

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123574.2 + phase: 0 /pseudo
(984 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
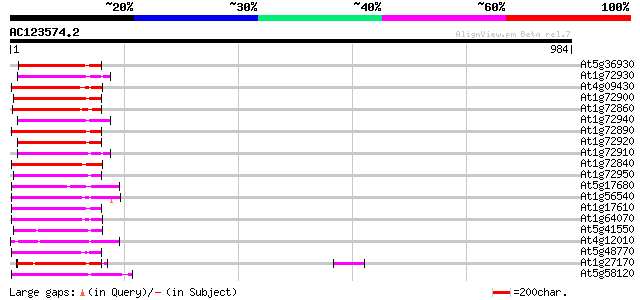
Score E
Sequences producing significant alignments: (bits) Value
At5g36930 disease resistance like protein 140 4e-33
At1g72930 putative protein 137 4e-32
At4g09430 putative protein 136 5e-32
At1g72900 unknown protein 136 7e-32
At1g72860 unknown protein 135 1e-31
At1g72940 unknown protein 135 1e-31
At1g72890 hypothetical protein 134 2e-31
At1g72920 hypothetical protein 134 3e-31
At1g72910 unknown protein 134 3e-31
At1g72840 hypothetical protein 134 3e-31
At1g72950 hypothetical protein 133 4e-31
At5g17680 disease resistance protein RPP1-WsB - like protein 130 4e-30
At1g56540 disease resistance protein, putative 126 7e-29
At1g17610 disease resistance protein, putative 125 9e-29
At1g64070 disease resistance protein, putative 125 1e-28
At5g41550 disease resistance protein-like 124 3e-28
At4g12010 like disease resistance protein (TMV N-like) 123 4e-28
At5g48770 disease resistance protein 123 6e-28
At1g27170 disease resistance protein, putative 123 6e-28
At5g58120 resistance protein - like 122 8e-28
>At5g36930 disease resistance like protein
Length = 1164
Score = 140 bits (353), Expect = 4e-33
Identities = 72/146 (49%), Positives = 101/146 (68%), Gaps = 6/146 (4%)
Query: 16 YQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIFIP 75
Y VF+SFRG+D R F +LY +L GI TF+DD ELQRG+ I+P L NAIE S+I I
Sbjct: 14 YDVFVSFRGADVRKNFLSHLYDSLRRCGISTFMDDVELQRGEYISPELLNAIETSKILIV 73
Query: 76 VFSANYASSSFCLDELVHIIHLYKQN-GRLVLPVFFGVDPSHVRHHRGSYGEALAKHEER 134
V + +YASS++CLDELVHI+ +K N +V P+F VDPS +R +GSY ++ +KH+
Sbjct: 74 VLTKDYASSAWCLDELVHIMKSHKNNPSHMVFPIFLYVDPSDIRWQQGSYAKSFSKHK-- 131
Query: 135 FQHNTDHMERLQKWKIALTQAANLSG 160
N+ + +L+ W+ ALT+ AN+SG
Sbjct: 132 ---NSHPLNKLKDWREALTKVANISG 154
>At1g72930 putative protein
Length = 176
Score = 137 bits (344), Expect = 4e-32
Identities = 77/162 (47%), Positives = 97/162 (59%), Gaps = 6/162 (3%)
Query: 15 KYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIFI 74
KY VFLSFRG DTR+ F LYK L + I TF DD EL+ G +P L + IE SR +
Sbjct: 8 KYDVFLSFRGHDTRHNFISFLYKELVRRSIRTFKDDKELENGQRFSPELKSPIEVSRFAV 67
Query: 75 PVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEER 134
V S NYA+SS+CLDELV I+ K+ V+P+F+GV+P+HVR G E KH R
Sbjct: 68 VVVSENYAASSWCLDELVTIMDFEKKGSITVMPIFYGVEPNHVRWQTGVLAEQFKKHASR 127
Query: 135 FQHNTDHMERLQKWKIALTQAANLSGDHRSPGYEYKLTGKIA 176
+ E++ KW+ ALT A LSGD S + KL KIA
Sbjct: 128 -----EDPEKVLKWRQALTNFAQLSGD-CSGDDDSKLVDKIA 163
>At4g09430 putative protein
Length = 1039
Score = 136 bits (343), Expect = 5e-32
