

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123593.9 + phase: 0 /pseudo
(409 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
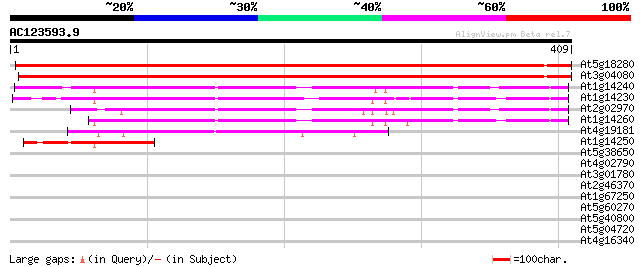
Score E
Sequences producing significant alignments: (bits) Value
At5g18280 apyrase (gb|AAF00612.1) 443 e-125
At3g04080 apyrase (Atapy1) 425 e-119
At1g14240 putative nucleoside triphosphatase 157 8e-39
At1g14230 putative nucleoside triphosphatase 152 3e-37
At2g02970 nucleoside triphosphatase like protein 147 8e-36
At1g14260 hypothetical protein, 5' partial 138 6e-33
At4g19181 putative protein 89 3e-18
At1g14250 putative nucleoside triphosphatase, 3' partial 56 4e-08
At5g38650 unknown protein 31 1.4
At4g02790 unknown protein 30 1.8
At3g01780 unknown protein 30 3.1
At2g46370 putative auxin-responsive protein 30 3.1
At1g67250 unknown protein 29 4.1
At5g60270 receptor like protein kinase 28 7.0
At5g40800 putative protein 28 7.0
At5g04720 unknown protein 28 7.0
At4g16340 unknown protein 28 9.1
>At5g18280 apyrase (gb|AAF00612.1)
Length = 472
Score = 443 bits (1139), Expect = e-125
Identities = 218/406 (53%), Positives = 290/406 (70%), Gaps = 2/406 (0%)
Query: 5 ITLITTVLLLLMPAITSSQYLGNNLLTNRKIFQKQETISSYAVVFDAGSTGSRIHVYHFD 64
I LI VLLL+ S + + NRK +YAV+FDAGS+GSR+HVY FD
Sbjct: 31 IVLIGLVLLLMPGRSISDSVVEEYSVHNRKGGPNSRGPKNYAVIFDAGSSGSRVHVYCFD 90
Query: 65 QNLDLLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHKTPIRLG 124
QNLDL+ +G ++E F ++ PGLS+Y DP QAA SL+ LL +AE VP +L KT +R+G
Sbjct: 91 QNLDLIPLGNELELFLQLKPGLSAYPTDPRQAANSLVSLLDKAEASVPRELRPKTHVRVG 150
Query: 125 ATAGLRLLNGDASEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYALGNL 184
ATAGLR L DASE ILQAVR++ +RS + +AV+++DGTQEGSY WVT+NY L NL
Sbjct: 151 ATAGLRTLGHDASENILQAVRELLRDRSMLKTEANAVTVLDGTQEGSYQWVTINYLLRNL 210
Query: 185 GKKYTKTVGVMDLGGGSVQMAYAVSKKTAKNAPKVADGVDPYIKKLVLKGKPYDLYVHSY 244
GK Y+ TVGV+DLGGGSVQMAYA+S++ A +APK +G D Y++++ LKG+ Y LYVHSY
Sbjct: 211 GKPYSDTVGVVDLGGGSVQMAYAISEEDAASAPKPLEGEDSYVREMYLKGRKYFLYVHSY 270
Query: 245 LHFGREASRAEIMKVTRSSPNPCLLAGFDGTYTYAGEEFKAKAPASGANFNGCKKIIRKA 304
LH+G A+RAEI+KV+ S NPC++AG+DG Y Y G+EFKA A SGA+ + C++I A
Sbjct: 271 LHYGLLAARAEILKVSEDSENPCIVAGYDGMYKYGGKEFKAPASQSGASLDECRRITINA 330
Query: 305 LKLNYP-CPYQNCTFGGIWNGGGGNGQKHLFASSSFFYLPEDVGMVDPKTPNFKIRPVDL 363
LK+N C + CTFGG+WNGG G GQK++F +S FF + G VDPK P +RP+D
