

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC123571.8 - phase: 0
(522 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
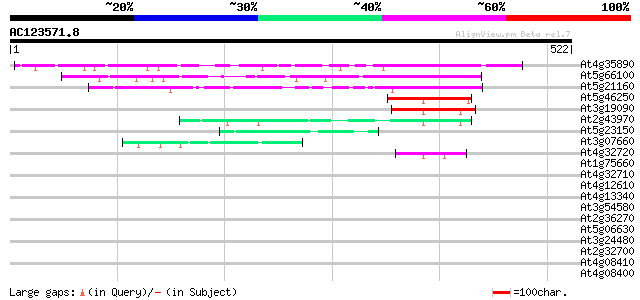
Score E
Sequences producing significant alignments: (bits) Value
At4g35890 unknown protein 235 5e-62
At5g66100 unknown protein 203 2e-52
At5g21160 unknown protein 113 2e-25
At5g46250 unknown protein 62 1e-09
At3g19090 RNA-binding protein, putative 53 5e-07
At2g43970 unknown protein 51 2e-06
At5g23150 transcription factor-like protein (gb|AAD31171.1) 42 6e-04
At3g07660 unknown protein 42 6e-04
At4g32720 putative RNA-binding protein LAH1 42 8e-04
At1g75660 Dhp1-like protein 42 0.001
At4g32710 putative protein kinase 40 0.002
At4g12610 putative protein 39 0.005
At4g13340 extensin-like protein 39 0.009
At3g54580 extensin precursor -like protein 39 0.009
At2g36270 bZip transcription factor AtbZip39 38 0.012
At5g06630 putative protein 37 0.020
At3g24480 disease resistance protein, putative 37 0.027
At2g32700 unknown protein 37 0.027
At4g08410 extensin-like protein 36 0.045
At4g08400 extensin-like protein 36 0.045
>At4g35890 unknown protein
Length = 523
Score = 235 bits (599), Expect = 5e-62
Identities = 176/520 (33%), Positives = 263/520 (49%), Gaps = 99/520 (19%)
Query: 5 GNHSSNHSPDNLQSRRLP------PWNQVVRGESESIAAVPAVSLSEESFPIVTAPVDDS 58
GNH S + QS R P PW Q+VRGESE IAA AV+ S P AP++
Sbjct: 21 GNHGSP-TASVAQSPRRPSRQVSSPWTQIVRGESEPIAAAAAVA--GPSSPQSRAPIEPI 77
Query: 59 TSAEVISDNA----------DNGGERNGG---TGKRPAWNRSSGNGGVSEVQPVMDAHSW 105
S V + A D E +GG GK+P W R S G SEV PVM A SW
Sbjct: 78 ASVSVAAPTAAVLTVEAAAGDEKSEASGGQDNAGKKPVWKRPSN--GASEVGPVMGASSW 135
Query: 106 PALSDSARGSTKSESSKGLLD----------GSSVSPWQGME--STPSSSMQRQVGDNVN 153
PALS++ + + SS L SSV QG+ S P+ + N
Sbjct: 136 PALSETTKAPSNKSSSDSLKSLGDVPSSSSASSSVPVTQGIANASVPAPKQAGRANPNPT 195
Query: 154 VNNMAPTRQKSIKH-NSSNASSNGGHTQQSAPQVSIAATGSHTSSSRDHTQSPRDHTQSP 212
N+ +RQ+S K N ++ S+NG T S S+ T+ P
Sbjct: 196 PNH---SRQRSFKQRNGASGSANG--------------TVSQPSAQGSFTELP------- 231
Query: 213 RDHAQSPRDHTQRSGFVPSDH-----PQQRNSFRHRNGGPHQRGDGSHHHHNYGNRRDQD 267
H SPR Q++GF +H P QR+S+R++NG HQ G + +GN Q+
Sbjct: 232 -SHNPSPRGQNQKNGFASQNHGGTENPSQRDSYRNQNGNHHQ-SHGGRRNQEHGN---QN 286
Query: 268 WNSRRNYNGRDMHVPP-RVSPRIIRPSLPPNSAPFIHPPP----LRPFGGHMGFHELAAP 322
W +R++NGR+ + R +P +R + +P + P P +PF H+ F P
Sbjct: 287 WTFQRSFNGREGNAQSQRGTPAFVR-----HPSPTVQPIPQFMAAQPFPSHIPF-----P 336
Query: 323 VVLFAGPPPPIDSLRGVPFVPPMP----LYYAGPDPQLHSKIVNQIDYYFSNENLVKDIF 378
L P +P++ P+P +Y DP LH K+ QI YYFS+ENL+ DI+
Sbjct: 337 TELAQSSYYP-----RMPYMTPIPHGPQFFYHYQDPPLHMKLHKQIQYYFSDENLITDIY 391
Query: 379 LRKNMDAQGWVPITLIAGFKKVMDLTDNIQLIIDAIRTSSVVEVQGDKIRRQNDWEKWIM 438
LR M+ +G+VP+ ++AGFKKV +LTDNIQ I++A++ S VEVQGD IR++++W+ W++
Sbjct: 392 LRGFMNNEGFVPLRVVAGFKKVAELTDNIQQIVEALQNSPHVEVQGDFIRKRDNWQNWVL 451
Query: 439 PSPVQFPNVTSPEVLNQ-DMLAEKMRNIALETTIYDGAGG 477
+ P + P+ +++ D +A+++ N++++ + D GG
Sbjct: 452 R---RNPTGSGPQSVDRADAVAKRLGNLSVDQSSADPIGG 488
>At5g66100 unknown protein
Length = 453
Score = 203 bits (516), Expect = 2e-52
Identities = 145/417 (34%), Positives = 218/417 (51%), Gaps = 67/417 (16%)
Query: 49 PIVTAPVDDSTSAEVISDNADNGGERNGGTGKRP--AWNRSSGNGGVSEVQPVMDA-HSW 105
P ++ DD SA +S N K+P WN S N S+V PVM A SW
Sbjct: 43 PTLSLSQDDPFSAPSVSPPTGNNSSDYDNADKKPPPVWNMPSSNSS-SDVGPVMGAAESW 101
Query: 106 PALSDSARGST----KSESSKGLLDGSSVS--PWQGMESTPS-----SSMQRQVGDNVNV 154
PALS SAR S+ ++SK DGSS S P Q +T + SS+ +N V
Sbjct: 102 PALSLSARSSSIKSPSLDASKPFPDGSSSSIPPPQATSNTSTNANAGSSVSATSSENSAV 161
Query: 155 NNMAPTRQKSIKHNSSNASSNGGHTQQSAPQVSIAATGSHTSSSRDHTQSPRDHTQSPRD 214
NN R+ ++N++++SS + +AP ++RD
Sbjct: 162 NNSQ--RKPFRRNNNTSSSSTSSNVSNAAPL-----------NTRD-------------- 194
Query: 215 HAQSPRDHTQRSGFVPSDHPQQRNSFRHRNGGPHQRGDGSHHHHNYGNRRD-----QDWN 269
