

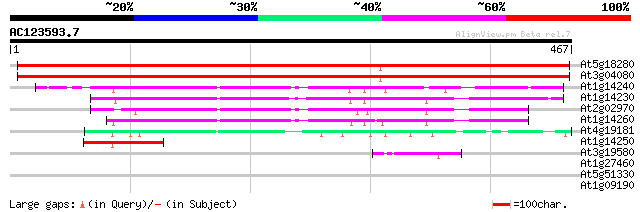
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123593.7 + phase: 0
(467 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g18280 apyrase (gb|AAF00612.1) 659 0.0
At3g04080 apyrase (Atapy1) 649 0.0
At1g14240 putative nucleoside triphosphatase 164 1e-40
At1g14230 putative nucleoside triphosphatase 154 1e-37
At2g02970 nucleoside triphosphatase like protein 142 5e-34
At1g14260 hypothetical protein, 5' partial 137 1e-32
At4g19181 putative protein 81 1e-15
At1g14250 putative nucleoside triphosphatase, 3' partial 53 3e-07
At3g19580 zinc finger protein, putative 42 7e-04
At1g27460 unknown protein 32 0.57
At5g51330 DYAD/SWITCH1 (SWI1) 28 8.3
At1g09190 hypothetical protein 28 8.3
>At5g18280 apyrase (gb|AAF00612.1)
Length = 472
Score = 659 bits (1699), Expect = 0.0
Identities = 334/463 (72%), Positives = 384/463 (82%), Gaps = 3/463 (0%)
Query: 7 RQESLSDKIYRFRGTLLVVSIPILLITFVLYMMPSSSNYDSA-GDYALLNRKLSPDKKSG 65
R ESL+DKI R RG +LV+S+PI+LI VL +MP S DS +Y++ NRK P+ +
Sbjct: 9 RHESLADKIQRHRGIILVISVPIVLIGLVLLLMPGRSISDSVVEEYSVHNRKGGPNSRGP 68
Query: 66 GSYAVIFDAGSSGSRVHVFHFDQNLDLVHIGKDLELFEQLKPGLSAYAQKPQQAAESLVS 125
+YAVIFDAGSSGSRVHV+ FDQNLDL+ +G +LELF QLKPGLSAY P+QAA SLVS
Sbjct: 69 KNYAVIFDAGSSGSRVHVYCFDQNLDLIPLGNELELFLQLKPGLSAYPTDPRQAANSLVS 128
Query: 126 LLEKAEGVVPRELRSKTPVRIGATAGLRALEGDASDKILRAVRDLLKHRSSFKSDADAVT 185
LL+KAE VPRELR KT VR+GATAGLR L DAS+ IL+AVR+LL+ RS K++A+AVT
Sbjct: 129 LLDKAEASVPRELRPKTHVRVGATAGLRTLGHDASENILQAVRELLRDRSMLKTEANAVT 188
Query: 186 VLDGTQEGAYQWVTINYLLGNLGKDYSKTVGVVDLGGGSVQMAYAISESEAAMAPQVMDG 245
VLDGTQEG+YQWVTINYLL NLGK YS TVGVVDLGGGSVQMAYAISE +AA AP+ ++G
Sbjct: 189 VLDGTQEGSYQWVTINYLLRNLGKPYSDTVGVVDLGGGSVQMAYAISEEDAASAPKPLEG 248
Query: 246 EDPYVKEMFLRGRKYYLYVHSYLRYGLLAARAEILKVAGDAENPCILSGSDGTYKYGGKS 305
ED YV+EM+L+GRKY+LYVHSYL YGLLAARAEILKV+ D+ENPCI++G DG YKYGGK
Sbjct: 249 EDSYVREMYLKGRKYFLYVHSYLHYGLLAARAEILKVSEDSENPCIVAGYDGMYKYGGKE 308
Query: 306 FK--ASSSGASLNECKSVAHKALKVNESTCTHMKCTFGGIWNGGGGDGQKNLFVASFFFD 363
FK AS SGASL+EC+ + ALKVN++ CTHMKCTFGG+WNGG G GQKN+FVASFFFD
Sbjct: 309 FKAPASQSGASLDECRRITINALKVNDTLCTHMKCTFGGVWNGGRGGGQKNMFVASFFFD 368
Query: 364 RAAEAGFVDPNSPVAIVRPADFEDAAKQACQTKLENAKSTYPRVEEGNLPYLCMDLVYQY 423