Sbjct: 331 LKVNDTLCTHMKCTFGGVWNGGRGGGQKNMFVASFFFDRAAEAGFVDPKQPVATVRPMDF 390
Query: 364 VSEAKKACALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGF 409
AKKAC++ E+ KST+P + ++N+ Y+CMDL+YQY LL+DGF
Sbjct: 391 EKAAKKACSMKLEEGKSTFPLVEEENL-PYLCMDLVYQYTLLIDGF 435
>At3g04080 apyrase (Atapy1)
Length = 471
Score = 425 bits (1092), Expect = e-119
Identities = 206/404 (50%), Positives = 281/404 (68%), Gaps = 2/404 (0%)
Query: 7 LITTVLLLLMPAITSSQYLGNNLLTNRKIFQKQETISSYAVVFDAGSTGSRIHVYHFDQN 66
++ ++LLLMP ++S + + N + +YAV+FDAGS+GSR+HVY FDQN
Sbjct: 32 VLIALVLLLMPGTSTSVSVIEYTMKNHEGGSNSRGPKNYAVIFDAGSSGSRVHVYCFDQN 91
Query: 67 LDLLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHKTPIRLGAT 126
LDL+ + ++E F ++ PGLS+Y NDP Q+A SL+ LL +AE VP +L KTP+R+GAT
Sbjct: 92 LDLVPLENELELFLQLKPGLSAYPNDPRQSANSLVTLLDKAEASVPRELRPKTPVRVGAT 151
Query: 127 AGLRLLNGDASEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYALGNLGK 186
AGLR L ASE ILQAVR++ RS + +AV+++DGTQEGSY WVT+NY L LGK
Sbjct: 152 AGLRALGHQASENILQAVRELLKGRSRLKTEANAVTVLDGTQEGSYQWVTINYLLRTLGK 211
Query: 187 KYTKTVGVMDLGGGSVQMAYAVSKKTAKNAPKVADGVDPYIKKLVLKGKPYDLYVHSYLH 246
Y+ TVGV+DLGGGSVQMAYA+ ++ A APK +G D Y++++ LKG+ Y LYVHSYLH
Sbjct: 212 PYSDTVGVVDLGGGSVQMAYAIPEEDAATAPKPVEGEDSYVREMYLKGRKYFLYVHSYLH 271
Query: 247 FGREASRAEIMKVTRSSPNPCLLAGFDGTYTYAGEEFKAKAPASGANFNGCKKIIRKALK 306
+G A+RAEI+KV+ S NPC+ G+ GTY Y G+ FKA A SGA+ + C+++ ALK
Sbjct: 272 YGLLAARAEILKVSEDSNNPCIATGYAGTYKYGGKAFKAAASPSGASLDECRRVAINALK 331
Query: 307 LNYP-CPYQNCTFGGIWNGGGGNGQKHLFASSSFFYLPEDVGMVDPKTPNFKIRPVDLVS 365
+N C + CTFGG+WNGGGG GQK +F +S FF + G VDP P ++RP+D
Sbjct: 332 VNNSLCTHMKCTFGGVWNGGGGGGQKKMFVASFFFDRAAEAGFVDPNQPVAEVRPLDFEK 391
Query: 366 EAKKACALNFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVDGF 409
A KAC + E+ KS +P + + N+ Y+C+DL+YQY LLVDGF
Sbjct: 392 AANKACNMRMEEGKSKFPRVEEDNL-PYLCLDLVYQYTLLVDGF 434
>At1g14240 putative nucleoside triphosphatase
Length = 483
Score = 157 bits (398), Expect = 8e-39
Identities = 118/425 (27%), Positives = 207/425 (47%), Gaps = 49/425 (11%)
Query: 4 LITLITTVLLLLMPAITSSQYLGNNLLTNRKIFQKQETISSYAVVFDAGSTGSRIHV--Y 61
++ +T L LL+ S+ + + L +R+ + Y+V+ DAGS+G+R+HV Y
Sbjct: 35 VVVSVTITLGLLLYVFNSNSVISSGSLLSRRCKLR------YSVLIDAGSSGTRVHVFGY 88
Query: 62 HFDQNLDLLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHKTPI 121
F+ + G+ K+TPGLSSYA++PE A+ S+ L++ A+ +P + ++ I