++H+QR G RNS R+RN + RG+G HH GNRR+ Q
Sbjct: 195 -----QNHSQRGGG-SFGSGNFRNSQRNRNSSSYPRGEGLHH----GNRRNYEHGNQSGF 244
Query: 270 SRRNYNGRDMHVPPRVSPRIIRP-------SLPPNSAPFIHPPPLRPFGGHMGFHELAAP 322
S RNY+GRDMH+ P+ +IRP S P +SA ++ P L +GG + + + A
Sbjct: 245 SHRNYSGRDMHLQPQRGVGMIRPQMLMGPPSFPASSAQYMAAPQLGSYGGPIIYPDYAQH 304
Query: 323 VVLFAGPPPPIDSLRGVPFVPPMPLYYAGPDPQLHSKIVNQIDYYFSNENLVKDIFLRKN 382
V + P P D + V P P+Y+ D L++KI+ Q++YYFS +NL +D LR
Sbjct: 305 VFM---PHPSPDPMGLVGPFPLQPMYFRNFDAILYNKILTQVEYYFSADNLSRDEHLRDQ 361
Query: 383 MDAQGWVPITLIAGFKKVMDLTDNIQLIIDAIRTSSVVEVQGDKIRRQNDWEKWIMP 439
M+ +GWVP+ +IA F+++ +LT+NIQ I++A+R+S VVE+QG+ +RR+ DW+K+++P
Sbjct: 362 MNDEGWVPVRVIAAFRRLAELTNNIQTILEALRSSEVVEIQGETLRRRGDWDKYLLP 418
>At5g21160 unknown protein
Length = 826
Score = 113 bits (283), Expect = 2e-25
Identities = 111/380 (29%), Positives = 161/380 (42%), Gaps = 48/380 (12%)
Query: 74 RNGGTGKRPAWNRSSGNGGVSEVQPVMDAHSWPALSDSARGST-KSESSKGLLDGSSVSP 132
R GG G + W ++ + PVM AHSWPAL+D+A+ K+ + S P
Sbjct: 18 REGGIGTKSPWKTTTSPVETIDA-PVMGAHSWPALADAAQQPRPKNPPAPAPAPPSKNIP 76
Query: 133 WQGMESTPSSSMQRQV--GDNVNVNNMAPTRQKSIKHNSSNASSNGGHTQQSAPQVSIAA 190
TP+ + Q + G N + P+ + S SN NG P + A
Sbjct: 77 TSIPIPTPAVTGQAKSKGGGKANPGHKNPSGRHSKPGPRSN--QNG----PPPPPYLVHA 130
Query: 191 TGSHTSSSRDHTQSPRDHTQSPR-DHAQSPRDHTQRSGFVPSDHPQQRNSFRHRNGGPHQ 249
H P H P +A P S + +Q + P
Sbjct: 131 VPYHPPPFPPMVPLP--HAAGPDFPYAPYPPYPVPVPPVTESGNEKQVQASPLPPVLPAP 188
Query: 250 RGDGSHHHHNYGNRRDQDWNSRRNYNGRDMHVPPRVSPRII-RPSLPPNSAPFIHPPPLR 308
+GD + W +R ++ R+M P PR RP PF+ P P
Sbjct: 189 QGDPG-----------KPWPHQRGFDPRNM--PQGAGPRNFGRP-------PFMGPAPGF 228
Query: 309 PFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFVPPMPLYYAGPDP--------QLHSKI 360
G GF PV GPPP ++RG P+ P Y P L ++
Sbjct: 229 LVGPGPGF---PGPVYYLPGPPP--GAIRG-PYPPRFAPYPVNQGPPILSPEKLDLRDRV 282
Query: 361 VNQIDYYFSNENLVKDIFLRKNMDAQGWVPITLIAGFKKVMDLTDNIQLIIDAIRTSSVV 420
+ Q++YYFS+ENL D +L MD +GWVP +IAGFK+V +T ++ I+ A+ S+ V
Sbjct: 283 LKQVEYYFSDENLENDHYLISLMDEEGWVPTKIIAGFKRVKAMTMDVDFIVYALGFSNSV 342
Query: 421 EVQGDKIRRQNDWEKWIMPS 440
EVQGD+IR+++ W WI S
Sbjct: 343 EVQGDQIRKRDKWSDWIPAS 362
Score = 30.8 bits (68), Expect = 1.9
Identities = 26/88 (29%), Positives = 34/88 (38%), Gaps = 14/88 (15%)
Query: 249 QRGDGSHHHHNYGNRRDQDWNSRRNYNGRDMHVPPRVSPRIIRPSLPPNSAPFIHPPPLR 308
++ G+H HH RR N R NG PP S S PP+S H P L
Sbjct: 577 KQNKGTHKHHTAHARRFFSSNIRN--NGNISESPPSSSIGFFFGSTPPDS----HGPRLS 630
Query: 309 PFGGHMGFHELAAPVVLFAGPPPPIDSL 336
++P +G PP+ SL
Sbjct: 631 KLS--------SSPQCTLSGSSPPVGSL 650
>At5g46250 unknown protein
Length = 422
Score = 61.6 bits (148), Expect = 1e-09
Identities = 35/82 (42%), Positives = 53/82 (63%), Gaps = 4/82 (4%)
Query: 352 PDPQLHSKIVNQIDYYFSNENLVKDIFLRKNM--DAQGWVPITLIAGFKKVMDLTDNIQL 409
P +L+ KI+ Q++YYFS+ENL D FL M + +G+VPI+ IA F K+ LT + L
Sbjct: 99 PIDELNQKIIRQVEYYFSDENLPTDKFLLNAMKRNKKGFVPISTIATFHKMKKLTRDHAL 158
Query: 410 IIDAIRTSSVVEVQGD--KIRR 429
I+ A++ SS + V D K++R
Sbjct: 159 IVSALKESSFLVVSADEKKVKR 180
>At3g19090 RNA-binding protein, putative
Length = 455
Score = 52.8 bits (125), Expect = 5e-07
Identities = 30/82 (36%), Positives = 52/82 (62%), Gaps = 4/82 (4%)
Query: 356 LHSKIVNQIDYYFSNENLVKDIFLRKNM--DAQGWVPITLIAGFKKVMDLTDNIQLIIDA 413
L KIV Q++Y F++ +L+ + + K++ D +G+VP++ IA KK+ LT N L+ A
Sbjct: 144 LRLKIVKQVEYQFTDMSLLANESISKHISKDPEGYVPVSYIASTKKIKALTSNHHLVSLA 203
Query: 414 IRTSS--VVEVQGDKIRRQNDW 433
+R+SS VV G K++R + +
Sbjct: 204 LRSSSKLVVSEDGKKVKRTSQF 225
>At2g43970 unknown protein
Length = 545
Score = 50.8 bits (120), Expect = 2e-06
Identities = 66/287 (22%), Positives = 104/287 (35%), Gaps = 46/287 (16%)
Query: 159 PTRQKSIKHNSSNASSNGGHTQQSAPQVSIAATGSHTSSSRDH----------TQSPRDH 208
PT+ + H+ S +S+ + S P + + + S ++ Q PR
Sbjct: 14 PTQSDDLSHSHSTSSTTSA-SSSSDPSLLRSLSLSRLNAGAPEFVPGRTTPPLPQPPRMI 72