RAAEAGFVDP PVA VRP DFE AAK+AC KLE KST+P VEE NLPYLCMDLVYQY
Sbjct: 369 RAAEAGFVDPKQPVATVRPMDFEKAAKKACSMKLEEGKSTFPLVEEENLPYLCMDLVYQY 428
Query: 424 TLLVDGFGIYPWQEITLVKKVKYDDALVEAAWPLGSAIEAVSS 466
TLL+DGFG+ P Q ITLVKKVKY D VEAAWPLGSAIEAVSS
Sbjct: 429 TLLIDGFGLEPSQTITLVKKVKYGDQAVEAAWPLGSAIEAVSS 471
>At3g04080 apyrase (Atapy1)
Length = 471
Score = 649 bits (1673), Expect = 0.0
Identities = 329/462 (71%), Positives = 379/462 (81%), Gaps = 2/462 (0%)
Query: 7 RQESLSDKIYRFRGTLLVVSIPILLITFVLYMMPSSSNYDSAGDYALLNRKLSPDKKSGG 66
R ESL+DK++R RG LLV+SIPI+LI VL +MP +S S +Y + N + + +
Sbjct: 9 RHESLADKVHRHRGLLLVISIPIVLIALVLLLMPGTSTSVSVIEYTMKNHEGGSNSRGPK 68
Query: 67 SYAVIFDAGSSGSRVHVFHFDQNLDLVHIGKDLELFEQLKPGLSAYAQKPQQAAESLVSL 126
+YAVIFDAGSSGSRVHV+ FDQNLDLV + +LELF QLKPGLSAY P+Q+A SLV+L
Sbjct: 69 NYAVIFDAGSSGSRVHVYCFDQNLDLVPLENELELFLQLKPGLSAYPNDPRQSANSLVTL 128
Query: 127 LEKAEGVVPRELRSKTPVRIGATAGLRALEGDASDKILRAVRDLLKHRSSFKSDADAVTV 186
L+KAE VPRELR KTPVR+GATAGLRAL AS+ IL+AVR+LLK RS K++A+AVTV
Sbjct: 129 LDKAEASVPRELRPKTPVRVGATAGLRALGHQASENILQAVRELLKGRSRLKTEANAVTV 188
Query: 187 LDGTQEGAYQWVTINYLLGNLGKDYSKTVGVVDLGGGSVQMAYAISESEAAMAPQVMDGE 246
LDGTQEG+YQWVTINYLL LGK YS TVGVVDLGGGSVQMAYAI E +AA AP+ ++GE
Sbjct: 189 LDGTQEGSYQWVTINYLLRTLGKPYSDTVGVVDLGGGSVQMAYAIPEEDAATAPKPVEGE 248
Query: 247 DPYVKEMFLRGRKYYLYVHSYLRYGLLAARAEILKVAGDAENPCILSGSDGTYKYGGKSF 306
D YV+EM+L+GRKY+LYVHSYL YGLLAARAEILKV+ D+ NPCI +G GTYKYGGK+F
Sbjct: 249 DSYVREMYLKGRKYFLYVHSYLHYGLLAARAEILKVSEDSNNPCIATGYAGTYKYGGKAF 308
Query: 307 K--ASSSGASLNECKSVAHKALKVNESTCTHMKCTFGGIWNGGGGDGQKNLFVASFFFDR 364
K AS SGASL+EC+ VA ALKVN S CTHMKCTFGG+WNGGGG GQK +FVASFFFDR
Sbjct: 309 KAAASPSGASLDECRRVAINALKVNNSLCTHMKCTFGGVWNGGGGGGQKKMFVASFFFDR 368
Query: 365 AAEAGFVDPNSPVAIVRPADFEDAAKQACQTKLENAKSTYPRVEEGNLPYLCMDLVYQYT 424
AAEAGFVDPN PVA VRP DFE AA +AC ++E KS +PRVEE NLPYLC+DLVYQYT
Sbjct: 369 AAEAGFVDPNQPVAEVRPLDFEKAANKACNMRMEEGKSKFPRVEEDNLPYLCLDLVYQYT 428
Query: 425 LLVDGFGIYPWQEITLVKKVKYDDALVEAAWPLGSAIEAVSS 466
LLVDGFG+ P Q ITLVKKVKY D VEAAWPLGSAIEAVSS
Sbjct: 429 LLVDGFGLKPSQTITLVKKVKYGDYAVEAAWPLGSAIEAVSS 470
>At1g14240 putative nucleoside triphosphatase
Length = 483
Score = 164 bits (414), Expect = 1e-40
Identities = 139/466 (29%), Positives = 215/466 (45%), Gaps = 62/466 (13%)