Identities = 76/160 (47%), Positives = 103/160 (63%), Gaps = 8/160 (5%)
Query: 3 SSSSSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPS 62
+SSS+SS + V +Y VFLSFRG+DTR L+KAL D GI TF DD EL+ GD I+
Sbjct: 2 ASSSTSSPTRVKEYDVFLSFRGADTRNNIVSYLHKALVDVGIRTFKDDKELEEGDIISEK 61
Query: 63 LDNAIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRG 122
L NAI+ S + V S Y +SS+CL+EL HI+ L Q+ +V+P+F+ V+PS VR+ +
Sbjct: 62 LVNAIQTSWFAVVVLSEKYVTSSWCLEELRHIMELSIQDDIIVVPIFYKVEPSDVRYQKN 121
Query: 123 SYGEALAKHEERFQHNTDHMERLQKWKIALTQAANLSGDH 162
S+ E + QH D E++ KWK ALTQ N+SG H
Sbjct: 122 SF-------EVKLQHYRD-PEKILKWKGALTQVGNMSGKH 153
>At1g72900 unknown protein
Length = 363
Score = 136 bits (342), Expect = 7e-32
Identities = 73/155 (47%), Positives = 96/155 (61%), Gaps = 5/155 (3%)
Query: 8 SSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAI 67
SS + + Y VFLSFRG DTR F LYK L + I TF DD EL+ G I+P L AI
Sbjct: 2 SSATATYNYDVFLSFRGPDTRRKFISFLYKELVGRDIRTFKDDKELENGQMISPELILAI 61
Query: 68 EESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRL-VLPVFFGVDPSHVRHHRGSYGE 126
E+SR + V S NYA+SS+CLDELV I+ + K G + V+P+F+GV+P H+R G E
Sbjct: 62 EDSRFAVVVVSVNYAASSWCLDELVKIMDIQKNKGSITVMPIFYGVNPCHLRRQIGDVAE 121
Query: 127 ALAKHEERFQHNTDHMERLQKWKIALTQAANLSGD 161
KHE R +E++ KW+ AL A++SGD
Sbjct: 122 QFKKHEAR----EKDLEKVLKWRQALAALADISGD 152
>At1g72860 unknown protein
Length = 1337
Score = 135 bits (340), Expect = 1e-31
Identities = 71/155 (45%), Positives = 101/155 (64%), Gaps = 7/155 (4%)
Query: 6 SSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDN 65
+SSS S ++KY VFLSFRG DTR +L+K L DKG+ TF DD +L+ GD I+ +
Sbjct: 2 ASSSSSPIWKYDVFLSFRGEDTRKNIVSHLHKQLVDKGVVTFKDDKKLELGDSISEEISR 61
Query: 66 AIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYG 125
AI+ S + + S NYASSS+CLDEL ++ L+ +N V+P+F+GVDPSHVRH GS+
Sbjct: 62 AIQNSTYALVILSENYASSSWCLDELRMVMDLHLKNKIKVVPIFYGVDPSHVRHQTGSF- 120
Query: 126 EALAKHEERFQHNTDHMERLQKWKIALTQAANLSG 160
K+++ N ++ W+ ALTQ A+L+G
Sbjct: 121 -TFDKYQDSKMPN-----KVTTWREALTQIASLAG 149
>At1g72940 unknown protein
Length = 371
Score = 135 bits (339), Expect = 1e-31
Identities = 77/163 (47%), Positives = 96/163 (58%), Gaps = 5/163 (3%)
Query: 15 KYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIFI 74
KY VFLSFRG DTR F LYK + I TF DD EL+ G I+P L AIEES+ +
Sbjct: 8 KYDVFLSFRGVDTRRNFISFLYKEFVRRKIRTFKDDKELENGRRISPELKRAIEESKFAV 67
Query: 75 PVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEER 134
V S NYA+S +CLDELV I+ + V+P+F+GVDP H+R G E KHE R
Sbjct: 68 VVVSVNYAASPWCLDELVKIMDFENKGSITVMPIFYGVDPCHLRRQIGDVAEQFKKHEAR 127
Query: 135 FQHNTDHMERLQKWKIALTQAANLSGDHRSP-GYEYKLTGKIA 176
DH E++ W+ ALT A++SGD S E KL +IA