Query: 209 TQSPRDHAQSPRDHTQRSGFVP--SDHPQQRNSFRHRNGGPHQRGDGSHHHHNYGNRRDQ 266
P H H Q P P Q + +N PH R HHH+ + N++
Sbjct: 73 IPPPPPHGMLHMYHHQPPFNTPVLGPVPIQPHLVPVQNHHPHHRFHQHHHHNRHQNQQYV 132
Query: 267 DWNSRRNYNGRDMHVPPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLF 326
+ Y R V P ++ E V
Sbjct: 133 PVRNHGEYQQRG-GVGGEQEPDLVSKKNDRRDH---------------SKRESKNDQVTE 176
Query: 327 AGPPPPIDSLRGVPFVPPMPLYYAGPDPQLHSKIVNQIDYYFSNENLVKDIFLRKNM--D 384
G IDS G+P KIVNQ++YYFS+ NL L + + D
Sbjct: 177 TGASVSIDSKTGLP-------------EDSIQKIVNQVEYYFSDLNLATTDHLMRFICKD 223
Query: 385 AQGWVPITLIAGFKKVMDLTDNIQLIIDAIRTSS--VVEVQGDKIRR 429
+G+VPI ++A FKK+ + +N + ++ S+ V G K+RR
Sbjct: 224 PEGYVPIHVVASFKKIKAVINNNSQLAAVLQNSAKLFVSEDGKKVRR 270
Score = 29.6 bits (65), Expect = 4.2
Identities = 14/44 (31%), Positives = 20/44 (44%), Gaps = 1/44 (2%)
Query: 230 PSDHPQQRNSFRHRNGGPHQRGDGSHHHHNYGNRRDQDWNSRRN 273
P + Q N+ H H G G+HHHH++ Q N+ N
Sbjct: 466 PHQNQNQNNNHSHNQNHNHN-GRGNHHHHHHHQVGTQPSNNPMN 508
>At5g23150 transcription factor-like protein (gb|AAD31171.1)
Length = 1392
Score = 42.4 bits (98), Expect = 6e-04
Identities = 38/148 (25%), Positives = 56/148 (37%), Gaps = 19/148 (12%)
Query: 196 SSSRDHTQSPRDHTQSPRDHAQSPRDHTQRSGFVPSDHPQQRNSFRHRNGGPHQRGDGSH 255
+SS++H P S R + P + + Q ++S+ H P QR D
Sbjct: 1241 ASSQNHRFQPSTPL-SQRPMVRLPSAPSSHFSYPSHIQSQSQHSYTHPYPFPPQRDDARR 1299
Query: 256 HHHNYGNRRDQDWNSRRNYNGRDMHVPPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMG 315
+ + R +S N NG +H R P LP + F PPP RP G M
Sbjct: 1300 YRNEEPWRIPSSGHSAENQNGAWIH------GRNSHPGLPRVTDSFFRPPPERPPSGTMN 1353
Query: 316 FHELAAPVVLFAGPPPPIDSLRGVPFVP 343
+ AA +L+ VP +P
Sbjct: 1354 YQPSAA------------SNLQAVPAIP 1369
>At3g07660 unknown protein
Length = 782
Score = 42.4 bits (98), Expect = 6e-04
Identities = 46/180 (25%), Positives = 70/180 (38%), Gaps = 18/180 (10%)
Query: 106 PALSDSARGSTKS----ESSKGLLDGSSVSPWQGMEST------PSSSMQRQVG-DNVNV 154
P+ S S TK S ++D S+V P G P+ S Q + G +V
Sbjct: 67 PSTSTSQEIKTKDIALVSSHSAVMDKSTVGPKSGSSDVRQSSNLPAGSGQLKAGRSSVPS 126
Query: 155 NNM--APTRQKSIKHNSSNASSNGGHTQQSAPQVSIAATGSHTSSSRDHTQSPRDHTQSP 212
NN+ AP+ + K N S HT+Q A S+ +S R + +P+ +P
Sbjct: 127 NNIDGAPSSVELSK-NRVAVGSRVVHTEQKAVD-SLPLPRPSSSEVRFTSSNPKSEQTAP 184
Query: 213 RDHAQSPRDHTQRSGFVPSDHPQQRNSFRHRNGGPHQRGDGSHHHHNYGNRRDQDWNSRR 272
H + HT+ G + +++ R H GS NY NR Q +R
Sbjct: 185 EQHIGESKSHTRSRGVGKT---AVNDTYGSRPASSHSNSTGSRPSSNYSNRSHQTVGPQR 241
>At4g32720 putative RNA-binding protein LAH1
Length = 433
Score = 42.0 bits (97), Expect = 8e-04
Identities = 24/80 (30%), Positives = 40/80 (50%), Gaps = 14/80 (17%)
Query: 360 IVNQIDYYFSNENLVKDIFLRKNM--DAQGWVPITLIAGFKKVMDL------------TD 405
++ Q+++YFS+ NL D FL+K + G V + LI F K+ D
Sbjct: 14 VLRQVEFYFSDSNLPIDDFLKKTVTESEDGLVSLALICSFSKMRGYLKLGDSKGDDIPED 73
Query: 406 NIQLIIDAIRTSSVVEVQGD 425
I+ + D +RTSS +++ D
Sbjct: 74 TIKAVADTLRTSSALKISDD 93
>At1g75660 Dhp1-like protein
Length = 1020
Score = 41.6 bits (96), Expect = 0.001
Identities = 28/96 (29%), Positives = 38/96 (39%), Gaps = 17/96 (17%)
Query: 182 SAPQVSIAATGSHTSSSRDHTQSPRDHTQSPRDHAQSPRDHTQRSGFVPSDHPQQRNSFR 241
S P S + G H S ++ + ++ Q + H Q + G+ H
Sbjct: 939 SEPYQSQSREGQHASRGGGYSGNHQNQHQQQQWHGQGGSEQNNPRGYNGQHH-------- 990
Query: 242 HRNGGPHQR---GDGSHHHHNYGNRRDQDWNSRRNY 274
H+ GG H R G GSHHHH DQ N R Y
Sbjct: 991 HQQGGDHDRRGRGRGSHHHH------DQGGNPRHRY 1020
Score = 29.3 bits (64), Expect = 5.5
Identities = 32/178 (17%), Positives = 55/178 (29%), Gaps = 6/178 (3%)
Query: 106 PALSDSARGSTKSESSKGLLDGSSVSPWQGMESTPSSSMQRQVGDNVN---VNNMAPTRQ 162
PAL G ++ G+ + + QG ++ P S R +G+ + N++
Sbjct: 817 PALWHEDNGRRPMHNNHGMHNNHGMHNNQGRQNPPGSVSGRHLGNAAHRLVSNSLQMGTD 876
Query: 163 KSIKHNSSNASSNGGHTQQSAPQVSIAATGSHTSSSRDHTQSPRDHTQSPRDHAQSPRDH 222
+ A G + Q P + G P + PR
