

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123573.8 + phase: 0 /pseudo
(548 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
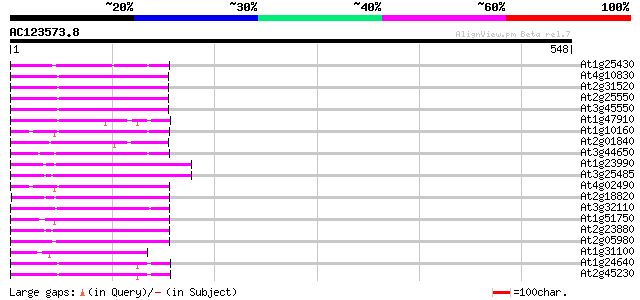
Score E
Sequences producing significant alignments: (bits) Value
At1g25430 hypothetical protein 80 3e-15
At4g10830 putative protein 80 4e-15
At2g31520 putative non-LTR retroelement reverse transcriptase 77 2e-14
At2g25550 putative non-LTR retroelement reverse transcriptase 77 2e-14
At3g45550 putative protein 76 5e-14
At1g47910 reverse transcriptase, putative 75 9e-14
At1g10160 putative reverse transcriptase 75 9e-14
At2g01840 putative non-LTR retroelement reverse transcriptase 74 2e-13
At3g44650 putative protein 73 4e-13
At1g23990 reverse transcriptase, putative 72 1e-12
At3g25485 unknown protein 71 2e-12
At4g02490 70 2e-12
At2g18820 putative non-LTR retroelement reverse transcriptase 70 2e-12
At3g32110 non-LTR reverse transcriptase, putative 70 3e-12
At1g51750 hypothetical protein 69 9e-12
At2g23880 putative non-LTR retroelement reverse transcriptase 68 1e-11
At2g05980 putative non-LTR retroelement reverse transcriptase 67 3e-11
At1g31100 hypothetical protein 66 4e-11
At1g24640 hypothetical protein 66 4e-11
At2g45230 putative non-LTR retroelement reverse transcriptase 65 7e-11
>At1g25430 hypothetical protein
Length = 1213
Score = 80.1 bits (196), Expect = 3e-15
Identities = 57/157 (36%), Positives = 84/157 (53%), Gaps = 5/157 (3%)
Query: 1 RGLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGINEPIVMSHLQFADDTLIVG 60
+GLRQGDP SP+LF+LA E F L+ S + + N + +SHL FADD +I
Sbjct: 648 KGLRQGDPLSPYLFVLAMEAFSNLLHSRYESGLIHYHPKASN--LSISHLMFADDVMIFF 705
Query: 61 EKS*ANVTGMRAALLLFETMSGLKVNFSKSLLVSVNVVGLWLSMVARVLNCRVGSLPFVY 120
+ ++ G+ L F + SGLKVN KS L + L S +G+LP Y
Sbjct: 706 DGGSFSLHGICETLDDFASWSGLKVNKDKSHLYLAGLNQL-ESNANAAYGFPIGTLPIRY 764
Query: 121 LGM-LIGGNVRCLSF*EPIIDRIKSRLSGWKSKHLSF 156
LG+ L+ +R + EP++++I +R W +K LSF
Sbjct: 765 LGLPLMNRKLRIAEY-EPLLEKITARFRSWVNKCLSF 800
>At4g10830 putative protein
Length = 1294
Score = 79.7 bits (195), Expect = 4e-15
Identities = 52/156 (33%), Positives = 82/156 (52%), Gaps = 2/156 (1%)
Query: 1 RGLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGINEPIVMSHLQFADDTLIVG 60
RG+RQGDP SP+LF+L A+ + L+++ G +IG P V +HLQFADD+L