Sbjct: 128 ---EEDH-EKVASWRRALTSLASISGDCSSKCEDEAKLVDEIA 166
>At1g72890 hypothetical protein
Length = 438
Score = 134 bits (338), Expect = 2e-31
Identities = 76/158 (48%), Positives = 97/158 (61%), Gaps = 5/158 (3%)
Query: 4 SSSSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSL 63
SSSSSS K+ VFLSFRG DTR F LYKAL KGI TF DD EL+RG I P L
Sbjct: 27 SSSSSSSFTPQKFDVFLSFRGKDTRKNFVSFLYKALVSKGIRTFKDDEELERGRPIPPEL 86
Query: 64 DNAIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGS 123
AI+ SRI + V S Y +SS+CL+EL I+ L K V+P+F+ ++PS VR G
Sbjct: 87 RQAIKGSRIAVVVVSVTYPASSWCLEELREILKLEKLGLLTVIPIFYEINPSDVRRQSGV 146
Query: 124 YGEALAKHEERFQHNTDHMERLQKWKIALTQAANLSGD 161
+ KHE+R ER++ W+ ALT+ A+LSG+
Sbjct: 147 VSKQFKKHEKR-----QSRERVKSWREALTKLASLSGE 179
>At1g72920 hypothetical protein
Length = 275
Score = 134 bits (337), Expect = 3e-31
Identities = 71/147 (48%), Positives = 90/147 (60%), Gaps = 4/147 (2%)
Query: 15 KYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIFI 74
KY VFLSFRG DTR F LYK L + I TF DD EL+ G I+P L AIEES+ +
Sbjct: 8 KYDVFLSFRGLDTRRNFISFLYKELVRRKIRTFKDDKELENGRRISPELKRAIEESKFAV 67
Query: 75 PVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEER 134
V S NYA+S +CLDELV I+ + V+P+F+GVDP H+R G E KHE R
Sbjct: 68 VVVSVNYAASPWCLDELVKIMDFENKGSITVMPIFYGVDPCHLRRQIGDVAEQFKKHEAR 127
Query: 135 FQHNTDHMERLQKWKIALTQAANLSGD 161
DH E++ W+ ALT A++SG+
Sbjct: 128 ---EEDH-EKVASWRRALTSLASISGE 150
>At1g72910 unknown protein
Length = 380
Score = 134 bits (336), Expect = 3e-31
Identities = 77/162 (47%), Positives = 94/162 (57%), Gaps = 6/162 (3%)
Query: 15 KYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIFI 74
KY VFLSFRG DTR F LYK L + I TF DD EL+ G I+ L IE SR +
Sbjct: 8 KYDVFLSFRGHDTRQNFISFLYKELVRRSIRTFKDDKELENGQRISSELKRTIEVSRFAV 67
Query: 75 PVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEER 134
V S YA+SS+CLDELV I+ K+ V+P+F+GV+P+HVR G E KH R
Sbjct: 68 VVVSETYAASSWCLDELVTIMDFEKKGSITVMPIFYGVEPNHVRWQTGVLAEQFKKHGSR 127
Query: 135 FQHNTDHMERLQKWKIALTQAANLSGDHRSPGYEYKLTGKIA 176
H E++ KW+ ALT A LSGD S + KL KIA
Sbjct: 128 EDH-----EKVLKWRQALTNFAQLSGD-CSGDDDSKLVDKIA 163
>At1g72840 hypothetical protein
Length = 1183
Score = 134 bits (336), Expect = 3e-31
Identities = 72/160 (45%), Positives = 102/160 (63%), Gaps = 4/160 (2%)
Query: 3 SSSSSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPS 62
+SSSSSS + + Y VFLSFRG DTR +LY AL + G+ TF DD +L+ GD I
Sbjct: 2 ASSSSSSATRLRHYDVFLSFRGVDTRQTIVSHLYVALRNNGVLTFKDDRKLEIGDTIADG 61
Query: 63 LDNAIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRG 122
L AI+ S + + S NYA+S++CL+EL I+ L+ + VLP+F+GV PS VR+ G
Sbjct: 62 LVKAIQTSWFAVVILSENYATSTWCLEELRLIMQLHSEEQIKVLPIFYGVKPSDVRYQEG 121
