

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123593.3 + phase: 0 /pseudo
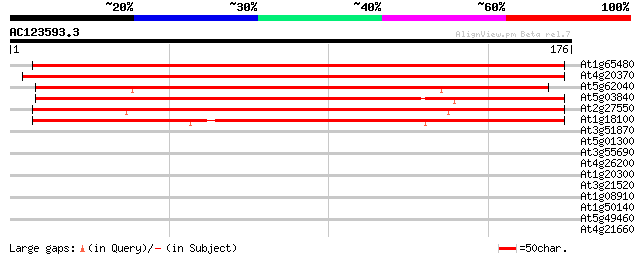
(176 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g65480 flowering time locus T (FT) 240 3e-64
At4g20370 TWIN SISTER OF FT (TSF) 233 3e-62
At5g62040 Fdr1 Cen - like protein 179 7e-46
At5g03840 Terminal flower1 (TFL1) 178 2e-45
At2g27550 Arabidopsis thaliana centroradialis (ATC) 175 1e-44
At1g18100 terminal Flower 1 (TFL1), putative 156 5e-39
At3g51870 carrier like protein 30 0.67
At5g01300 unknown protein 28 2.5
At3g55690 unknown protein 28 2.5
At4g26200 1-aminocyclopropane-1-carboxylate synthase -like protein 27 4.3
At1g20300 putative protein 27 4.3
At3g21520 unknown protein 27 5.7
At1g08910 hypothetical protein 27 5.7
At1g50140 hypothetical protein, 5' partial 27 7.4
At5g49460 ATP-citrate lyase subunit B 26 9.6
At4g21660 spliceosome associated protein - like 26 9.6
>At1g65480 flowering time locus T (FT)
Length = 175
Score = 240 bits (612), Expect = 3e-64
Identities = 114/167 (68%), Positives = 128/167 (76%)
Query: 8 NPLAVGRVIGDVLDPFESTIPLLVTYGNRTVTNGGELKPSQVANQPQVIIGVNDPTALYT 67
+PL V RV+GDVLDPF +I L VTYG R VTNG +L+PSQV N+P+V IG D YT
Sbjct: 7 DPLIVSRVVGDVLDPFNRSITLKVTYGQREVTNGLDLRPSQVQNKPRVEIGGEDLRNFYT 66
Query: 68 LVLVDPDAPSPSYPSFREYLHWMVTDIPATNAASFGNEVVSYEKPRPNLGIHRFVFVLLH 127
LV+VDPD PSPS P REYLHW+VTDIPAT +FGNE+V YE P P GIHR VF+L
Sbjct: 67 LVMVDPDVPSPSNPHLREYLHWLVTDIPATTGTTFGNEIVCYENPSPTAGIHRVVFILFR 126
Query: 128 QQCRQRVYAPGWRQNFNTREFIEFYNLGSPVAAVFFNCQRETGSGGR 174
Q RQ VYAPGWRQNFNTREF E YNLG PVAAVF+NCQRE+G GGR
Sbjct: 127 QLGRQTVYAPGWRQNFNTREFAEIYNLGLPVAAVFYNCQRESGCGGR 173
>At4g20370 TWIN SISTER OF FT (TSF)
Length = 175
Score = 233 bits (595), Expect = 3e-62
Identities = 115/170 (67%), Positives = 127/170 (74%)
Query: 5 SRPNPLAVGRVIGDVLDPFESTIPLLVTYGNRTVTNGGELKPSQVANQPQVIIGVNDPTA 64
SR +PL VG V+GDVLDPF + L VTYG+R VTNG +L+PSQV N+P V IG +D
Sbjct: 4 SRRDPLVVGSVVGDVLDPFTRLVSLKVTYGHREVTNGLDLRPSQVLNKPIVEIGGDDFRN 63
Query: 65 LYTLVLVDPDAPSPSYPSFREYLHWMVTDIPATNAASFGNEVVSYEKPRPNLGIHRFVFV 124
YTLV+VDPD PSPS P REYLHW+VTDIPAT +FGNEVV YE PRP GIHR V V
Sbjct: 64 FYTLVMVDPDVPSPSNPHQREYLHWLVTDIPATTGNAFGNEVVCYESPRPPSGIHRIVLV 123
Query: 125 LLHQQCRQRVYAPGWRQNFNTREFIEFYNLGSPVAAVFFNCQRETGSGGR 174
L Q RQ VYAPGWRQ FNTREF E YNLG PVAA +FNCQRE G GGR
Sbjct: 124 LFRQLGRQTVYAPGWRQQFNTREFAEIYNLGLPVAASYFNCQRENGCGGR 173