Sbjct: 877 RYQTPTDVPAPGYGYNPPQYVPPIPYQHGGYMAPPGAQGYAQPAPYQNRG---GYQPRGP 933
Query: 223 TQRSGFVPSDHPQQRNSFRHRNGGPHQRGDGSHHHHNYGNRRDQDWNSRRNYNGRDMH 280
+ R P + R GG H + + + N+ R YNG+ H
Sbjct: 934 SGRFPSEPYQSQSREGQHASRGGGYSGNHQNQHQQQQWHGQGGSEQNNPRGYNGQHHH 991
>At4g32710 putative protein kinase
Length = 731
Score = 40.4 bits (93), Expect = 0.002
Identities = 28/81 (34%), Positives = 36/81 (43%), Gaps = 7/81 (8%)
Query: 281 VPPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDS----- 335
+PP +S PS PP SAP PPPL E P +L + PPPP++S
Sbjct: 40 LPPPLSSPPPLPSPPPLSAPTASPPPLPVESPPSPPIESPPPPLLESPPPPPLESPSPPS 99
Query: 336 --LRGVPFVPPMPLYYAGPDP 354
+ PP+P A P P
Sbjct: 100 PHVSAPSGSPPLPFLPAKPSP 120
>At4g12610 putative protein
Length = 649
Score = 39.3 bits (90), Expect = 0.005
Identities = 38/129 (29%), Positives = 62/129 (47%), Gaps = 16/129 (12%)
Query: 60 SAEVISDNADNGGERNGGTGKRPAWNRSSGNGGVSEVQPVMDAHSWPALSDSARGSTKSE 119
S E D++D+ E N GT +S ++ +PV +A + PA S RG+ ++
Sbjct: 357 SDEDDDDDSDDEEETNYGTVT------NSKQKEAAKEEPVDNAPAKPAPSGPPRGTPPAK 410
Query: 120 SSKG---LLDGSSVSPWQGM------ESTPSSSMQRQVGDNVNVNNMAPTRQKSIKHNSS 170
SKG L DG S P + E+ P SS++ + + V+ +N PT+ + S+
Sbjct: 411 PSKGKRKLNDGDSKKPSSSVQKKVKTENDPKSSLKEERANTVSKSN-TPTKAVKAEPASA 469
Query: 171 NASSNGGHT 179
ASS+ T
Sbjct: 470 PASSSSAAT 478
>At4g13340 extensin-like protein
Length = 760
Score = 38.5 bits (88), Expect = 0.009
Identities = 28/79 (35%), Positives = 31/79 (38%), Gaps = 6/79 (7%)
Query: 280 HVPPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMG----FHELAAPVVLFAGPPPPIDS 335
H PP SP PP SAP PP P H H P V+ PPPP
Sbjct: 680 HSPPP-SPVHYSSPPPPPSAPCEESPPPAPVVHHSPPPPMVHHSPPPPVIHQSPPPPSPE 738
Query: 336 LRGVPFVPPMPLYYAGPDP 354
G P P + + YA P P
Sbjct: 739 YEG-PLPPVIGVSYASPPP 756
Score = 35.4 bits (80), Expect = 0.077
Identities = 26/74 (35%), Positives = 31/74 (41%), Gaps = 12/74 (16%)
Query: 282 PPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF 341
PP SP PS PP P PPP P P +++ PPPP+ S P
Sbjct: 477 PPVYSPP--PPSPPPPPPPVYSPPPPPP---------PPPPPPVYSPPPPPVYSSPPPPP 525
Query: 342 VP-PMPLYYAGPDP 354
P P P+Y P P
Sbjct: 526 SPAPTPVYCTRPPP 539
Score = 35.0 bits (79), Expect = 0.10
Identities = 23/73 (31%), Positives = 28/73 (37%), Gaps = 11/73 (15%)
Query: 282 PPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF 341
PP P PP+S+P H PP H P+ + PPPP V
Sbjct: 552 PPPPEPYYYSSPPPPHSSPPPHSPPPP--------HSPPPPIYPYLSPPPPPTP---VSS 600
Query: 342 VPPMPLYYAGPDP 354
PP P+Y P P
Sbjct: 601 PPPTPVYSPPPPP 613
Score = 35.0 bits (79), Expect = 0.10
Identities = 21/60 (35%), Positives = 26/60 (43%), Gaps = 5/60 (8%)
Query: 295 PPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFVPPMPLYYAGPDP 354
PP P I PPP P + + P V+ PPP P PP P+YY+ P P
Sbjct: 609 PPPPPPCIEPPPPPPC---IEYSPPPPPPVVHYSSPPPPPVYYSSP--PPPPVYYSSPPP 663
Score = 34.7 bits (78), Expect = 0.13
Identities = 24/77 (31%), Positives = 29/77 (37%), Gaps = 18/77 (23%)
Query: 282 PPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF 341
PP SP P PP + PPP P PPPP+ S P
Sbjct: 461 PPVYSPPPPPPPPPPPPPVYSPPPPSPP------------------PPPPPVYSPPPPPP 502
Query: 342 VPPMPLYYAGPDPQLHS 358
PP P Y+ P P ++S
Sbjct: 503 PPPPPPVYSPPPPPVYS 519
Score = 34.7 bits (78), Expect = 0.13
Identities = 22/73 (30%), Positives = 29/73 (39%), Gaps = 15/73 (20%)
Query: 282 PPRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF 341
PP P + P PP P ++ PP P P +++ PPPP P
Sbjct: 428 PPSPPPPVYSPPPPPPPPPPVYSPPPPP--------PPPPPPPVYSPPPPP-------PP 472
Query: 342 VPPMPLYYAGPDP 354
PP P Y+ P P
Sbjct: 473 PPPPPPVYSPPPP 485
Score = 34.3 bits (77), Expect = 0.17
Identities = 27/76 (35%), Positives = 30/76 (38%), Gaps = 15/76 (19%)
Query: 282 PPRVSPRIIRPSLPP-NSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVP 340
PP P + P PP S+P PPP P PV PPPP S