Query: 22 LLVVSIPILLITFVLYMMPSSSNYDSAGDYALLNRKLSPDKKSGGSYAVIFDAGSSGSRV 81
L+VVS+ I L +LY+ S+S S +LL+R+ Y+V+ DAGSSG+RV
Sbjct: 34 LVVVSVTITL-GLLLYVFNSNSVISSG---SLLSRRCKL------RYSVLIDAGSSGTRV 83
Query: 82 HVFH--FDQNLDLVHIGKDLELFEQLKPGLSAYAQKPQQAAESLVSLLEKAEGVVPRELR 139
HVF F+ + G+ +L PGLS+YA P+ A+ S+ L+E A+ +P+ +
Sbjct: 84 HVFGYWFESGKPVFDFGEKHYANLKLTPGLSSYADNPEGASVSVTKLVEFAKQRIPKRMF 143
Query: 140 SKTPVRIGATAGLRALEGDASDKILRAVRDLLKHRSSFKSDADAVTVLDGTQEGAYQWVT 199
++ +R+ ATAG+R LE ++IL R +L+ S F + V+ G+ EG Y W+T
Sbjct: 144 RRSDIRLMATAGMRLLEVPVQEQILEVTRRVLRS-SGFMFRDEWANVISGSDEGIYSWIT 202
Query: 200 INYLLGNLGKDYSKTVGVVDLGGGSVQMAYAISESEAAMAPQVMDGEDPYVKEMFLRGRK 259
NY LG+LG D +T G+V+LGG S Q+ + SE + P+ Y + +
Sbjct: 203 ANYALGSLGTDPLETTGIVELGGASAQVTFVSSEH---VPPE-------YSRTIAYGNIS 252
Query: 260 YYLYVHSYLRYGLLAARAEILK---------VAGDAENPCILSG--SDGTYKYGGKSFKA 308
Y +Y HS+L YG AA ++L+ V G E+PC G D K F A
Sbjct: 253 YTIYSHSFLDYGKDAALKKLLEKLQNSANSTVDGVVEDPCTPKGYIYDTNSKNYSSGFLA 312
Query: 309 SSS--------GASLNECKSVAHKALKVNESTCTHMKCTFGGIWNGGGGDGQKN-LFVAS 359
S + ++C+S LK + C + C+ G + D Q + L AS
Sbjct: 313 DESKLKGSLQAAGNFSKCRSATFALLKEGKENCLYEHCSIGSTFT---PDLQGSFLATAS 369
Query: 360 FF----FDRAAEAGFVDPNSPVAIVRPADFEDAAKQACQTKLENAKSTYPRVEEGNLPYL 415
F+ F E G++ P A K+ C + YP +E L
Sbjct: 370 FYYTAKFFELEEKGWLSELIP-----------AGKRYCGEEWSKLILEYPTTDEEYLRGY 418
Query: 416 CMDLVYQYTLLVDGFGIYPWQEITLVKKVKYDDALVEAAWPLGSAI 461
C Y ++L D GI + ++ K + + W LG+ I
Sbjct: 419 CFSAAYTISMLHDSLGI-ALDDESITYASKAGEKHIPLDWALGAFI 463
>At1g14230 putative nucleoside triphosphatase
Length = 503
Score = 154 bits (389), Expect = 1e-37
Identities = 120/420 (28%), Positives = 190/420 (44%), Gaps = 51/420 (12%)
Query: 68 YAVIFDAGSSGSRVHVFHF--DQNLDLVHIGKDLELFEQLKPGLSAYAQKPQQAAESLVS 125
Y+VI D GSSG+RVHVF + + + G++ +L PGLSAYA P+ +ES+
Sbjct: 81 YSVIIDGGSSGTRVHVFGYRIESGKPVFDFGEENYASLKLSPGLSAYADNPEGVSESVTE 140
Query: 126 LLEKAEGVVPRELRSKTPVRIGATAGLRALEGDASDKILRAVRDLLKHRSSFKSDADAVT 185
L+E A+ V + K+ +R+ ATAG+R LE ++IL R +L+ S F + +
Sbjct: 141 LVEFAKKRVHKGKLKKSDIRLMATAGMRLLELPVQEQILDVTRRVLRS-SGFDFRDEWAS 199
Query: 186 VLDGTQEGAYQWVTINYLLGNLGKDYSKTVGVVDLGGGSVQMAYAISESEAAMAPQVMDG 245
V+ G+ EG Y WV N+ LG+LG + KT G+V+LGG S Q+ + +E + P