Query: 123 SYGEALAKHEERFQHNTDHMERLQKWKIALTQAANLSGDH 162
S+ A +R++ + + E++ KW+ ALTQ ANLSG H
Sbjct: 122 SFATAF----QRYEADPEMEEKVSKWRRALTQVANLSGKH 157
>At1g72950 hypothetical protein
Length = 379
Score = 133 bits (335), Expect = 4e-31
Identities = 73/155 (47%), Positives = 91/155 (58%), Gaps = 5/155 (3%)
Query: 7 SSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNA 66
S S + + K+ VFLSFRG DTR F LYK L + I TF DD EL+ G ITP L A
Sbjct: 2 SDSSNTLPKFDVFLSFRGLDTRRSFISFLYKELIRRNIRTFKDDKELKNGRRITPELIRA 61
Query: 67 IEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGE 126
IE SR + V S NYA+S +CL+ELV I+ V+P+F+GVDP HVR G E
Sbjct: 62 IEGSRFAVVVVSVNYAASRWCLEELVKIMDFENMGSLKVMPIFYGVDPCHVRRQIGEVAE 121
Query: 127 ALAKHEERFQHNTDHMERLQKWKIALTQAANLSGD 161
KHE R H E++ W+ ALT A++SGD
Sbjct: 122 QFKKHEGREDH-----EKVLSWRQALTNLASISGD 151
>At5g17680 disease resistance protein RPP1-WsB - like protein
Length = 1294
Score = 130 bits (327), Expect = 4e-30
Identities = 82/192 (42%), Positives = 112/192 (57%), Gaps = 14/192 (7%)
Query: 4 SSSSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSL 63
SSSSSS S V+K VF+SFRG D R F +L+ GI F DD +LQRG I+P L
Sbjct: 6 SSSSSSSSTVWKTDVFVSFRGEDVRKTFVSHLFCEFDRMGIKAFRDDLDLQRGKSISPEL 65
Query: 64 DNAIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGS 123
+AI+ SR I V S NYA+SS+CLDEL+ I+ K ++P+F+ VDPS VR RGS
Sbjct: 66 IDAIKGSRFAIVVVSRNYAASSWCLDELLKIMECNKDT---IVPIFYEVDPSDVRRQRGS 122
Query: 124 YGEALAKHEERFQHNTDHMERLQKWKIALTQAANLSGDHRSPGYEYKLTGKIAFNQTPDL 183
+GE + H ++ E++ KWK AL + A +SG+ + KL KI + + L
Sbjct: 123 FGEDVESHSDK--------EKVGKWKEALKKLAAISGEDSRNWDDSKLIKKIVKDISDKL 174
Query: 184 SS---DCSQSLV 192
S D S+ L+
Sbjct: 175 VSTSWDDSKGLI 186
Score = 34.3 bits (77), Expect = 0.36
Identities = 14/28 (50%), Positives = 18/28 (64%)
Query: 585 HFSNGLKYLPNSLRVLKWKGCLLESLSS 612
H NGL YLP LR L+W G L+++ S
Sbjct: 570 HLPNGLSYLPRKLRYLRWDGYPLKTMPS 597
>At1g56540 disease resistance protein, putative
Length = 1096
Score = 126 bits (316), Expect = 7e-29
Identities = 83/199 (41%), Positives = 108/199 (53%), Gaps = 13/199 (6%)
Query: 3 SSSSSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPS 62
+SSSSSS ++Y VF SF G D R F +L K GI T DD ++R I P+
Sbjct: 2 ASSSSSSSRRTWRYNVFTSFHGPDVRKTFLSHLRKQFNYNGI-TMFDDQRIERSQIIAPA 60
Query: 63 LDNAIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRG 122
L AI ESRI I + S NYASSS+CLDEL+ I+ +Q G++V+ VF+GV PS VR G
Sbjct: 61 LTEAIRESRIAIVLLSKNYASSSWCLDELLEILDCKEQLGQIVMTVFYGVHPSDVRKQTG 120
Query: 123 SYGEALAKHEERFQHNTDHMERLQKWKIALTQAANLSGDH-RSPGYEYKLTGKIA----- 176
+G A E T+ E+ QKW ALT N++G+H ++ E K+ KIA
Sbjct: 121 DFGIAF---NETCARKTE--EQRQKWSQALTYVGNIAGEHFQNWDNEAKMIEKIASDVSD 175