Sbjct: 89 WFESGKPVFDFGEKHYANLKLTPGLSSYADNPEGASVSVTKLVEFAKQRIPKRMFRRSDI 148
Query: 122 RLGATAGLRLLNGDASEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYAL 181
RL ATAG+RLL E+IL+ R + + S F + + ++I G+ EG Y W+T NYAL
Sbjct: 149 RLMATAGMRLLEVPVQEQILEVTRRVLRS-SGFMFRDEWANVISGSDEGIYSWITANYAL 207
Query: 182 GNLGKKYTKTVGVMDLGGGSVQMAYAVSKKTAKNAPKVADGVDPYIKKLVLKGK-PYDLY 240
G+LG +T G+++LGG S Q+ + S + V P + + G Y +Y
Sbjct: 208 GSLGTDPLETTGIVELGGASAQVTFVSS-----------EHVPPEYSRTIAYGNISYTIY 256
Query: 241 VHSYLHFGREASRAEIMKVTRSSPN---------PCLLAGF-------DGTYTYAGEEFK 284
HS+L +G++A+ ++++ ++S N PC G+ + + + +E K
Sbjct: 257 SHSFLDYGKDAALKKLLEKLQNSANSTVDGVVEDPCTPKGYIYDTNSKNYSSGFLADESK 316
Query: 285 AKAPASGA-NFNGCKKIIRKALKLNYP-CPYQNCTFGGIWNGGGGNGQKHLFASSSFFYL 342
K A NF+ C+ LK C Y++C+ G + + Q A++SF+Y
Sbjct: 317 LKGSLQAAGNFSKCRSATFALLKEGKENCLYEHCSIGSTFT---PDLQGSFLATASFYYT 373
Query: 343 PEDVGMVDPKTPNFKIRPVDLVSEAKKACALNFEDAKSTYPFLAKKNIASYVCMDLIYQY 402
+ + + K +L+ K+ C + YP ++ + Y C Y
Sbjct: 374 AKFFELEE------KGWLSELIPAGKRYCGEEWSKLILEYPTTDEEYLRGY-CFSAAYTI 426
Query: 403 VLLVD 407
+L D
Sbjct: 427 SMLHD 431
>At1g14230 putative nucleoside triphosphatase
Length = 503
Score = 152 bits (384), Expect = 3e-37
Identities = 117/428 (27%), Positives = 206/428 (47%), Gaps = 56/428 (13%)
Query: 3 FLITLITTVLLLLMPAITSSQYLGNNLLTNRKIFQKQETISSYAVVFDAGSTGSRIHV-- 60
F+I T+ L L+ ++G ++L + + + Y+V+ D GS+G+R+HV
Sbjct: 49 FVIVACVTIALGLL-------FIGYSILRSGR---NRRVSLHYSVIIDGGSSGTRVHVFG 98
Query: 61 YHFDQNLDLLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHKTP 120
Y + + G++ K++PGLS+YA++PE ++S+ L++ A+ V K+
Sbjct: 99 YRIESGKPVFDFGEENYASLKLSPGLSAYADNPEGVSESVTELVEFAKKRVHKGKLKKSD 158
Query: 121 IRLGATAGLRLLNGDASEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYA 180
IRL ATAG+RLL E+IL R + + S F+ + + S+I G+ EG Y WV N+A
Sbjct: 159 IRLMATAGMRLLELPVQEQILDVTRRVLRS-SGFDFRDEWASVISGSDEGVYAWVVANHA 217
Query: 181 LGNLGKKYTKTVGVMDLGGGSVQMAYAVSKKTAKNAPKVADGVDPYIKKLVLKGKPYDLY 240
LG+LG + KT G+++LGG S Q+ + ++ + + L Y+LY
Sbjct: 218 LGSLGGEPLKTTGIVELGGASAQVTFVSTELVPSE----------FSRTLAYGNVSYNLY 267
Query: 241 VHSYLHFGREASRAEIMKVTRSS----------PNPCLLAGF----------DGTYTYAG 280
HS+L FG++A++ ++ + +S P+PC+ G+ G G
Sbjct: 268 SHSFLDFGQDAAQEKLSESLYNSAANSTGEGIVPDPCIPKGYILETNLQKDLPGFLADKG 327
Query: 281 EEFKAKAPASGANFNGCKKIIRKALKLNY-PCPYQNCTFGGIWNGGGGNGQKHLFASSSF 339