Sbjct: 993 RGIRQGDPLSPYLFILCADILNHLIKNRVAEGDIRGIRIGNGVPGV-THLQFADDSLFFC 1051
Query: 61 EKS*ANVTGMRAALLLFETMSGLKVNFSKSLLV-SVNVVGLWLSMVARVLNCRVGSLPFV 119
+ + N ++ ++E SG K+N SKS++ V G + + +L +
Sbjct: 1052 QSNVRNCQALKDVFDVYEYYSGQKINMSKSMITFGSRVHGTTQNRLKNILGIQSHGGGGK 1111
Query: 120 YLGMLIGGNVRCLSF*EPIIDRIKSRLSGWKSKHLS 155
YLG+ + II+R+K R S W +K+LS
Sbjct: 1112 YLGLPEQFGRKKRDMFNYIIERVKKRTSSWSAKYLS 1147
>At2g31520 putative non-LTR retroelement reverse transcriptase
Length = 1524
Score = 77.0 bits (188), Expect = 2e-14
Identities = 51/156 (32%), Positives = 79/156 (49%), Gaps = 2/156 (1%)
Query: 1 RGLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGINEPIVMSHLQFADDTLIVG 60
RG+RQGDP SP+LF+L + L+ + + G +IG P + +HLQFADD+L
Sbjct: 787 RGIRQGDPLSPYLFILCGDILSHLINGRASSGDLRGVRIGNGAPAI-THLQFADDSLFFC 845
Query: 61 EKS*ANVTGMRAALLLFETMSGLKVNFSKSLLV-SVNVVGLWLSMVARVLNCRVGSLPFV 119
+ + N ++ ++E SG K+N KS++ V G S + ++L
Sbjct: 846 QANVRNCQALKDVFDVYEYYSGQKINVQKSMITFGSRVYGSTQSKLKQILEIPNQGGGGK 905
Query: 120 YLGMLIGGNVRCLSF*EPIIDRIKSRLSGWKSKHLS 155
YLG+ + E IIDR+K R S W ++ LS
Sbjct: 906 YLGLPEQFGRKKKEMFEYIIDRVKKRTSTWSARFLS 941
>At2g25550 putative non-LTR retroelement reverse transcriptase
Length = 1750
Score = 77.0 bits (188), Expect = 2e-14
Identities = 51/156 (32%), Positives = 79/156 (49%), Gaps = 2/156 (1%)
Query: 1 RGLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGINEPIVMSHLQFADDTLIVG 60
RG+RQGDP SP+LF+L + L+ + + G +IG P + +HLQFADD+L
Sbjct: 1013 RGIRQGDPLSPYLFILCGDILSHLINGRASSGDLRGVRIGNGAPAI-THLQFADDSLFFC 1071
Query: 61 EKS*ANVTGMRAALLLFETMSGLKVNFSKSLLV-SVNVVGLWLSMVARVLNCRVGSLPFV 119
+ + N ++ ++E SG K+N KS++ V G S + ++L
Sbjct: 1072 QANVRNCQALKDVFDVYEYYSGQKINVQKSMITFGSRVYGSTQSRLKQILEIPNQGGGGK 1131
Query: 120 YLGMLIGGNVRCLSF*EPIIDRIKSRLSGWKSKHLS 155
YLG+ + E IIDR+K R S W ++ LS
Sbjct: 1132 YLGLPEQFGRKKKEMFEYIIDRVKKRTSTWSARFLS 1167
>At3g45550 putative protein
Length = 851
Score = 75.9 bits (185), Expect = 5e-14
Identities = 51/156 (32%), Positives = 79/156 (49%), Gaps = 2/156 (1%)
Query: 1 RGLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGINEPIVMSHLQFADDTLIVG 60
RG+RQGDP SP+LF+L + L++ + + G +IG P + +HLQFADD+L
Sbjct: 252 RGIRQGDPLSPYLFILCGDILSHLIKVKASSGDIRGVRIGNGAPAI-THLQFADDSLFFC 310
Query: 61 EKS*ANVTGMRAALLLFETMSGLKVNFSKSLLV-SVNVVGLWLSMVARVLNCRVGSLPFV 119
+ + N ++ ++E SG K+N KSL+ V G + + +LN