Sbjct: 502 PPPPPPPVYSPPPPPVYSSP---PPPPSP---------APTPVYCTRPPPPPPHSPPPPQ 549
Query: 341 F--VPPMPLYYAGPDP 354
F PP P YY+ P P
Sbjct: 550 FSPPPPEPYYYSSPPP 565
Score = 32.7 bits (73), Expect = 0.50
Identities = 23/75 (30%), Positives = 28/75 (36%), Gaps = 10/75 (13%)
Query: 282 PPRVSPRIIRPSL--PPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGV 339
PP P I P L PP P PPP + P PPPP +
Sbjct: 577 PPHSPPPPIYPYLSPPPPPTPVSSPPPTPVYS--------PPPPPPCIEPPPPPPCIEYS 628
Query: 340 PFVPPMPLYYAGPDP 354
P PP ++Y+ P P
Sbjct: 629 PPPPPPVVHYSSPPP 643
Score = 32.0 bits (71), Expect = 0.86
Identities = 24/87 (27%), Positives = 34/87 (38%), Gaps = 19/87 (21%)
Query: 282 PPRVSPRIIRPSL---PPNSAPFIH---PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDS 335
PP + P P + PP P +H PPP + + P V ++ PPPP
Sbjct: 613 PPCIEPPPPPPCIEYSPPPPPPVVHYSSPPPPPVY-----YSSPPPPPVYYSSPPPPPPV 667
Query: 336 LRGVP--------FVPPMPLYYAGPDP 354
P PP P++Y+ P P
Sbjct: 668 HYSSPPPPEVHYHSPPPSPVHYSSPPP 694
Score = 30.8 bits (68), Expect = 1.9
Identities = 22/72 (30%), Positives = 29/72 (39%), Gaps = 6/72 (8%)
Query: 283 PRVSPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFV 342
P P I P PP + PPP P H + P ++ PPP P
Sbjct: 609 PPPPPPCIEPPPPPPCIEYSPPPP--PPVVH---YSSPPPPPVYYSSPPPPPVYYSSP-P 662
Query: 343 PPMPLYYAGPDP 354
PP P++Y+ P P
Sbjct: 663 PPPPVHYSSPPP 674
Score = 30.4 bits (67), Expect = 2.5
Identities = 20/68 (29%), Positives = 25/68 (36%), Gaps = 18/68 (26%)
Query: 287 PRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFVPPMP 346
P + P P P PPP P P +++ PPPP P PP P
Sbjct: 421 PTLTSPPPPSPPPPVYSPPPPPP-----------PPPPVYSPPPPP-------PPPPPPP 462
Query: 347 LYYAGPDP 354
+Y P P
Sbjct: 463 VYSPPPPP 470
Score = 28.5 bits (62), Expect = 9.5
Identities = 21/84 (25%), Positives = 31/84 (36%), Gaps = 7/84 (8%)
Query: 279 MHVPPRVSPRIIRPSLPPNSAPFIHPPPLRPFG------GHMGFHELAAPVVLFAGPPPP 332
+H P + S PP + PPP P + +H V ++ PPPP
Sbjct: 636 VHYSSPPPPPVYYSSPPPPPVYYSSPPPPPPVHYSSPPPPEVHYHSPPPSPVHYSSPPPP 695
Query: 333 IDSLRGVPFVPPMPLYYAGPDPQL 356
S PP P+ + P P +
Sbjct: 696 -PSAPCEESPPPAPVVHHSPPPPM 718
>At3g54580 extensin precursor -like protein
Length = 951
Score = 38.5 bits (88), Expect = 0.009
Identities = 25/77 (32%), Positives = 40/77 (51%), Gaps = 7/77 (9%)
Query: 286 SPRIIRPSLPPNSAPFIH--PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRG-VPFV 342
SP+++ S PP P+++ PPP +++ P +++ PPPP S V +
Sbjct: 705 SPKVVYKSPPP---PYVYSSPPPPHYSPSPKVYYKSPPPPYVYSSPPPPYYSPSPKVHYK 761
Query: 343 PPMPLYYAGPDPQLHSK 359
P P YYA P P++H K
Sbjct: 762 SPPPPYYA-PTPKVHYK 777
Score = 34.7 bits (78), Expect = 0.13
Identities = 22/75 (29%), Positives = 33/75 (43%), Gaps = 12/75 (16%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFV--P 343
SP++ S PP P+ P P +++ P +++ PPPP S + P
Sbjct: 538 SPKVYYKSPPP---PYYSPSP-------KVYYKSPPPPYVYSSPPPPYYSPSPKVYYKSP 587
Query: 344 PMPLYYAGPDPQLHS 358
P P Y+ P P HS
Sbjct: 588 PPPYVYSSPPPPYHS 602
Score = 34.3 bits (77), Expect = 0.17
Identities = 24/79 (30%), Positives = 37/79 (46%), Gaps = 11/79 (13%)
Query: 286 SPRIIRPSLPPNSAPFIH--PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFV- 342
SP++ S PP P+++ PPP +++ P +++ PPPP S P V
Sbjct: 554 SPKVYYKSPPP---PYVYSSPPPPYYSPSPKVYYKSPPPPYVYSSPPPPYHSPS--PKVQ 608
Query: 343 ---PPMPLYYAGPDPQLHS 358
PP P Y+ P P +S
Sbjct: 609 YKSPPPPYVYSSPPPPYYS 627
Score = 33.9 bits (76), Expect = 0.23
Identities = 22/77 (28%), Positives = 36/77 (46%), Gaps = 7/77 (9%)
Query: 286 SPRIIRPSLPPNSAPFIH--PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFV- 342
SP++ S PP P+++ PPP +++ P +++ PPPP S +
Sbjct: 604 SPKVQYKSPPP---PYVYSSPPPPYYSPSPKVYYKSPPPPYVYSSPPPPYYSPSPKVYYK 660
Query: 343 -PPMPLYYAGPDPQLHS 358
PP P Y+ P P +S
Sbjct: 661 SPPPPYVYSSPPPPYYS 677
Score = 33.5 bits (75), Expect = 0.29