+F A A+G NF+ C+ L+ C Y+ C+ G I+ N Q A+ +F
Sbjct: 328 -KFTATLQAAG-NFSECRSAAFAMLQEEKGKCTYKRCSIGSIFT---PNLQGSFLATENF 382
Query: 340 FYLPEDVGMVDPKTPNFKIRPVDLVSEAKKACALNFEDAKSTYPFLAKKNIASYVCMDLI 399
F+ + G+ + + + +++ K+ C + K YP +N+ Y C
Sbjct: 383 FHTSKFFGLGEKEWLS------EMILAGKRFCGEEWSKLKVKYPTFKDENLLRY-CFSSA 435
Query: 400 YQYVLLVD 407
Y +L D
Sbjct: 436 YIISMLHD 443
>At2g02970 nucleoside triphosphatase like protein
Length = 555
Score = 147 bits (372), Expect = 8e-36
Identities = 117/394 (29%), Positives = 192/394 (48%), Gaps = 58/394 (14%)
Query: 45 YAVVFDAGSTGSRIHVYHFDQNLDLLHIGKDVEFFN-------KITPGLSSYANDPEQAA 97
Y+VV D GSTG+RIHV+ + + GK V F K+ PGLS++A+DP+ A+
Sbjct: 87 YSVVIDGGSTGTRIHVFGYR-----IESGKPVFEFRGANYASLKLHPGLSAFADDPDGAS 141
Query: 98 KSLIPLLQQAENVVPIDLHHKTPIRLGATAGLRLLNGDASEKILQAVRDMFSNRSTFNVQ 157
SL L++ A+ VP + +T +RL ATAG+RLL EKIL R + + S F +
Sbjct: 142 VSLTELVEFAKGRVPKGMWIETEVRLMATAGMRLLELPVQEKILGVARRVLKS-SGFLFR 200
Query: 158 PDAVSIIDGTQEGSYLWVTVNYALGNLGKKYTKTVGVMDLGGGSVQMAYAVSKKTAKNAP 217
+ S+I G+ EG Y WV N+ALG+LG KT G+++LGG S Q+ + S
Sbjct: 201 DEWASVISGSDEGVYAWVVANFALGSLGGDPLKTTGIVELGGASAQVTFVSS-------- 252
Query: 218 KVADGVDPYIKKLVLKGK-PYDLYVHSYLHFGREASRAEI------------MKVTRSS- 263
+ + P + + G Y+LY HS+LHFG+ A+ ++ ++ TR
Sbjct: 253 ---EPMPPEFSRTISFGNVTYNLYSHSFLHFGQNAAHDKLWGSLLSRDHNSAVEPTREKI 309
Query: 264 -PNPCLLAGFD---GTYTY-----AGEEFKAKAPASGANFNGCKKIIRKALK-LNYPCPY 313
+PC G++ T + A E + + +G N++ C+ L+ N C Y
Sbjct: 310 FTDPCAPKGYNLDANTQKHLSGLLAEESRLSDSFQAGGNYSQCRSAALTILQDGNEKCSY 369
Query: 314 QNCTFGGIWNGGGGNGQKHLFASSSFFYLPEDVGMVDPKTPNFKIRPVDLVSEAKKACAL 373
Q+C+ G + + A+ +FFY + G+ + K +++S ++ C
Sbjct: 370 QHCSIGSTFT---PKLRGRFLATENFFYTSKFFGLGE------KAWLSNMISAGERFCGE 420
Query: 374 NFEDAKSTYPFLAKKNIASYVCMDLIYQYVLLVD 407
++ + P L ++++ Y C Y LL D
Sbjct: 421 DWSKLRVKDPSLHEEDLLRY-CFSSAYIVSLLHD 453
>At1g14260 hypothetical protein, 5' partial
Length = 669
Score = 138 bits (347), Expect = 6e-33
Identities = 106/374 (28%), Positives = 177/374 (46%), Gaps = 46/374 (12%)
Query: 58 IHV--YHFDQNLDLLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPLLQQAENVVPIDL 115
IHV Y F+ + G++ K++PGLSSYA++PE A+ S+ L++ A+ +P
Sbjct: 1 IHVFGYWFESGKPVFDFGEEHYASLKLSPGLSSYADNPEGASVSVTKLVEFAKGRIPKGK 60
Query: 116 HHKTPIRLGATAGLRLLNGDASEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQEGSYLWV 175