Sbjct: 311 QANVRNCQALKDVFDVYEYYSGQKINVQKSLITFGSRVYGSTQTRLKTLLNIPNQGGGGK 370
Query: 120 YLGMLIGGNVRCLSF*EPIIDRIKSRLSGWKSKHLS 155
YLG+ + IIDR+K R + W +K LS
Sbjct: 371 YLGLPEQFGRKKKEMFNYIIDRVKERTASWSAKFLS 406
>At1g47910 reverse transcriptase, putative
Length = 1142
Score = 75.1 bits (183), Expect = 9e-14
Identities = 56/165 (33%), Positives = 86/165 (51%), Gaps = 16/165 (9%)
Query: 1 RGLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGINEPIVMSHLQFADDTLIVG 60
RGLRQGDP SP+LF+L E + N TG ++ P V SHL FADD+L
Sbjct: 415 RGLRQGDPLSPYLFILCTEVLIANIRKAERQNLITGIKVATPSPAV-SHLLFADDSLFFC 473
Query: 61 EKS*ANVTGMRAALLLFETMSGLKVNFSKSLL----VSVNVVGLWLSMVARVLNC-RVGS 115
+ + + L +E++SG ++NFSKS + + + + ++ + N +GS
Sbjct: 474 KANKEQCGIILEILKQYESVSGQQINFSKSSIQFGHKVEDSIKADIKLILGIHNLGGMGS 533
Query: 116 LPFVYLGM---LIGGNVRCLSF*EPIIDRIKSRLSGWKSKHLSFG 157
YLG+ L G + SF + DR++SR++GW +K LS G
Sbjct: 534 ----YLGLPESLGGSKTKVFSF---VRDRLQSRINGWSAKFLSKG 571
>At1g10160 putative reverse transcriptase
Length = 1118
Score = 75.1 bits (183), Expect = 9e-14
Identities = 55/159 (34%), Positives = 86/159 (53%), Gaps = 8/159 (5%)
Query: 1 RGLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGIN--EPIVMSHLQFADDTLI 58
+G+RQGDP S LF+L + VL +SL + + + N PI+ +HL FADD L+
Sbjct: 648 KGIRQGDPMSSHLFVLVMD---VLSKSLDLGALNGLFNLHPNCLAPII-THLSFADDVLV 703
Query: 59 VGEKS*ANVTGMRAALLLFETMSGLKVNFSKSLLVSVNVVGLWLSMVARVLNCRVGSLPF 118
+ + +++ G+ L F SGL +N K+ L+ +A L GSLP
Sbjct: 704 FSDGAASSIAGILTILDDFRQGSGLGINREKTELLLDGGNFARNRSLADNLGITHGSLPV 763
Query: 119 VYLGM-LIGGNVRCLSF*EPIIDRIKSRLSGWKSKHLSF 156
YLG+ L+ +R + +P++DRI SR + W ++HLSF
Sbjct: 764 RYLGVPLMSQKMRRQDY-QPLVDRINSRFTSWTARHLSF 801
>At2g01840 putative non-LTR retroelement reverse transcriptase
Length = 1715
Score = 73.9 bits (180), Expect = 2e-13
Identities = 51/159 (32%), Positives = 77/159 (48%), Gaps = 8/159 (5%)
Query: 1 RGLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGINEPIVMSHLQFADDTLIVG 60
RG+RQGDP SP+LFL AE ++ VN G +I + + +SHL FADD+L
Sbjct: 993 RGIRQGDPLSPYLFLFCAEVLSNMLRKAEVNKQIHGMKI-TKDCLAISHLLFADDSLFFC 1051
Query: 61 EKS*ANVTGMRAALLLFETMSGLKVNFSKSLLVSVNVVGLW----LSMVARVLNCRVGSL 116
S N+ + +E SG K+N++KS ++ + L + + N R G
Sbjct: 1052 RASNQNIEQLALIFKKYEEASGQKINYAKSSIIFGQKIPTMRRQRLHRLLGIDNVRGGG- 1110
Query: 117 PFVYLGMLIGGNVRCLSF*EPIIDRIKSRLSGWKSKHLS 155
YLG+ R + E I+ ++K R GW +LS