Identities = 22/71 (30%), Positives = 33/71 (45%), Gaps = 12/71 (16%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPP-IDSLRGVPFVPP 344
SP++ S PP P+++ P P+ +P V + PPPP + S P+ P
Sbjct: 579 SPKVYYKSPPP---PYVYSSPPPPY-------HSPSPKVQYKSPPPPYVYSSPPPPYYSP 628
Query: 345 MP-LYYAGPDP 354
P +YY P P
Sbjct: 629 SPKVYYKSPPP 639
Score = 32.7 bits (73), Expect = 0.50
Identities = 20/75 (26%), Positives = 34/75 (44%), Gaps = 12/75 (16%)
Query: 286 SPRIIRPSLPPNSAPFIH--PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFVP 343
SP++ S PP P+++ PPP +++ P +++ PPPP + P
Sbjct: 488 SPKVYYKSPPP---PYVYSSPPPPYYSPSPKVYYKSPPPPYVYSSPPPPY-------YSP 537
Query: 344 PMPLYYAGPDPQLHS 358
+YY P P +S
Sbjct: 538 SPKVYYKSPPPPYYS 552
Score = 32.0 bits (71), Expect = 0.86
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 288 SPKVDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPTYSPSPKVEYKSPP 347
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 348 PPYVYSSPPPPTYS 361
Score = 31.6 bits (70), Expect = 1.1
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 413 SPKVEYKSPPPPYVYSSPPPPYYSPSPKVEYKSPPPPYVYSSPPPPTYSPSPKVDYKSPP 472
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 473 PPYVYSSPPPPYYS 486
Score = 31.6 bits (70), Expect = 1.1
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 363 SPKVEYKSPPPPYVYSSPPPPTYSPSPKVEYKSPPPPYVYSSPPPPTYSPSPKVEYKSPP 422
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 423 PPYVYSSPPPPYYS 436
Score = 31.6 bits (70), Expect = 1.1
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 88 SPKVEYKSPPPPYVYSSPPPPTYSPSPKVEYKSPPPPYVYSSPPPPTYSPSPKVEYKSPP 147
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 148 PPYVYSSPPPPTYS 161
Score = 31.6 bits (70), Expect = 1.1
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 238 SPKVDYKSPPPPYVYSSPPPPYYSPSPKVEYKSPPPPYVYSSPPPPTYSPSPKVDYKSPP 297
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 298 PPYVYSSPPPPYYS 311
Score = 31.6 bits (70), Expect = 1.1
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 113 SPKVEYKSPPPPYVYSSPPPPTYSPSPKVEYKSPPPPYVYSSPPPPTYSPSPKVEYKSPP 172
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 173 PPYVYSSPPPPTYS 186
Score = 31.6 bits (70), Expect = 1.1
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 338 SPKVEYKSPPPPYVYSSPPPPTYSPSPKVEYKSPPPPYVYSSPPPPTYSPSPKVEYKSPP 397
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 398 PPYVYSSPPPPTYS 411
Score = 31.6 bits (70), Expect = 1.1
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 188 SPKVEYKSPPPPYVYSSPPPPYYSPSPKVEYKSPPPPYVYSSPPPPTYSPSPKVDYKSPP 247
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 248 PPYVYSSPPPPYYS 261
Score = 31.6 bits (70), Expect = 1.1
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 313 SPKVDYKSPPPPYVYSSPPPPTYSPSPKVEYKSPPPPYVYSSPPPPTYSPSPKVEYKSPP 372
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 373 PPYVYSSPPPPTYS 386
Score = 31.6 bits (70), Expect = 1.1
Identities = 21/73 (28%), Positives = 31/73 (41%), Gaps = 11/73 (15%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPP-IDSLRGVPFVPP 344
SP++ S PP P+++ P P+ +P V + PPPP + S P+ P
Sbjct: 629 SPKVYYKSPPP---PYVYSSPPPPYYS-------PSPKVYYKSPPPPYVYSSPPPPYYSP 678
Query: 345 MPLYYAGPDPQLH 357
P Y P H
Sbjct: 679 SPKVYYKSPPHPH 691
Score = 31.6 bits (70), Expect = 1.1
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 138 SPKVEYKSPPPPYVYSSPPPPTYSPSPKVEYKSPPPPYVYSSPPPPTYSPSPKVEYKSPP 197
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 198 PPYVYSSPPPPYYS 211
Score = 31.2 bits (69), Expect = 1.5
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 263 SPKVEYKSPPPPYVYSSPPPPTYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPP 322
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 323 PPYVYSSPPPPTYS 336
Score = 31.2 bits (69), Expect = 1.5