K+ IRL ATAG+RLL+ E+IL R + + S F Q + ++I GT EG Y WV
Sbjct: 61 LKKSDIRLMATAGMRLLDVPVQEQILDVTRRVLRS-SGFKFQDEWATVISGTDEGIYAWV 119
Query: 176 TVNYALGNLGKKYTKTVGVMDLGGGSVQMAYAVSKKTAKNAPKVADGVDPYIKKLVLKGK 235
N+ALG+LG KT G+++LGG S Q+ + S + V P + + G
Sbjct: 120 VANHALGSLGGDPLKTTGIVELGGASAQVTFVPS-----------EHVPPEFSRTISYGN 168
Query: 236 -PYDLYVHSYLHFGREASRAEIMKVTRSS----------PNPCLLAGF-------DGTYT 277
Y +Y HS+L FG++A+ ++++ ++S +PC G+ +
Sbjct: 169 VSYTIYSHSFLDFGQDAAEDKLLESLQNSVAASTGDGIVEDPCTPKGYIYDTHSQKDSSG 228
Query: 278 YAGEEFKAKAP---ASGANFNGCKKIIRKALKLNYP-CPYQNCTFGGIWNGGGGNGQKHL 333
+ EE K KA + +F C+ L+ C Y++C+ G + N Q
Sbjct: 229 FLSEESKFKASLQVQAAGDFTKCRSATLAMLQEGKENCAYKHCSIGSTFT---PNIQGSF 285
Query: 334 FASSSFFYLPEDVGMVDPKTPNFKIRPVDLVSEAKKACALNFEDAKSTYPFLAKKNIASY 393
A+ +FF+ + G+ + + + +++ K+ C + K YP K + Y
Sbjct: 286 LATENFFHTSKFFGLGEKEWLS------EMILAGKRFCGEEWSKLKEKYPTTKDKYLHRY 339
Query: 394 VCMDLIYQYVLLVD 407
C Y +L D
Sbjct: 340 -CFSSAYIISMLHD 352
>At4g19181 putative protein
Length = 740
Score = 89.4 bits (220), Expect = 3e-18
Identities = 73/257 (28%), Positives = 119/257 (45%), Gaps = 24/257 (9%)
Query: 43 SSYAVVFDAGSTGSRIHVYHF------DQNLDLLHIGKDVEFFNK----------ITPGL 86
S Y VVFD GSTG+R +VY D +L ++ K PG
Sbjct: 143 SRYYVVFDCGSTGTRAYVYQASINYKKDSSLPIVMKSLTEGISRKSRGRAYDRMETEPGF 202
Query: 87 SSYANDPEQAAKSLIPLLQQAENVVPIDLHHKTPIRLGATAGLRLLNGDASEKILQAVRD 146
N+ ++ PL+Q AE +P + H T + + ATAG+R L S IL V
Sbjct: 203 DKLVNNRTGLKTAIKPLIQWAEKQIPKNAHRTTSLFVYATAGVRRLRPADSSWILGNVWS 262
Query: 147 MFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYALGNLGKKYTK-TVGVMDLGGGSVQMA 205
+ + +S F + + V II GT+E + W +NY LG K T G +DLGG S+Q+
Sbjct: 263 ILA-KSPFTCRREWVKIISGTEEAYFGWTALNYQTSMLGALPKKATFGALDLGGSSLQVT 321
Query: 206 YAVSKKT--AKNAPKVADGVDPYIKKLVLKGKPY-DLYVHSYLHFGRE---ASRAEIMKV 259
+ ++T N V+ ++ L G D + S +H ++ +++++++
Sbjct: 322 FENEERTHNETNLNLRIGSVNHHLSAYSLAGYGLNDAFDRSVVHLLKKLPNVNKSDLIEG 381
Query: 260 TRSSPNPCLLAGFDGTY 276
+PCL +G++G Y
Sbjct: 382 KLEMKHPCLNSGYNGQY 398
>At1g14250 putative nucleoside triphosphatase, 3' partial
Length = 128
Score = 55.8 bits (133), Expect = 4e-08
Identities = 35/97 (36%), Positives = 60/97 (61%), Gaps = 7/97 (7%)
Query: 11 VLLLLMPAITSSQYLGNNLLTNRKIFQKQETISSYAVVFDAGSTGSRIHV--YHFDQNLD 68
+L++ AIT LG+N + F ++ ++ Y+V+ DAGS+G+RIHV Y F+
Sbjct: 36 MLIVASLAIT----LGSNSVMFSASFLRRSSLH-YSVIIDAGSSGTRIHVFGYWFESGKP 90