>At5g62040 Fdr1 Cen - like protein
Length = 177
Score = 179 bits (454), Expect = 7e-46
Identities = 91/163 (55%), Positives = 115/163 (69%), Gaps = 2/163 (1%)
Query: 9 PLAVGRVIGDVLDPFESTIPLLVTYGNRT-VTNGGELKPSQVANQPQVIIGVNDPTALYT 67
PL VGRVIGDVL+ F ++ + VT+ + T V+NG EL PS + ++P+V IG D + +T
Sbjct: 7 PLIVGRVIGDVLEMFNPSVTMRVTFNSNTIVSNGHELAPSLLLSKPRVEIGGQDLRSFFT 66
Query: 68 LVLVDPDAPSPSYPSFREYLHWMVTDIPATNAASFGNEVVSYEKPRPNLGIHRFVFVLLH 127
L+++DPDAPSPS P REYLHWMVTDIP T ASFG E+V YE P+P GIHR+VF L
Sbjct: 67 LIMMDPDAPSPSNPYMREYLHWMVTDIPGTTDASFGREIVRYETPKPVAGIHRYVFALFK 126
Query: 128 QQCRQRV-YAPGWRQNFNTREFIEFYNLGSPVAAVFFNCQRET 169
Q+ RQ V AP R+ FNT F ++ L PVAAV+FN QRET
Sbjct: 127 QRGRQAVKAAPETRECFNTNAFSSYFGLSQPVAAVYFNAQRET 169
>At5g03840 Terminal flower1 (TFL1)
Length = 177
Score = 178 bits (451), Expect = 2e-45
Identities = 92/168 (54%), Positives = 120/168 (70%), Gaps = 3/168 (1%)
Query: 9 PLAVGRVIGDVLDPFESTIPLLVTYGNRTVTNGGELKPSQVANQPQVIIGVNDPTALYTL 68
PL +GRV+GDVLD F T + V+Y + V+NG EL PS V+++P+V I D + +TL
Sbjct: 11 PLIMGRVVGDVLDFFTPTTKMNVSYNKKQVSNGHELFPSSVSSKPRVEIHGGDLRSFFTL 70
Query: 69 VLVDPDAPSPSYPSFREYLHWMVTDIPATNAASFGNEVVSYEKPRPNLGIHRFVFVLLHQ 128
V++DPD P PS P +E+LHW+VT+IP T A+FG EVVSYE PRP++GIHRFVFVL Q
Sbjct: 71 VMIDPDVPGPSDPFLKEHLHWIVTNIPGTTDATFGKEVVSYELPRPSIGIHRFVFVLFRQ 130
Query: 129 QCRQRVYAPG--WRQNFNTREFIEFYNLGSPVAAVFFNCQRETGSGGR 174
+ ++RV P R +FNTR+F Y+LG PVAAVFFN QRET + R
Sbjct: 131 K-QRRVIFPNIPSRDHFNTRKFAVEYDLGLPVAAVFFNAQRETAARKR 177
>At2g27550 Arabidopsis thaliana centroradialis (ATC)
Length = 175
Score = 175 bits (444), Expect = 1e-44
Identities = 90/169 (53%), Positives = 113/169 (66%), Gaps = 2/169 (1%)
Query: 8 NPLAVGRVIGDVLDPFESTIPLLVTYGN-RTVTNGGELKPSQVANQPQVIIGVNDPTALY 66
+PL VGRVIGDV+D + + VTY + + V NG EL PS V +P+V + D + +
Sbjct: 7 DPLMVGRVIGDVVDNCLQAVKMTVTYNSDKQVYNGHELFPSVVTYKPKVEVHGGDMRSFF 66
Query: 67 TLVLVDPDAPSPSYPSFREYLHWMVTDIPATNAASFGNEVVSYEKPRPNLGIHRFVFVLL 126
TLV+ DPD P PS P RE+LHW+VTDIP T SFG E++ YE PRPN+GIHRFV++L
Sbjct: 67 TLVMTDPDVPGPSDPYLREHLHWIVTDIPGTTDVSFGKEIIGYEMPRPNIGIHRFVYLLF 126
Query: 127 HQQCRQRVYA-PGWRQNFNTREFIEFYNLGSPVAAVFFNCQRETGSGGR 174
Q R V + P +R FNTREF +LG PVAAVFFNCQRET + R
Sbjct: 127 KQTRRGSVVSVPSYRDQFNTREFAHENDLGLPVAAVFFNCQRETAARRR 175
>At1g18100 terminal Flower 1 (TFL1), putative
Length = 173
Score = 156 bits (395), Expect = 5e-39