Query: 177 -FNQTPDLSSDCSQSLVEH 194
N TP D L H
Sbjct: 176 KLNTTPSRDFDGMIGLEAH 194
>At1g17610 disease resistance protein, putative
Length = 836
Score = 125 bits (315), Expect = 9e-29
Identities = 70/157 (44%), Positives = 95/157 (59%), Gaps = 5/157 (3%)
Query: 4 SSSSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSL 63
SSSSSS + K+ VFLSFRG DTR F LYK L + GI TF DD ELQ G I L
Sbjct: 459 SSSSSSSATPQKFDVFLSFRGKDTRRTFISFLYKELIEMGIRTFKDDVELQSGRRIASDL 518
Query: 64 DNAIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGS 123
AIE S+I + + S NY++S +CL ELV I+ + K+ +V+P+F+ V+P+HVR
Sbjct: 519 LAAIENSKIAVVIISKNYSASPWCLQELVMIMDVEKKGSIIVMPIFYNVEPAHVRRQIEQ 578
Query: 124 YGEALAKHEERFQHNTDHMERLQKWKIALTQAANLSG 160
+ KHE R ++ E + W+ ALT A++SG
Sbjct: 579 VAKQFRKHEGR-----ENYETVVSWRQALTNLASISG 610
Score = 36.2 bits (82), Expect = 0.094
Identities = 40/154 (25%), Positives = 64/154 (40%), Gaps = 27/154 (17%)
Query: 13 VFKYQVFLSFRGS---DTRYGFTGNLYKALTDKGIHTFIDDSELQRGDE-ITPSLDNAIE 68
V K VFLSF G D +G+ L+ GI F +S + + I A+
Sbjct: 20 VTKLDVFLSFSGKIALDVDFGYD------LSRNGIKAFKSESWKESSFKPIDLRTLEALT 73
Query: 69 ESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEAL 128
ES++ + + S SS L+EL+ II ++ V+PVF P V
Sbjct: 74 ESKVAVVMTSDEEVSSVGFLEELIVIIEFQEKRSLTVIPVFLTKHPLDVE---------- 123
Query: 129 AKHEERFQHNTDHMERLQKWKIALTQAANLSGDH 162
K + F ER + W+ A+ + N++ +
Sbjct: 124 -KVSQIFP------ERAKIWRTAIAKLDNIAAQY 150
>At1g64070 disease resistance protein, putative
Length = 1134
Score = 125 bits (314), Expect = 1e-28
Identities = 71/159 (44%), Positives = 94/159 (58%), Gaps = 6/159 (3%)
Query: 4 SSSSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSL 63
+SSSSS S ++Y+VF SF GSD R F + K + GI T DD + RG+ I+P+L
Sbjct: 192 ASSSSSASRTWRYRVFTSFHGSDVRTSFLSHFRKQFNNNGI-TMFDDQRILRGETISPAL 250
Query: 64 DNAIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGS 123
AI ESRI I + S NYASS +CLDEL+ I+ G++V+ VF+GVDPS VR G
Sbjct: 251 TQAIRESRISIVLLSKNYASSGWCLDELLEILKCKDDMGQIVMTVFYGVDPSDVRKQTGE 310
Query: 124 YGEALAKHEERFQHNTDHMERLQKWKIALTQAANLSGDH 162
+G A E T+ E QKW AL N++G+H
Sbjct: 311 FGIAF---NETCACRTE--EERQKWSQALNYVGNIAGEH 344
>At5g41550 disease resistance protein-like
Length = 1085
Score = 124 bits (311), Expect = 3e-28
Identities = 69/156 (44%), Positives = 92/156 (58%), Gaps = 6/156 (3%)
Query: 7 SSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNA 66
SSS S + +Y VF SF G D R GF +L+ KGI TF D E+++G+ I P L NA
Sbjct: 4 SSSSSNIRRYHVFPSFHGPDVRKGFLSHLHYHFASKGITTF-KDQEIEKGNTIGPELVNA 62
Query: 67 IEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGE 126
I ESR+ I + S YASSS+CLDELV I+ + G++V+ +F+ VDPS VR +G +G