Query: 69 LLHIGKDVEFFNKITPGLSSYANDPEQAAKSLIPLLQ 105
+ G++ K++PGLSSYA++PE A+ S+ L++
Sbjct: 91 VFDFGEEHYASLKLSPGLSSYADNPEGASVSVTKLVE 127
>At5g38650 unknown protein
Length = 141
Score = 30.8 bits (68), Expect = 1.4
Identities = 14/44 (31%), Positives = 21/44 (46%), Gaps = 11/44 (25%)
Query: 85 GLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHKTPIRLGATAG 128
G Y NDP +++E + P+D HH +RLG + G
Sbjct: 102 GFEDYLNDP-----------RESETLKPVDFHHGMEVRLGLSKG 134
>At4g02790 unknown protein
Length = 372
Score = 30.4 bits (67), Expect = 1.8
Identities = 13/27 (48%), Positives = 18/27 (66%)
Query: 182 GNLGKKYTKTVGVMDLGGGSVQMAYAV 208
GN GKK+ KT+G+ GG S Q A+ +
Sbjct: 327 GNCGKKFVKTLGLNLFGGDSHQAAFRI 353
>At3g01780 unknown protein
Length = 1192
Score = 29.6 bits (65), Expect = 3.1
Identities = 51/228 (22%), Positives = 89/228 (38%), Gaps = 39/228 (17%)
Query: 109 NVVPIDLHHKTPIRLGATAGLRLLNGDASEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQ 168
N+ ++L+ + IR+G + L ++G QAVR + R+ + P S+ G
Sbjct: 818 NLTELELN-RVDIRVGLSGALYFMDGSP-----QAVRQL---RNLVSQDPVQCSVTVGVS 868
Query: 169 --EGSYLWVTVNYALGNLGKKYTKTVGVMDLGGGSVQMAYAVSKKTAKNAPKVADGVDPY 226
E WV V Y Y + G ++ + K+ + ++ + V
Sbjct: 869 QFERCGFWVQVLY--------YPFRGARGEYDGDYIEEDPQIMKQKRGSKAELGEPV--- 917
Query: 227 IKKLVLKGKPYDLYVHSYLHFGREASRAEIMKVTRSSPNPCLLAGFDGTYTYAGEEFKAK 286
+L+ +PY + + L + S E ++ S P +A + GTY Y G F A
Sbjct: 918 ----ILRCQPYKIPLTELL-LPHKISPVEFFRLWPSLP---AVAEYTGTYMYEGSGFMAT 969
Query: 287 APAS-GAN--FNGCKKIIRKALK------LNYPCPYQNCTFGGIWNGG 325
A GA+ +G K + K + +Q C W+GG
Sbjct: 970 AAQQYGASPFLSGLKSLSSKPFHRVCSHIIRTVAGFQLCYAAKTWHGG 1017
>At2g46370 putative auxin-responsive protein
Length = 575
Score = 29.6 bits (65), Expect = 3.1
Identities = 12/24 (50%), Positives = 17/24 (70%)
Query: 85 GLSSYANDPEQAAKSLIPLLQQAE 108
GL+ A DPE+A KS++PL+ E
Sbjct: 49 GLNGNATDPEEAFKSMVPLVTDVE 72
>At1g67250 unknown protein
Length = 141
Score = 29.3 bits (64), Expect = 4.1
Identities = 14/44 (31%), Positives = 19/44 (42%), Gaps = 11/44 (25%)
Query: 85 GLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHKTPIRLGATAG 128
G Y NDP + +E P+D HH +RLG + G
Sbjct: 102 GFEDYLNDP-----------RDSETFKPVDFHHGMEVRLGISKG 134
>At5g60270 receptor like protein kinase
Length = 668
Score = 28.5 bits (62), Expect = 7.0
Identities = 24/97 (24%), Positives = 42/97 (42%), Gaps = 7/97 (7%)