Identities = 85/170 (50%), Positives = 109/170 (64%), Gaps = 5/170 (2%)
Query: 8 NPLAVGRVIGDVLDPFESTIPLLVTYGNRTVTNGGELKPSQVANQPQV-IIGVNDPTALY 66
+PL VGRVIGDVLD F T + V +G + +TNG E+KPS N P+V I G +D LY
Sbjct: 6 DPLVVGRVIGDVLDMFIPTANMSVYFGPKHITNGCEIKPSTAVNPPKVNISGHSDE--LY 63
Query: 67 TLVLVDPDAPSPSYPSFREYLHWMVTDIPATNAASFGNEVVSYEKPRPNLGIHRFVFVLL 126
TLV+ DPDAPSPS P+ RE++HW+V DIP S G E++ Y +PRP +GIHR++ VL
Sbjct: 64 TLVMTDPDAPSPSEPNMREWVHWIVVDIPGGTNPSRGKEILPYMEPRPPVGIHRYILVLF 123
Query: 127 HQQ--CRQRVYAPGWRQNFNTREFIEFYNLGSPVAAVFFNCQRETGSGGR 174
Q V P R NF+TR F ++LG PVA V+FN Q+E S R
Sbjct: 124 RQNSPVGLMVQQPPSRANFSTRMFAGHFDLGLPVATVYFNAQKEPASRRR 173
>At3g51870 carrier like protein
Length = 381
Score = 30.0 bits (66), Expect = 0.67
Identities = 17/55 (30%), Positives = 25/55 (44%)
Query: 90 MVTDIPATNAASFGNEVVSYEKPRPNLGIHRFVFVLLHQQCRQRVYAPGWRQNFN 144
+ T PA + A V R + G+ RF + L ++C QR +AP Q N
Sbjct: 21 ITTSSPAKSGAEQFRRRVLRNPARGDFGLGRFACISLVEKCEQREFAPTTAQLLN 75
>At5g01300 unknown protein
Length = 162
Score = 28.1 bits (61), Expect = 2.5
Identities = 16/37 (43%), Positives = 20/37 (53%), Gaps = 1/37 (2%)
Query: 59 VNDPTALYTLVLVDPDAPSPSYPSFREYLHWMVTDIP 95
V + T LV+ D DAP PS P + W+V DIP
Sbjct: 43 VPEGTKTLALVVEDIDAPDPSGP-LVPWTVWVVVDIP 78
>At3g55690 unknown protein
Length = 293
Score = 28.1 bits (61), Expect = 2.5
Identities = 19/78 (24%), Positives = 33/78 (41%), Gaps = 4/78 (5%)
Query: 38 VTNGGELKPSQVANQPQVIIGVNDPTALYTLVLVDPDAPSPSYPSFREYLHWMVTDIPAT 97
VT+ ++KP + G+ P ++ V P +PS S + L ++
Sbjct: 85 VTDKSQMKPKVTESTQS---GLGSPNGPFSQVPSPPTSPSREEDSLK-VLSAAAGEVAKI 140
Query: 98 NAASFGNEVVSYEKPRPN 115
A+F + +SY P PN
Sbjct: 141 KKANFDAKPISYPNPNPN 158
>At4g26200 1-aminocyclopropane-1-carboxylate synthase -like protein
Length = 447
Score = 27.3 bits (59), Expect = 4.3
Identities = 13/36 (36%), Positives = 16/36 (44%), Gaps = 4/36 (11%)
Query: 58 GVNDPTALYTLVLVDPD----APSPSYPSFREYLHW 89
G L T +L DP+ P+P YP F L W
Sbjct: 134 GATAANELLTFILADPNDALLVPTPYYPGFDRDLRW 169
>At1g20300 putative protein
Length = 537
Score = 27.3 bits (59), Expect = 4.3
Identities = 30/119 (25%), Positives = 47/119 (39%), Gaps = 7/119 (5%)
Query: 15 VIGDVLDPFESTIPLLVTYGNRT---VTNGGELKPSQVANQPQVIIGVNDPTALYTLVLV 71
V D+LD P +T+ N V G K QV NQ + + G T Y L+
Sbjct: 312 VFADMLD--SGCAPNAITFNNLMRVHVKAGRTEKVLQVYNQMKKL-GCEPDTITYNF-LI 367
Query: 72 DPDAPSPSYPSFREYLHWMVTDIPATNAASFGNEVVSYEKPRPNLGIHRFVFVLLHQQC 130