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 213 SPKVEYKSPPPPYVYSSPPPPTYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKVEYKSPP 272
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 273 PPYVYSSPPPPTYS 286
Score = 31.2 bits (69), Expect = 1.5
Identities = 24/84 (28%), Positives = 31/84 (36%), Gaps = 3/84 (3%)
Query: 278 DMHVPPRVSP--RIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDS 335
D PP SP + S PP PPP + + P V + PPP
Sbjct: 53 DSSPPPTYSPAPEVEYKSPPPPYVYSSPPPPTYSPSPKVEYKSPPPPYVYSSPPPPTYSP 112
Query: 336 LRGVPF-VPPMPLYYAGPDPQLHS 358
V + PP P Y+ P P +S
Sbjct: 113 SPKVEYKSPPPPYVYSSPPPPTYS 136
Score = 31.2 bits (69), Expect = 1.5
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 821 SPKVEYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPP 880
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 881 PPYVYSSPPPPYYS 894
Score = 30.8 bits (68), Expect = 1.9
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 163 SPKVEYKSPPPPYVYSSPPPPTYSPSPKVEYKSPPPPYVYSSPPPPYYSPSPKVEYKSPP 222
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 223 PPYVYSSPPPPTYS 236
Score = 30.8 bits (68), Expect = 1.9
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 846 SPKVDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPVVDYKSPP 905
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 906 PPYVYSSPPPPYYS 919
Score = 30.8 bits (68), Expect = 1.9
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 796 SPKVHYKSPPPPYVYSSPPPPYYSPSPKVEYKSPPPPYVYSSPPPPYYSPSPKVDYKSPP 855
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 856 PPYVYSSPPPPYYS 869
Score = 30.8 bits (68), Expect = 1.9
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 388 SPKVEYKSPPPPYVYSSPPPPTYSPSPKVEYKSPPPPYVYSSPPPPYYSPSPKVEYKSPP 447
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 448 PPYVYSSPPPPTYS 461
Score = 30.4 bits (67), Expect = 2.5
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 438 SPKVEYKSPPPPYVYSSPPPPTYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKVYYKSPP 497
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 498 PPYVYSSPPPPYYS 511
Score = 29.6 bits (65), Expect = 4.2
Identities = 20/74 (27%), Positives = 29/74 (39%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
+P++ S PP PPP + + P V + PPP V + PP
Sbjct: 771 TPKVHYKSPPPPYVYSSPPPPYYSPSPKVHYKSPPPPYVYSSPPPPYYSPSPKVEYKSPP 830
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 831 PPYVYSSPPPPYYS 844
>At2g36270 bZip transcription factor AtbZip39
Length = 442
Score = 38.1 bits (87), Expect = 0.012
Identities = 48/238 (20%), Positives = 84/238 (35%), Gaps = 41/238 (17%)
Query: 39 PAVSLSEESFPIVTAPVDDSTSAEVISDNADNGGERNGGTGKRPAWNRSSGNGGVSEVQP 98
P SL +S I + +D+ A + +N N G N WN N +
Sbjct: 33 PFTSLGRQS-SIYSLTLDEFQHA--LCENGKNFGSMNMDEFLVSIWNAEENNNNQQQAAA 89
Query: 99 VMDAHSWPA-------------------LSDSARGSTKSESSKGLLDGSSVSPWQGMEST 139
+HS PA S +RG+ + + +G+ + SS+ P QG +
Sbjct: 90 AAGSHSVPANHNGFNNNNNNGGEGGVGVFSGGSRGNEDANNKRGIANESSL-PRQGSLTL 148
Query: 140 PSSSMQRQVGD-------NVNVNNMAPTRQKSIKHNSSNASSNGGHTQQSAP---QVSIA 189
P+ ++ V + N + +S N N + NGG T P ++++
Sbjct: 149 PAPLCRKTVDEVWSEIHRGGGSGNGGDSNGRSSSSNGQNNAQNGGETAARQPTFGEMTLE 208
Query: 190 ATGSHTSSSRDHTQSPRDHTQSPRDHAQSPRDHTQRSGFVPSDHPQQRNSFRHRNGGP 247
R+H +P+ + +P + S +P+ QQ G P
Sbjct: 209 DFLVKAGVVREHPTNPKPN--------PNPNQNQNPSSVIPAAAQQQLYGVFQGTGDP 258
>At5g06630 putative protein
Length = 434
Score = 37.4 bits (85), Expect = 0.020
Identities = 23/77 (29%), Positives = 37/77 (47%), Gaps = 7/77 (9%)
Query: 286 SPRIIRPSLPPNSAPFIH--PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFV- 342
SP++ S PP P+++ PPPL +++ P +++ PPPP S +
Sbjct: 115 SPKVYYKSPPP---PYVYSSPPPLYYSPSPKVYYKSPPPPYVYSSPPPPYYSPSPKVYYK 171
Query: 343 -PPMPLYYAGPDPQLHS 358
PP P Y+ P P +S
Sbjct: 172 SPPPPYVYSSPPPPYYS 188
Score = 35.0 bits (79), Expect = 0.10