Sbjct: 200 VISGSDEGVYAWVVANHALGSLGGEPLKTTGIVELGGASAQVTFVSTE----LVP----- 250
Query: 246 EDPYVKEMFLRGRKYYLYVHSYLRYGLLAARAEILKV----------AGDAENPCILSG- 294
+ + + Y LY HS+L +G AA+ ++ + G +PCI G
Sbjct: 251 -SEFSRTLAYGNVSYNLYSHSFLDFGQDAAQEKLSESLYNSAANSTGEGIVPDPCIPKGY 309
Query: 295 ---------SDGTYKYGGKSFKASSSGASLNECKSVAHKALKVNESTCTHMKCTFGGIWN 345
G GK + + +EC+S A L+ + CT+ +C+ G I+
Sbjct: 310 ILETNLQKDLPGFLADKGKFTATLQAAGNFSECRSAAFAMLQEEKGKCTYKRCSIGSIFT 369
Query: 346 ---GGGGDGQKNLFVASFFFDRAAEAGFVDPNSPVAIVRPADFEDAAKQACQTKLENAKS 402
G +N F S FF + ++ A K+ C + K
Sbjct: 370 PNLQGSFLATENFFHTSKFFGLGEKEWL------------SEMILAGKRFCGEEWSKLKV 417
Query: 403 TYPRVEEGNLPYLCMDLVYQYTLLVDGFGI-YPWQEITLVKKVKYDDALVEAAWPLGSAI 461
YP ++ NL C Y ++L D G+ + I K +D + W LG+ I
Sbjct: 418 KYPTFKDENLLRYCFSSAYIISMLHDSLGVALDDERIKYASKAGEED--IPLDWALGAFI 475
>At2g02970 nucleoside triphosphatase like protein
Length = 555
Score = 142 bits (357), Expect = 5e-34
Identities = 114/399 (28%), Positives = 177/399 (43%), Gaps = 62/399 (15%)
Query: 68 YAVIFDAGSSGSRVHVFHFDQNLDLVHIGKDLELFE-------QLKPGLSAYAQKPQQAA 120
Y+V+ D GS+G+R+HVF + + GK + F +L PGLSA+A P A+
Sbjct: 87 YSVVIDGGSTGTRIHVFGYR-----IESGKPVFEFRGANYASLKLHPGLSAFADDPDGAS 141
Query: 121 ESLVSLLEKAEGVVPRELRSKTPVRIGATAGLRALEGDASDKILRAVRDLLKHRSSFKSD 180
SL L+E A+G VP+ + +T VR+ ATAG+R LE +KIL R +LK S F
Sbjct: 142 VSLTELVEFAKGRVPKGMWIETEVRLMATAGMRLLELPVQEKILGVARRVLKS-SGFLFR 200
Query: 181 ADAVTVLDGTQEGAYQWVTINYLLGNLGKDYSKTVGVVDLGGGSVQMAYAISESEAAMAP 240
+ +V+ G+ EG Y WV N+ LG+LG D KT G+V+LGG S Q+ + SE M P
Sbjct: 201 DEWASVISGSDEGVYAWVVANFALGSLGGDPLKTTGIVELGGASAQVTFVSSEP---MPP 257
Query: 241 QVMDGEDPYVKEMFLRGRKYYLYVHSYLRYGLLAARAEILKVAGDAEN------------ 288
+ + + + Y LY HS+L +G AA ++ ++
Sbjct: 258 E-------FSRTISFGNVTYNLYSHSFLHFGQNAAHDKLWGSLLSRDHNSAVEPTREKIF 310
Query: 289 --PCILSGSD----------GTYKYGGKSFKASSSGASLNECKSVAHKALKVNESTCTHM 336
PC G + G + + +G + ++C+S A L+ C++
Sbjct: 311 TDPCAPKGYNLDANTQKHLSGLLAEESRLSDSFQAGGNYSQCRSAALTILQDGNEKCSYQ 370
Query: 337 KCTFGGIWN---GGGGDGQKNLFVASFFFDRAAEAGFVDPNSPVAIVRPADFEDAAKQAC 393
C+ G + G +N F S FF +A ++ A ++ C
Sbjct: 371 HCSIGSTFTPKLRGRFLATENFFYTSKFFGLGEKAWL------------SNMISAGERFC 418
Query: 394 QTKLENAKSTYPRVEEGNLPYLCMDLVYQYTLLVDGFGI 432