+ + + + L+ M+ NA++F EK R G HR ++ +C
Sbjct: 368 EAHCRDENLENAVKVLNTMIKKKCEVNASTFNTIFRYIEKKRDVNGAHRMYSKMMEAKC 426
>At3g21520 unknown protein
Length = 207
Score = 26.9 bits (58), Expect = 5.7
Identities = 15/51 (29%), Positives = 22/51 (42%)
Query: 67 TLVLVDPDAPSPSYPSFREYLHWMVTDIPATNAASFGNEVVSYEKPRPNLG 117
TLVL+D + S YP FRE +V +P + + R +G
Sbjct: 136 TLVLLDANTASCFYPRFRETQKTLVMALPPAVGVASATIFALFPSKRSGIG 186
>At1g08910 hypothetical protein
Length = 600
Score = 26.9 bits (58), Expect = 5.7
Identities = 21/59 (35%), Positives = 24/59 (40%), Gaps = 14/59 (23%)
Query: 48 QVANQPQVIIGVNDPTALYTLVLVDPDAPSPSYPSFREYLHWMVTDIPATNAASFGNEV 106
Q N PQVI D A L + PSP DI ATNAA+FG +
Sbjct: 215 QFVNLPQVI-NTRDSPASQALPMTFSPTPSPQ-------------DILATNAANFGTSM 259
>At1g50140 hypothetical protein, 5' partial
Length = 601
Score = 26.6 bits (57), Expect = 7.4
Identities = 15/48 (31%), Positives = 27/48 (56%), Gaps = 4/48 (8%)
Query: 9 PLAVGRVIGDVLDPFESTIPLLVTYGNRTVTNGGELKPSQVANQPQVI 56
PL+ G+ G+V + + + ++ YG+ + G E KP A QPQ++
Sbjct: 1 PLSSGQR-GEVYEVIGNRVAVIFEYGDDKTSEGSEKKP---AEQPQML 44
>At5g49460 ATP-citrate lyase subunit B
Length = 608
Score = 26.2 bits (56), Expect = 9.6
Identities = 12/38 (31%), Positives = 20/38 (52%)
Query: 31 VTYGNRTVTNGGELKPSQVANQPQVIIGVNDPTALYTL 68
V +G+ +GGE++ +Q NQ + G PT+ L
Sbjct: 269 VQFGHAGAKSGGEMESAQAKNQALIDAGAIVPTSFEAL 306
>At4g21660 spliceosome associated protein - like
Length = 584
Score = 26.2 bits (56), Expect = 9.6
Identities = 21/75 (28%), Positives = 31/75 (41%), Gaps = 14/75 (18%)
Query: 43 ELKPSQVANQPQVIIGVNDPTALYTLVLVDPDAPSPSYPSFREYLHWMVTDIPATNA--- 99
E KP ++N + +G+ + L+ + P PSYP + IP NA
Sbjct: 306 ETKPGFLSNDLKEALGMPEGAPPPWLINMQRYGPPPSYPHLK---------IPGLNAPIP 356
Query: 100 --ASFGNEVVSYEKP 112
ASFG + KP
Sbjct: 357 IGASFGFHAGGWGKP 371
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.139 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,373,667
Number of Sequences: 26719
Number of extensions: 194394
Number of successful extensions: 336
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 320
Number of HSP's gapped (non-prelim): 16
length of query: 176
length of database: 11,318,596
effective HSP length: 93
effective length of query: 83
effective length of database: 8,833,729
effective search space: 733199507
effective search space used: 733199507
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC123593.3


