIITSKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKT-TTTTPPTKTLPTPPTITTT 62
KP + +PP + P + +K KP T PPTK PP T
Sbjct: 109 KPPVSPPAKPPVKPPVYPPTKAPVKPPTKPPVKPPVYPPTKAPVKPPTKPPVKPPVYPPT 168
Query: 63 HNLKKPQTSSLPALLFNTFDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDEL-PPTQCKV 121
KP T + + PP KP V P V APV + PPT+ V
Sbjct: 169 KAPVKPPTK-----------PPVKPPVSPPAKPPVKP-PVYPPTKAPVKPPVSPPTKPPV 216
Query: 122 IKGTLPPSLN 131
PP N
Sbjct: 217 TPPVYPPKFN 226
>At5g14920 unknown protein
Length = 275
Score = 37.0 bits (84), Expect = 0.030
Identities = 35/134 (26%), Positives = 51/134 (37%), Gaps = 13/134 (9%)
Query: 3 PKPIIITSKQPPTTSSIPPFNISSIKTKE-KPQTSQSQTLKTTTTTPPTKTLPTPPTITT 61
P I +T +PP S PP + ++ KP T T+K + PPT PTP
Sbjct: 80 PPTIPVTPVKPPV--STPPIKLPPVQPPTYKPPTP---TVKPPSVQPPTYKPPTPTVKPP 134
Query: 62 THNLKKPQTS---SLPALLFNTF----DDIINTFIDPPIKPSVDPRHVLSQNFAPVLDEL 114
T + KP T+ P + T+ + PP+KP V + P +
Sbjct: 135 TTSPVKPPTTPPVQSPPVQPPTYKPPTSPVKPPTTTPPVKPPTTTPPVQPPTYNPPTTPV 194
Query: 115 PPTQCKVIKGTLPP 128
P +K PP
Sbjct: 195 KPPTAPPVKPPTPP 208
Score = 35.4 bits (80), Expect = 0.088
Identities = 45/156 (28%), Positives = 55/156 (34%), Gaps = 23/156 (14%)
Query: 4 KPIIITSKQP--PTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPT------ 55
KP + K P PTT PP + K KP T +K +TPP K P
Sbjct: 53 KPPTPSYKPPTLPTTPIKPP----TTKPPVKPPTIPVTPVKPPVSTPPIKLPPVQPPTYK 108
Query: 56 PPTITTTHNLKKPQTSSLPALLFNTFDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDEL- 114
PPT T +P T P T + + PP P V V + P +
Sbjct: 109 PPTPTVKPPSVQPPTYKPPT---PTVKPPTTSPVKPPTTPPVQSPPVQPPTYKPPTSPVK 165
Query: 115 PPTQCKVIK--GTLPPSLNGVYIRNGPNPQFLPRGP 148
PPT +K T PP Y NP P P
Sbjct: 166 PPTTTPPVKPPTTTPPVQPPTY-----NPPTTPVKP 196
Score = 34.3 bits (77), Expect = 0.20
Identities = 37/124 (29%), Positives = 47/124 (37%), Gaps = 9/124 (7%)
Query: 7 IITSKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPT-PPTITTTHNL 65
+++ P S P S K + + TL TT PPT P PPTI T
Sbjct: 30 LVSLPTPTLPSPSPATKPPSPALKPPTPSYKPPTLPTTPIKPPTTKPPVKPPTIPVTP-- 87
Query: 66 KKPQTSSLPALLFNTFDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDEL-PPTQCKVIKG 124
KP S+ P L + PP P+V P V + P + PPT V
Sbjct: 88 VKPPVSTPPIKL----PPVQPPTYKPP-TPTVKPPSVQPPTYKPPTPTVKPPTTSPVKPP 142
Query: 125 TLPP 128
T PP
Sbjct: 143 TTPP 146
>At5g15780 proline-rich protein
Length = 401
Score = 36.2 bits (82), Expect = 0.052
Identities = 38/133 (28%), Positives = 54/133 (40%), Gaps = 13/133 (9%)
Query: 4 KPIIITSKQPPTTSSIPPFN--ISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTITT 61
KP P SI P N I SI T P + + + + PP +PTPPT+ T
Sbjct: 256 KPNFFFPPNPLNPPSIIPPNPLIPSIPT---PTLPPNPLIPSPPSLPPIPLIPTPPTLPT 312
Query: 62 THNLKKPQTSSLPAL-LFNTFDDI----INTFIDPPIKPSVDPRHVLSQNFAPVLDELPP 116
L P T +LP + T + ++PP P P + P L +PP
Sbjct: 313 IPLLPTPPTPTLPPIPTIPTLPPLPVLPPVPIVNPPSLPPPPPSFPVPLPPVPGLPGIPP 372
Query: 117 TQCKVIKGTLPPS 129
+I G +PP+
Sbjct: 373 V--PLIPG-IPPA 382
>At1g68725 putative arabinogalactan protein AGP19
Length = 248
Score = 36.2 bits (82), Expect = 0.052
Identities = 36/148 (24%), Positives = 50/148 (33%), Gaps = 14/148 (9%)
Query: 5 PIIITSKQPPTTSSIPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTITTTHN 64
P+ T+ PP T++ PP + T P S +Q + T PP T +PP
Sbjct: 31 PVTSTTTAPPPTTAAPPTTAAPPPTTTTPPVSAAQPPASPVTPPPAVTPTSPPAPKVAPV 90
Query: 65 LK----KPQTSSLPALLFNTFDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQCK 120
+ PQ P T + + PP P+ P S AP P
Sbjct: 91 ISPATPPPQPPQSPPASAPT---VSPPPVSPPPAPTSPPPTPASPPPAPASPPPAPASPP 147
Query: 121 VIKGTLPPSLNGVYIRNGPNPQFLPRGP 148
+ PP P+P LP P
Sbjct: 148 PAPVSPPPV-------QAPSPISLPPAP 168
Score = 33.9 bits (76), Expect = 0.26
Identities = 32/131 (24%), Positives = 47/131 (35%), Gaps = 19/131 (14%)
Query: 19 IPPFNISSIKTKEKPQTSQSQTLKTTTTTPPTKTLPTPPTITTTHNLKKPQTSSLPALLF 78
I F++++ P TS + TT PPT P P T T + +P S
Sbjct: 17 ISSFSVNAQGPAASPVTSTTTAPPPTTAAPPTTAAPPPTTTTPPVSAAQPPASP------ 70
Query: 79 NTFDDIINTFIDPPIKPSVDPRHVLSQNFAPVLD-ELPPTQCKVIKGTLPPSLNGVYIRN 137
+ PP P+V P + APV+ PP Q P+++ +
Sbjct: 71 ----------VTPP--PAVTPTSPPAPKVAPVISPATPPPQPPQSPPASAPTVSPPPVSP 118
Query: 138 GPNPQFLPRGP 148
P P P P
Sbjct: 119 PPAPTSPPPTP 129
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.137 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,696,828
Number of Sequences: 26719
Number of extensions: 731772
Number of successful extensions: 2738
Number of sequences better than 10.0: 86
Number of HSP's better than 10.0 without gapping: 28
Number of HSP's successfully gapped in prelim test: 61
Number of HSP's that attempted gapping in prelim test: 2309
Number of HSP's gapped (non-prelim): 250
length of query: 581
length of database: 11,318,596
effective HSP length: 105
effective length of query: 476
effective length of database: 8,513,101
effective search space: 4052236076
effective search space used: 4052236076
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC123596.4