Sbjct: 1111 --KYLGLPEQLGRRKVELFEYIVTKVKERTEGWAYNYLS 1147
>At3g44650 putative protein
Length = 762
Score = 73.2 bits (178), Expect = 4e-13
Identities = 48/157 (30%), Positives = 81/157 (51%), Gaps = 4/157 (2%)
Query: 1 RGLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGINEPIVMSHLQFADDTLIVG 60
RGLRQG SP+LF++ + L++ + V GY + ++HL FADD +I+
Sbjct: 207 RGLRQGCALSPYLFVICMDVLSKLLDKV-VGIGRIGYHPHCKR-MGLTHLSFADDLMILT 264
Query: 61 EKS*ANVTGMRAALLLFETMSGLKVNFSKSLLVSVNVVGLWLSMVARVLNCRVGSLPFVY 120
+ ++ G+ LF SGLK++ KS + S + + + VG LP Y
Sbjct: 265 DGQCRSIEGIIEVFDLFSKWSGLKISMEKSTIFSAGLSSTSRAQLHTHFPFEVGELPIRY 324
Query: 121 LGM-LIGGNVRCLSF*EPIIDRIKSRLSGWKSKHLSF 156
LG+ L+ + + + P+I++I+ R+ W S+ LSF
Sbjct: 325 LGLPLVTKRLSSVDY-APLIEQIRKRIGSWSSRFLSF 360
>At1g23990 reverse transcriptase, putative
Length = 657
Score = 71.6 bits (174), Expect = 1e-12
Identities = 50/179 (27%), Positives = 88/179 (48%), Gaps = 5/179 (2%)
Query: 1 RGLRQGDPFSPFLFLLAAEGFHVLME-SLSVNNFYTGYQIGINEPIVMSHLQFADDTLIV 59
RGLRQG SP+LF+++ + L++ + S F GY E + ++HL FADD +++
Sbjct: 103 RGLRQGCSLSPYLFVMSMDVLSKLLDQAASAKKF--GYHSRCKE-LSLTHLSFADDLMVL 159
Query: 60 GEKS*ANVTGMRAALLLFETMSGLKVNFSKSLLVSVNVVGLWLSMVARVLNCRVGSLPFV 119
+ ++ G+ +F SGLK++ KS + V + VG LP
Sbjct: 160 SDGKVRSIDGIVEVFDIFAKFSGLKISMEKSTIYLAGVTEDVYHEIQNRYQFDVGQLPVR 219
Query: 120 YLGMLIGGNVRCLSF*EPIIDRIKSRLSGWKSKHLSF-GVV*FC*SMSCHLCLFMLFPF 177
YLG+ + + P+++ IK ++ W +++LS+ G + S+ +C F L F
Sbjct: 220 YLGLPLVTKRLTATDYSPLLEHIKKKIGTWTTRYLSYAGRLNLITSVLWSICNFWLAAF 278
>At3g25485 unknown protein
Length = 979
Score = 70.9 bits (172), Expect = 2e-12
Identities = 48/178 (26%), Positives = 90/178 (49%), Gaps = 3/178 (1%)
Query: 1 RGLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGINEPIVMSHLQFADDTLIVG 60
RGLRQG SP+LF++A + +++ + + GY + I ++HL FADD +++
Sbjct: 570 RGLRQGCSLSPYLFVIAMDVLSKMLDRAAGFKKF-GYHPRC-QTIGLTHLTFADDLMVLS 627
Query: 61 EKS*ANVTGMRAALLLFETMSGLKVNFSKSLLVSVNVVGLWLSMVARVLNCRVGSLPFVY 120
+ +V G+ + F SGLK++ KS + ++ + VGSLP Y
Sbjct: 628 DGKVRSVEGIVSVFDEFAKKSGLKISMEKSTIYLAGSANMYRREIEERFKFAVGSLPVRY 687
Query: 121 LGMLIGGNVRCLSF*EPIIDRIKSRLSGWKSKHLSF-GVV*FC*SMSCHLCLFMLFPF 177
LG+ + + P++++IK ++ W +++LS+ G + S+ +C F + F
Sbjct: 688 LGLPLVTKRFSSADYRPLVEQIKRKIGTWTARYLSYAGRLNLISSVIWSICNFWIAAF 745
>At4g02490
Length = 657
Score = 70.5 bits (171), Expect = 2e-12