+ P + E +L C Y +LL D GI
Sbjct: 419 GEDWSKLRVKDPSLHEEDLLRYCFSSAYIVSLLHDTLGI 457
>At1g14260 hypothetical protein, 5' partial
Length = 669
Score = 137 bits (345), Expect = 1e-32
Identities = 107/380 (28%), Positives = 169/380 (44%), Gaps = 52/380 (13%)
Query: 81 VHVFH--FDQNLDLVHIGKDLELFEQLKPGLSAYAQKPQQAAESLVSLLEKAEGVVPREL 138
+HVF F+ + G++ +L PGLS+YA P+ A+ S+ L+E A+G +P+
Sbjct: 1 IHVFGYWFESGKPVFDFGEEHYASLKLSPGLSSYADNPEGASVSVTKLVEFAKGRIPKGK 60
Query: 139 RSKTPVRIGATAGLRALEGDASDKILRAVRDLLKHRSSFKSDADAVTVLDGTQEGAYQWV 198
K+ +R+ ATAG+R L+ ++IL R +L+ S FK + TV+ GT EG Y WV
Sbjct: 61 LKKSDIRLMATAGMRLLDVPVQEQILDVTRRVLRS-SGFKFQDEWATVISGTDEGIYAWV 119
Query: 199 TINYLLGNLGKDYSKTVGVVDLGGGSVQMAYAISESEAAMAPQVMDGEDPYVKEMFLRGR 258
N+ LG+LG D KT G+V+LGG S Q+ + SE + P+ + + +
Sbjct: 120 VANHALGSLGGDPLKTTGIVELGGASAQVTFVPSEH---VPPE-------FSRTISYGNV 169
Query: 259 KYYLYVHSYLRYGLLAARAEILKVA----------GDAENPCILSG----------SDGT 298
Y +Y HS+L +G AA ++L+ G E+PC G S G
Sbjct: 170 SYTIYSHSFLDFGQDAAEDKLLESLQNSVAASTGDGIVEDPCTPKGYIYDTHSQKDSSGF 229
Query: 299 YKYGGKSFKAS---SSGASLNECKSVAHKALKVNESTCTHMKCTFGGIWN---GGGGDGQ 352
K FKAS + +C+S L+ + C + C+ G + G
Sbjct: 230 LSEESK-FKASLQVQAAGDFTKCRSATLAMLQEGKENCAYKHCSIGSTFTPNIQGSFLAT 288
Query: 353 KNLFVASFFFDRAAEAGFVDPNSPVAIVRPADFEDAAKQACQTKLENAKSTYPRVEEGNL 412
+N F S FF + ++ A K+ C + K YP ++ L
Sbjct: 289 ENFFHTSKFFGLGEKEWL------------SEMILAGKRFCGEEWSKLKEKYPTTKDKYL 336
Query: 413 PYLCMDLVYQYTLLVDGFGI 432
C Y ++L D G+
Sbjct: 337 HRYCFSSAYIISMLHDSLGV 356
>At4g19181 putative protein
Length = 740
Score = 80.9 bits (198), Expect = 1e-15
Identities = 111/468 (23%), Positives = 187/468 (39%), Gaps = 101/468 (21%)
Query: 63 KSGGSYAVIFDAGSSGSRVHVF----HFDQNLDLVHIGKDL----------ELFEQLK-- 106
+ Y V+FD GS+G+R +V+ ++ ++ L + K L +++++
Sbjct: 140 RGASRYYVVFDCGSTGTRAYVYQASINYKKDSSLPIVMKSLTEGISRKSRGRAYDRMETE 199
Query: 107 PGLSAYAQKPQQAAESLVSLLEKAEGVVPRELRSKTPVRIGATAGLRALEGDASDKILRA 166
PG ++ L++ AE +P+ T + + ATAG+R L S IL
Sbjct: 200 PGFDKLVNNRTGLKTAIKPLIQWAEKQIPKNAHRTTSLFVYATAGVRRLRPADSSWILGN 259
Query: 167 VRDLLKHRSSFKSDADAVTVLDGTQEGAYQWVTINYLLGNLGKDYSK-TVGVVDLGGGSV 225
V +L +S F + V ++ GT+E + W +NY LG K T G +DLGG S+
Sbjct: 260 VWSILA-KSPFTCRREWVKIISGTEEAYFGWTALNYQTSMLGALPKKATFGALDLGGSSL 318