Sbjct: 63 IRESRVSIVLLSKKYASSSWCLDELVEILKCKEDQGQIVMTIFYDVDPSSVRKQKGDFGS 122
Query: 127 ALAKHEERFQHNTDHMERLQKWKIALTQAANLSGDH 162
K E E Q+W ALT AN+ G+H
Sbjct: 123 TFMKTCEGKSE-----EVKQRWTKALTHVANIKGEH 153
>At4g12010 like disease resistance protein (TMV N-like)
Length = 1219
Score = 123 bits (309), Expect = 4e-28
Identities = 85/197 (43%), Positives = 113/197 (57%), Gaps = 14/197 (7%)
Query: 1 MQSSSSSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEIT 60
M+SSS SS+ ++ VFLSFRG DTR FTG+L KAL +GI +FIDD L+RGD +T
Sbjct: 1 MESSSPSSA-----EFDVFLSFRGFDTRNNFTGHLQKALRLRGIDSFIDD-RLRRGDNLT 54
Query: 61 PSLDNAIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHH 120
D IE+S+I I VFS NYA+S++CL ELV I+ N +LV+P+F+ VD S V
Sbjct: 55 ALFDR-IEKSKIAIIVFSTNYANSAWCLRELVKILECRNSNQQLVVPIFYKVDKSDVEKQ 113
Query: 121 RGSYGEALAKHEERFQHNTDHMERLQKWKIALTQAANLSG--DHRSPGYEYKLTGKIA-- 176
R S+ E F T E + WK AL A+N+ G E KL +IA
Sbjct: 114 RNSFAVPFKLPELTFPGVTP--EEISSWKAALASASNILGYVVKEISTSEAKLVDEIAVD 171
Query: 177 -FNQTPDLSSDCSQSLV 192
F + DL+ ++ LV
Sbjct: 172 TFKKLNDLAPSGNEGLV 188
Score = 30.8 bits (68), Expect = 3.9
Identities = 20/54 (37%), Positives = 25/54 (46%), Gaps = 12/54 (22%)
Query: 569 AFKKMTKLKTLIIENGHFSNG------------LKYLPNSLRVLKWKGCLLESL 610
AF+ M LK L I + H S G L +LPN L L W G L+S+
Sbjct: 560 AFQGMYNLKYLKIYDSHCSRGCEAEFKLHLRRGLSFLPNELTYLHWHGYPLQSI 613
>At5g48770 disease resistance protein
Length = 1190
Score = 123 bits (308), Expect = 6e-28
Identities = 73/157 (46%), Positives = 93/157 (58%), Gaps = 7/157 (4%)
Query: 4 SSSSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSL 63
+SSS S Y VFLSFRG DTR +LY AL KGI TF DD +L+ GD I+ L
Sbjct: 2 ASSSLSSPPPCNYDVFLSFRGEDTRRTIVSHLYAALGAKGIITFKDDQDLEVGDHISSHL 61
Query: 64 DNAIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGS 123
AIE S+ + V S Y +S +CL EL I+ LY VLP+F+ VDPS VRH RGS
Sbjct: 62 RRAIEGSKFAVVVLSERYTTSRWCLMELQLIMELYNLGKLKVLPLFYEVDPSDVRHQRGS 121
Query: 124 YGEALAKHEERFQHNTDHMERLQKWKIALTQAANLSG 160
+G ER+Q + + +Q+W++AL ANLSG
Sbjct: 122 FG------LERYQ-GPEFADIVQRWRVALCMVANLSG 151
>At1g27170 disease resistance protein, putative
Length = 1560
Score = 123 bits (308), Expect = 6e-28
Identities = 65/149 (43%), Positives = 92/149 (61%), Gaps = 4/149 (2%)
Query: 12 YVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESR 71
Y KY VFLSFRG+DTR F +LYKAL DK + F D+ ++RGDEI+ SL +E+S
Sbjct: 157 YRLKYDVFLSFRGADTRDNFGDHLYKALKDK-VRVFRDNEGMERGDEISSSLKAGMEDSA 215
Query: 72 IFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKH 131
+ V S NY+ S +CLDEL + + R +LP+F+ VDPSHVR + +H
Sbjct: 216 ASVIVISRNYSGSRWCLDELAMLCKMKSSLDRRILPIFYHVDPSHVRKQSDHIKKDFEEH 275
Query: 132 EERFQHNTDHMERLQKWKIALTQAANLSG 160