Identities = 50/158 (31%), Positives = 82/158 (51%), Gaps = 6/158 (3%)
Query: 1 RGLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGIN--EPIVMSHLQFADDTLI 58
+G+RQGDP S LF+L + +L SL + + + P++ +HL FADD L+
Sbjct: 195 KGIRQGDPMSSHLFVLVMD---ILARSLDLGAVEGRFVLHPKCLAPMI-THLSFADDILV 250
Query: 59 VGEKS*ANVTGMRAALLLFETMSGLKVNFSKSLLVSVNVVGLWLSMVARVLNCRVGSLPF 118
+ S +++ + L +F+ SGL +N K+ L+ ++A L GSLP
Sbjct: 251 FCDGSLSSLVAILDILDVFKKGSGLGINLQKTALLLDGGNFERNRIMAASLGVSQGSLPV 310
Query: 119 VYLGMLIGGNVRCLSF*EPIIDRIKSRLSGWKSKHLSF 156
YLG+ + +P++DRI SR + W ++HLSF
Sbjct: 311 RYLGVPLMSQKMKKHDYQPLVDRINSRFTSWTARHLSF 348
>At2g18820 putative non-LTR retroelement reverse transcriptase
Length = 1412
Score = 70.5 bits (171), Expect = 2e-12
Identities = 45/155 (29%), Positives = 79/155 (50%), Gaps = 2/155 (1%)
Query: 2 GLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGINEPIVMSHLQFADDTLIVGE 61
GLRQG SP+LF++ +++ +V + GY + ++HL FADD ++
Sbjct: 910 GLRQGCSLSPYLFVICMNVLSAMLDKGAVEKRF-GYHPRCRN-MGLTHLCFADDIMVFSA 967
Query: 62 KS*ANVTGMRAALLLFETMSGLKVNFSKSLLVSVNVVGLWLSMVARVLNCRVGSLPFVYL 121
S ++ G+ A F SGL ++ KS L ++ + + GSLP YL
Sbjct: 968 GSAHSLEGVLAIFKDFAAFSGLNISLEKSTLFMASISSETCASILARFPFDSGSLPVRYL 1027
Query: 122 GMLIGGNVRCLSF*EPIIDRIKSRLSGWKSKHLSF 156
G+ + L+ P++++I+SR+S WK++ LS+
Sbjct: 1028 GLPLMTKRMTLADCLPLLEKIRSRISSWKNRFLSY 1062
>At3g32110 non-LTR reverse transcriptase, putative
Length = 1911
Score = 70.1 bits (170), Expect = 3e-12
Identities = 55/158 (34%), Positives = 81/158 (50%), Gaps = 4/158 (2%)
Query: 1 RGLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGINEPIVMSHLQFADDTLIVG 60
RGLRQGDP SP+LF+L E L+ES + +I + P +SH+ FADD ++
Sbjct: 1170 RGLRQGDPLSPYLFVLCIERLCHLIESSIAAKKWKPIKISQSGP-RLSHICFADDLILFA 1228
Query: 61 EKS*ANVTGMRAALLLFETMSGLKVNFSKS-LLVSVNVVGLWLSMVARVLNCRVGSLPFV 119
E S + +R L F SG KV+ KS + S NV+ ++ +
Sbjct: 1229 EASIDQIRVLRGVLEKFCGASGQKVSLEKSKIYFSKNVLRDLGKRISEESGMKATKDLGK 1288
Query: 120 YLGM-LIGGNVRCLSF*EPIIDRIKSRLSGWKSKHLSF 156
YLG+ ++ + +F E +I R SRL+GWK + LSF
Sbjct: 1289 YLGVPILQKRINKETFGE-VIKRFSSRLAGWKGRMLSF 1325
>At1g51750 hypothetical protein
Length = 629
Score = 68.6 bits (166), Expect = 9e-12
Identities = 45/159 (28%), Positives = 82/159 (51%), Gaps = 8/159 (5%)
Query: 1 RGLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGIN---EPIVMSHLQFADDTL 57