Query: 226 QMAYAISESEAAMAPQVMDGEDPYVKEMFLRGR----KYYLYVHSYLRYGLLAA------ 275
Q+ + + E+ E L R ++L +S YGL A
Sbjct: 319 QVTF--------------ENEERTHNETNLNLRIGSVNHHLSAYSLAGYGLNDAFDRSVV 364
Query: 276 ----------RAEILKVAGDAENPCILSGSDGTY--------KYGGKSFKASSS-----G 312
++++++ + ++PC+ SG +G Y GGK K+ S
Sbjct: 365 HLLKKLPNVNKSDLIEGKLEMKHPCLNSGYNGQYICSQCASSVQGGKKGKSGVSIKLVGA 424
Query: 313 ASLNECKSVAHKALKVNEST-------CTHMKCTFGGIWNGGGGD--GQKNLFVASFFFD 363
+ EC ++A A+ +E + C C + G FV FF+
Sbjct: 425 PNWGECSALAKNAVNSSEWSNAKHGVDCDLQPCALPDGYPRPHGQFYAVSGFFVVYRFFN 484
Query: 364 RAAEAGFVDPNSPVAIVRPADFEDAAKQACQTKLENAKSTYPRVEEGNLPYLCMDLVYQY 423
+AEA D + + +F D A Q +T + S P +E+ C Y
Sbjct: 485 LSAEASLDD-----VLEKGREFCDKAWQVARTSV----SPQPFIEQ-----YCFRAPYIV 530
Query: 424 TLLVDGFGIYPWQEITLVKKVKYDDALVEAAWPLGSAI----EAVSST 467
+LL +G I Q I + W LG A+ +A+SST
Sbjct: 531 SLLREGLYITDKQIIIGSGSI---------TWTLGVALLESGKALSST 569
>At1g14250 putative nucleoside triphosphatase, 3' partial
Length = 128
Score = 53.1 bits (126), Expect = 3e-07
Identities = 28/69 (40%), Positives = 43/69 (61%), Gaps = 2/69 (2%)
Query: 62 KKSGGSYAVIFDAGSSGSRVHVF--HFDQNLDLVHIGKDLELFEQLKPGLSAYAQKPQQA 119
++S Y+VI DAGSSG+R+HVF F+ + G++ +L PGLS+YA P+ A
Sbjct: 59 RRSSLHYSVIIDAGSSGTRIHVFGYWFESGKPVFDFGEEHYASLKLSPGLSSYADNPEGA 118
Query: 120 AESLVSLLE 128
+ S+ L+E
Sbjct: 119 SVSVTKLVE 127
>At3g19580 zinc finger protein, putative
Length = 273
Score = 42.0 bits (97), Expect = 7e-04
Identities = 24/78 (30%), Positives = 40/78 (50%), Gaps = 6/78 (7%)
Query: 303 GKSFKASSSGASLNECKSVAHKALKVNESTCTHMKCTFGGIWNGGGGDGQKNL----FVA 358
G+ ++SG ++EC S+ HK ++ H +C + G GGGG G K++ V+
Sbjct: 153 GEKHPIAASG-KIHEC-SICHKVFPTGQALGGHKRCHYEGNLGGGGGGGSKSISHSGSVS 210
Query: 359 SFFFDRAAEAGFVDPNSP 376
S + + GF+D N P
Sbjct: 211 STVSEERSHRGFIDLNLP 228
>At1g27460 unknown protein
Length = 694
Score = 32.3 bits (72), Expect = 0.57
Identities = 50/233 (21%), Positives = 92/233 (39%), Gaps = 53/233 (22%)
Query: 253 MFLRGRKYYLYVHSYLRYGLLAARAEILKVA-GDAEN------PCILSGSDGTYKYGGKS 305
++ RG ++YL Y G+ A +LK+A G +E+ P +L G+ K K
Sbjct: 303 VYTRGERWYLLSLCYSAAGIDKAAINLLKMALGPSESRQIPHIPLLLFGA----KLCSKD 358
Query: 306 FKASSSGASLNECKSVAHKALKVNESTCTHMKC---TFGGIWNGGGGDGQK----NLFV- 357
K S G + AH+ L + S H+ F G+ G K +F+