+ RF ++ E++Q+W+ ALT NL+G
Sbjct: 276 QVRF---SEEKEKVQEWREALTLVGNLAG 301
Score = 94.7 bits (234), Expect = 2e-19
Identities = 55/157 (35%), Positives = 85/157 (54%), Gaps = 12/157 (7%)
Query: 15 KYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPSLDNAIEESRIFI 74
++ VFLSF+ D R+ FT LY+ L + + + +D + E+ SL A+E+S +
Sbjct: 15 EWDVFLSFQ-RDARHKFTERLYEVLVKEQVRVWNNDDVERGNHELGASLVEAMEDSVALV 73
Query: 75 PVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRGSYGEALAKHEER 134
V S NYA S +CL+EL + L GRLVLP+F+ V+P +R G Y +H +R
Sbjct: 74 VVLSPNYAKSHWCLEELAMLCDLKSSLGRLVLPIFYEVEPCMLRKQNGPYEMDFEEHSKR 133
Query: 135 FQHNTDHMERLQKWKIALTQAANLSGDHRSPGYEYKL 171
F E++Q+W+ AL N+ PG+ Y+L
Sbjct: 134 FSE-----EKIQRWRRALNIIGNI------PGFVYRL 159
Score = 45.8 bits (107), Expect = 1e-04
Identities = 22/53 (41%), Positives = 31/53 (57%)
Query: 569 AFKKMTKLKTLIIENGHFSNGLKYLPNSLRVLKWKGCLLESLSSSILSKASEV 621
+F MTKL+ L I N LK LP+ L+ ++WKGC LE+L L++ V
Sbjct: 744 SFAPMTKLRLLQINNVELEGNLKLLPSELKWIQWKGCPLENLPPDFLARQLSV 796
>At5g58120 resistance protein - like
Length = 1046
Score = 122 bits (307), Expect = 8e-28
Identities = 84/214 (39%), Positives = 117/214 (54%), Gaps = 16/214 (7%)
Query: 3 SSSSSSSISYVFKYQVFLSFRGSDTRYGFTGNLYKALTDKGIHTFIDDSELQRGDEITPS 62
+SSSSSS S ++Y+VF +F G D R F +L K + GI F D S ++R I P+
Sbjct: 2 ASSSSSSRSRTWRYRVFTNFHGPDVRKTFLSHLRKQFSYNGISMFNDQS-IERSQTIVPA 60
Query: 63 LDNAIEESRIFIPVFSANYASSSFCLDELVHIIHLYKQNGRLVLPVFFGVDPSHVRHHRG 122
L AI+ESRI I V S NYASS +CLDEL+ I+ + G++V+ VF+GVDPS VR G
Sbjct: 61 LTGAIKESRISIVVLSKNYASSRWCLDELLEILKCREDIGQIVMTVFYGVDPSDVRKQTG 120
Query: 123 SYGEALAKHEERFQHNTDHMERLQKWKIALTQAANLSGDH-RSPGYEYKLTGKIAFNQTP 181
+G A K E + E QKW AL N++G+H + E K+ KIA + +
Sbjct: 121 EFGIAFNKTCEGKTN-----EETQKWSKALNDVGNIAGEHFFNWDNEAKMIEKIARDVSN 175
Query: 182 DLSSDCSQSLVEHEQI*I*FYWRHSQIHLQQDQS 215
L++ S + I + HLQ+ QS
Sbjct: 176 KLNATISWDFEDMVGI---------EAHLQKMQS 200
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.355 0.155 0.549
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,557,084
Number of Sequences: 26719
Number of extensions: 697464
Number of successful extensions: 4425
Number of sequences better than 10.0: 135
Number of HSP's better than 10.0 without gapping: 125
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 3955
Number of HSP's gapped (non-prelim): 258
length of query: 984
length of database: 11,318,596
effective HSP length: 109
effective length of query: 875
effective length of database: 8,406,225
effective search space: 7355446875
effective search space used: 7355446875
T: 11
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.6 bits)
S2: 65 (29.6 bits)
Medicago: description of AC123574.2