RG+RQG SP+LF+++ E +++ + G + G + + + ++HL FADD +
Sbjct: 59 RGIRQGCALSPYLFVISMEVLSKMLDQAA-----GGKRFGFHPKCKNLGLTHLCFADDLM 113
Query: 58 IVGEKS*ANVTGMRAALLLFETMSGLKVNFSKSLLVSVNVVGLWLSMVARVLNCRVGSLP 117
I+ + +V G+ + LF SGL++N K+ L + V M+ +G LP
Sbjct: 114 ILTDGKVRSVDGIVEVMNLFAKRSGLQINMEKTTLYTAGVSDHNRYMMISRYPFGLGQLP 173
Query: 118 FVYLGMLIGGNVRCLSF*EPIIDRIKSRLSGWKSKHLSF 156
YLG+ + P+ ++I++R+ W S++LSF
Sbjct: 174 VRYLGLPLVTKRLTKEDLSPLFEQIRNRIGTWTSRYLSF 212
>At2g23880 putative non-LTR retroelement reverse transcriptase
Length = 1216
Score = 67.8 bits (164), Expect = 1e-11
Identities = 47/156 (30%), Positives = 80/156 (51%), Gaps = 2/156 (1%)
Query: 1 RGLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGINEPIVMSHLQFADDTLIVG 60
RGLRQG SP+LF+++ + +++ + + GY + + ++HL FADD +I+
Sbjct: 375 RGLRQGCSLSPYLFVISMDVLSRMLDKAAGAREF-GYHPRC-KTLGLTHLCFADDLMILT 432
Query: 61 EKS*ANVTGMRAALLLFETMSGLKVNFSKSLLVSVNVVGLWLSMVARVLNCRVGSLPFVY 120
+ +V G+ L F GLK+ K+ L V +++ + VG LP Y
Sbjct: 433 DGKIRSVDGIVKVLNQFAAKLGLKICMEKTTLYLAGVSDHSRQLMSSRYSFGVGKLPVRY 492
Query: 121 LGMLIGGNVRCLSF*EPIIDRIKSRLSGWKSKHLSF 156
LG+ + S P+ID+I+ R+ W S++LSF
Sbjct: 493 LGLPLVTKRLTTSDYSPLIDQIRRRIGMWTSRYLSF 528
>At2g05980 putative non-LTR retroelement reverse transcriptase
Length = 1352
Score = 67.0 bits (162), Expect = 3e-11
Identities = 40/156 (25%), Positives = 80/156 (50%), Gaps = 2/156 (1%)
Query: 1 RGLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGINEPIVMSHLQFADDTLIVG 60
RGLRQG SP+L+++ +++ +V + + N + ++HL FADD ++
Sbjct: 801 RGLRQGCSLSPYLYVICMNVLSCMLDKAAVEKKISYHPRCRN--MNLTHLCFADDIMVFS 858
Query: 61 EKS*ANVTGMRAALLLFETMSGLKVNFSKSLLVSVNVVGLWLSMVARVLNCRVGSLPFVY 120
+ + ++ G A F MS LK++ KS + + + + + +G+LP Y
Sbjct: 859 DGTSKSIQGTLAIFEKFAAMSWLKISLEKSTIFMAGISPNAKTSILQQFPFELGTLPVKY 918
Query: 121 LGMLIGGNVRCLSF*EPIIDRIKSRLSGWKSKHLSF 156
LG+ + S P++++I++R++ W ++ LSF
Sbjct: 919 LGLPLLTKRMTQSDYLPLVEKIRARITSWTNRFLSF 954
>At1g31100 hypothetical protein
Length = 1090
Score = 66.2 bits (160), Expect = 4e-11
Identities = 50/137 (36%), Positives = 69/137 (49%), Gaps = 7/137 (5%)
Query: 1 RGLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGY--QIGINEPIVMSHLQFADDTLI 58
RGLRQG+P SPFLF+LA E F L+ S F GY P+ +SHL FADD ++
Sbjct: 563 RGLRQGNPLSPFLFVLAMEVFSSLLNS----RFQAGYIHYHPKTSPLSISHLMFADDIMV 618
Query: 59 VGEKS*ANVTGMRAALLLFETMSGLKVNFSKSLLVSVNVVGLWLSMVARVLN-CRVGSLP 117