Sbjct: 359 PKHSRDGINF------AHRLLDLGNSQSEHLLSQAHKFLGVCYGNAARSSKLDSERVFLQ 412
Query: 358 --ASFFFDRAAEAGFVDPNSPVAIVRPADFEDAAKQACQTKLENAKSTYPRVEEGNLPYL 415
+ F + AA+ G DP + ++ E+A ++ Q L+ A
Sbjct: 413 KKSLFSLNEAAKRGKADPE--LDVIFNLSVENAVQRNVQAALDGA--------------- 455
Query: 416 CMDLVYQYTLLVDGFGIYPWQEITLV----KKVKYDDALVEAAWPLGSAIEAV 464
+Y+ +V G W+ + +V K++K +++++ IE +
Sbjct: 456 -----VEYSSMVGGVSTKGWKHLAIVLSAEKRLKDAESILDFTMEEAGDIEKI 503
>At5g51330 DYAD/SWITCH1 (SWI1)
Length = 639
Score = 28.5 bits (62), Expect = 8.3
Identities = 16/53 (30%), Positives = 26/53 (48%)
Query: 21 TLLVVSIPILLITFVLYMMPSSSNYDSAGDYALLNRKLSPDKKSGGSYAVIFD 73
++ VV + + + V PS + S DY+ +NR K+SGG +FD
Sbjct: 58 SIRVVMVSKITASDVSLRYPSMFSLRSHFDYSRMNRNKPMKKRSGGGLLPVFD 110
>At1g09190 hypothetical protein
Length = 999
Score = 28.5 bits (62), Expect = 8.3
Identities = 48/222 (21%), Positives = 89/222 (39%), Gaps = 30/222 (13%)
Query: 100 ELFEQLKPGLSAYAQKPQQAAESLVSLLEKAEGVVPRELRSKTPVRIGATAGLRALEG-- 157
E+F + G +++ QA S S + P L ++P I + + RAL+
Sbjct: 13 EVFTSIDLGTNSFKLLIVQAEPSGRSFVPIERLKEPVVLSRESPTSISSQSQARALQSLR 72
Query: 158 ----------DASDKILRAVRDLLKHRSSFKSDADAV--------TVLDGTQEGAYQWVT 199
+ D+I + L+ + + DAV VL G +E ++
Sbjct: 73 KFKSLILSHHVSGDRIRCVATEALRRAENQEEFVDAVFEDVGLKVDVLSGEEEARLVYLG 132
Query: 200 INYLLGNLGKDYSKTVGVVDLGGGSVQMAYAISESEAAMAPQVMDGEDPYVKEMFLRGRK 259
+ L + K+V VD+GGGS + + +A + G + +MF++
Sbjct: 133 VLQFL----PVFEKSVLCVDIGGGSTEFVVG-KCGDVKLAVSLKLGH-VNLTQMFMKNGI 186
Query: 260 YYLYVHSYLRYGLLAAR-AEILKVAGDAENPCILSGSDGTYK 300
+ Y+R+ + +R AE L + E ++ GS GT +
Sbjct: 187 GLKEMRDYIRHAIDESRLAEKLNESDGFE---VVVGSSGTIR 225
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.135 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,278,089
Number of Sequences: 26719
Number of extensions: 444477
Number of successful extensions: 1208
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 1175
Number of HSP's gapped (non-prelim): 14
length of query: 467
length of database: 11,318,596
effective HSP length: 103
effective length of query: 364
effective length of database: 8,566,539
effective search space: 3118220196
effective search space used: 3118220196
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC123593.7