Query: 5 ITLITTVLLLLMPAITSSQYLGNNLLTNRKIFQKQETISSYAVVFDAGSTGSRIHVYHFD 64
++ T + L+PA S + +++ F+ E + + GST +R+ D
Sbjct: 79 VSFSTYFVCALLPAGDPSGHGMTFFVSHSTDFKGAEATRYFGIFNRNGSTSTRVLAVELD 138
Query: 65 QNL-----DLL--HIGKDVEFFNKITPGLSSYANDPE 94
+L D+ H+G DV IT +SY +D E
Sbjct: 139 TSLASDVKDISDNHVGIDVNSAESITSANASYFSDKE 175
>At5g40800 putative protein
Length = 272
Score = 28.5 bits (62), Expect = 7.0
Identities = 44/171 (25%), Positives = 65/171 (37%), Gaps = 38/171 (22%)
Query: 83 TPGLSSYANDPEQAAKSLIPLLQQAENVVPIDLHHKTPIRLGATAGLRLLNGDASEKILQ 142
T G S + DPE + ++ L E P+ L H L T + +NGD +
Sbjct: 73 TNGFRSSSTDPENGREDIVTLQLLPERSTPLSLDHN---NLDPT--VETINGDET----- 122
Query: 143 AVRDMFSNRST---FNVQPDAVSIIDGTQEGSYLWVTVNYALGNLGKKYTKTVGVMDLGG 199
N T FN DA+ Q WVTV L + VG+ D
Sbjct: 123 ------CNTDTWLKFNGGDDALQ-----QVPVETWVTVESVNSGL---VSHAVGLTD--- 165
Query: 200 GSVQMAYAVSKKTAKNAPKVADGVDPYIKKLVLKGKPYDLYVHSYLHFGRE 250
++ YA+ K T ++DG + ++V+ + Y V FGRE
Sbjct: 166 --EELTYALDKDTCPGF--ISDGSN----RVVMVNEAYRRIVTGDGGFGRE 208
>At5g04720 unknown protein
Length = 811
Score = 28.5 bits (62), Expect = 7.0
Identities = 16/39 (41%), Positives = 22/39 (56%), Gaps = 6/39 (15%)
Query: 202 VQMAYAVSKKT------AKNAPKVADGVDPYIKKLVLKG 234
V+ YAVS+KT AKN + DG+ P IK++ G
Sbjct: 14 VRQLYAVSQKTLRCRGIAKNLATMIDGLQPTIKEIQYSG 52
>At4g16340 unknown protein
Length = 880
Score = 28.1 bits (61), Expect = 9.1
Identities = 20/93 (21%), Positives = 40/93 (42%), Gaps = 16/93 (17%)
Query: 137 SEKILQAVRDMFSNRSTFNVQPDAVSIIDGTQEGSYLWVTVNYALGNLGKKYTKTVGVMD 196
SE++ A+ + S S V+ ++GT + YLW VN L + + Y+
Sbjct: 287 SERLSPAINNYLSEASRQEVR------LEGTPDNGYLWQRVNSQLASPSQPYSLREA--- 337
Query: 197 LGGGSVQMAYAVSKKTAKNAPKVADGVDPYIKK 229
+A A S + +A + + + P +++
Sbjct: 338 -------LAQAQSSRIGASAQALRESLHPILRQ 363
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,334,867
Number of Sequences: 26719
Number of extensions: 422921
Number of successful extensions: 1078
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 1047
Number of HSP's gapped (non-prelim): 18
length of query: 409
length of database: 11,318,596
effective HSP length: 102
effective length of query: 307
effective length of database: 8,593,258
effective search space: 2638130206
effective search space used: 2638130206
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC123593.9