+ +++ G+ AL F SGL +N K+ L + + S +AR L G L
Sbjct: 619 FFDGGSSSLHGISEALEDFAFWSGLVLNREKTHLYLAGLDRIEASTIARKLRIAEYGPLL 678
Query: 118 FVYLGMLIGGNVRCLSF 134
+V+CLSF
Sbjct: 679 EKLAKRFRSWSVKCLSF 695
>At1g24640 hypothetical protein
Length = 1270
Score = 66.2 bits (160), Expect = 4e-11
Identities = 53/161 (32%), Positives = 74/161 (45%), Gaps = 8/161 (4%)
Query: 1 RGLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGINEPIVMSHLQFADDTLIVG 60
RGLRQGDP SPFLF+L EG L+ G Q N P+V HL FADD+L +
Sbjct: 594 RGLRQGDPLSPFLFVLCTEGLTHLLNKAQWEGALEGIQFSENGPMV-HHLLFADDSLFLC 652
Query: 61 EKS*ANVTGMRAALLLFETMSGLKVNFSKSLLVSVNVVGLWLSMVARV-LNCRVGSLPFV 119
+ S ++ L ++ +G +N +KS + V L R L
Sbjct: 653 KASREQSLVLQKILKVYGNATGQTINLNKSSITFGEKVDEQLKGTIRTCLGIFTEGGAGT 712
Query: 120 YLGM---LIGGNVRCLSF*EPIIDRIKSRLSGWKSKHLSFG 157
YLG+ G V L + + DR+K +L W ++ LS G
Sbjct: 713 YLGLPECFSGSKVDMLHY---LKDRLKEKLDVWFTRCLSQG 750
>At2g45230 putative non-LTR retroelement reverse transcriptase
Length = 1374
Score = 65.5 bits (158), Expect = 7e-11
Identities = 50/161 (31%), Positives = 80/161 (49%), Gaps = 8/161 (4%)
Query: 1 RGLRQGDPFSPFLFLLAAEGFHVLMESLSVNNFYTGYQIGINEPIVMSHLQFADDTLIVG 60
RGLRQGDP SP+LF++ E +++S N TG ++ P + SHL FADD++
Sbjct: 631 RGLRQGDPLSPYLFVICTEMLVKMLQSAEQKNQITGLKVARGAPPI-SHLLFADDSMFYC 689
Query: 61 EKS*ANVTGMRAALLLFETMSGLKVNFSK-SLLVSVNVVGLWLSMVARVLNCRVGSLPFV 119
+ + + + + + SG +VN+ K S+ ++ +V R L V
Sbjct: 690 KVNDEALGQIIRIIEEYSLASGQRVNYLKSSIYFGKHISEERRCLVKRKLGIEREGGEGV 749
Query: 120 YLGM---LIGGNVRCLSF*EPIIDRIKSRLSGWKSKHLSFG 157
YLG+ G V LS+ + DR+ ++ GW+S LS G
Sbjct: 750 YLGLPESFQGSKVATLSY---LKDRLGKKVLGWQSNFLSPG 787
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.351 0.160 0.556
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,918,269
Number of Sequences: 26719
Number of extensions: 425443
Number of successful extensions: 1553
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 46
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 1417
Number of HSP's gapped (non-prelim): 63
length of query: 548
length of database: 11,318,596
effective HSP length: 104
effective length of query: 444
effective length of database: 8,539,820
effective search space: 3791680080
effective search space used: 3791680080
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC123573.8