Identities = 23/81 (28%), Positives = 37/81 (45%), Gaps = 7/81 (8%)
Query: 286 SPRIIRPSLPPNSAPFIH--PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFV- 342
SP++ S PP P+++ PPP +++ P +++ PPPP S +
Sbjct: 265 SPKVYYKSPPP---PYVYSSPPPPYYSPSPKVYYKSPPPPYVYSSPPPPYYSPSPKVYYK 321
Query: 343 -PPMPLYYAGPDPQLHSKIVN 362
PP P Y+ P P +S N
Sbjct: 322 SPPPPYVYSSPPPPYYSPSPN 342
Score = 34.7 bits (78), Expect = 0.13
Identities = 22/77 (28%), Positives = 36/77 (46%), Gaps = 7/77 (9%)
Query: 286 SPRIIRPSLPPNSAPFIH--PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFV- 342
SP++ S PP P+++ PPP +++ P +++ PPPP S +
Sbjct: 140 SPKVYYKSPPP---PYVYSSPPPPYYSPSPKVYYKSPPPPYVYSSPPPPYYSPSPKVYYK 196
Query: 343 -PPMPLYYAGPDPQLHS 358
PP P Y+ P P +S
Sbjct: 197 SPPPPYVYSSPPPPYYS 213
Score = 34.7 bits (78), Expect = 0.13
Identities = 22/77 (28%), Positives = 36/77 (46%), Gaps = 7/77 (9%)
Query: 286 SPRIIRPSLPPNSAPFIH--PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFV- 342
SP++ S PP P+++ PPP +++ P +++ PPPP S +
Sbjct: 190 SPKVYYKSPPP---PYVYSSPPPPYYSPSPKVYYKSPPPPYVYSSPPPPYYSPSPKVYYK 246
Query: 343 -PPMPLYYAGPDPQLHS 358
PP P Y+ P P +S
Sbjct: 247 SPPPPYVYSSPPPPYYS 263
Score = 33.1 bits (74), Expect = 0.38
Identities = 24/79 (30%), Positives = 37/79 (46%), Gaps = 11/79 (13%)
Query: 286 SPRIIRPSLPPNSAPFIH--PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFV- 342
SP++ S PP P+++ PPP +++ P +++ PPPP S P V
Sbjct: 315 SPKVYYKSPPP---PYVYSSPPPPYYSPSPNVYYKSPPPPYVYSSPPPPYYS--PSPKVH 369
Query: 343 ---PPMPLYYAGPDPQLHS 358
PP P Y+ P P +S
Sbjct: 370 YKSPPPPYVYSSPPPPYYS 388
Score = 31.6 bits (70), Expect = 1.1
Identities = 24/86 (27%), Positives = 34/86 (38%), Gaps = 8/86 (9%)
Query: 280 HVPPRVSPRIIR--PSLPPNSAPFIH---PPPLRPFGGHMGFHELAAPVVLFAGPPPPID 334
H P SP P P+ P+++ PPP + + P V + PPPP
Sbjct: 29 HTPAYDSPSYEHKGPKYAPHPKPYVYSSPPPPYYTPSPKVNYKSPPPPYV-YNSPPPPYY 87
Query: 335 SLRGVPFV--PPMPLYYAGPDPQLHS 358
S + PP P Y+ P P +S
Sbjct: 88 SPSPKVYYKSPPPPYVYSSPPPPYYS 113
Score = 30.4 bits (67), Expect = 2.5
Identities = 21/74 (28%), Positives = 28/74 (37%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP + S PP PPP + + P V + PPP V + PP
Sbjct: 340 SPNVYYKSPPPPYVYSSPPPPYYSPSPKVHYKSPPPPYVYSSPPPPYYSPSPKVHYKSPP 399
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 400 PPYVYSSPPPPYYS 413
>At3g24480 disease resistance protein, putative
Length = 494
Score = 37.0 bits (84), Expect = 0.027
Identities = 26/79 (32%), Positives = 32/79 (39%), Gaps = 7/79 (8%)
Query: 282 PPRVSPRIIRPSL-PPNSAPFIHPPPLRPFGG-----HMGFHELAAPVVLFAGPPPPIDS 335
PP SP + P PP S P PPP P + + P V + PPPP
Sbjct: 413 PPPPSPPLPPPVYSPPPSPPVFSPPPSPPVYSPPPPPSIHYSSPPPPPVHHSSPPPPSPE 472
Query: 336 LRGVPFVPPMPLYYAGPDP 354
G P P + + YA P P
Sbjct: 473 FEG-PLPPVIGVSYASPPP 490
Score = 32.0 bits (71), Expect = 0.86
Identities = 21/69 (30%), Positives = 29/69 (41%), Gaps = 4/69 (5%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFVPPM 345
SP I+ PP +P + PP P F +P V PPP I PP
Sbjct: 404 SPPIVALPPPPPPSPPLPPPVYSPPPSPPVFSPPPSPPVYSPPPPPSIH----YSSPPPP 459
Query: 346 PLYYAGPDP 354
P++++ P P
Sbjct: 460 PVHHSSPPP 468
>At2g32700 unknown protein
Length = 787
Score = 37.0 bits (84), Expect = 0.027
Identities = 46/186 (24%), Positives = 66/186 (34%), Gaps = 12/186 (6%)
Query: 121 SKGLLDGSSVSPWQGMESTPSSSMQRQVGDNVNVNNMAPTRQKSIKHNSSNASSNGGHTQ 180
S GL G S P +G T M+ +G P QKS N S + Q
Sbjct: 236 SAGLNPGVSGLPLKGWPLTGIEQMRPGLG--------GPQVQKSFLQNQSQFQLSP-QQQ 286
Query: 181 QSAPQVSIAATGSHTSSSR-DHTQSPRDHTQSPRDHAQSPRDHTQRSGFVPSDHPQQRNS 239
Q + A G+ T+S PR T PR + +P+D Q + P Q +S
Sbjct: 287 QHQMLAQVQAQGNMTNSPMYGGDMDPRRFTGLPRGNL-NPKDGQQNANDGSIGSPMQSSS 345
Query: 240 FRHRNGGPHQRGDGSHHHHNYGNRRDQDWNSRRNYNGRDMHVPPRVSPRIIRPSLPPNSA 299
+H + P Q+ H + Q+ N +R + + PS S
Sbjct: 346 SKHISMPPVQQSSSQQQDHLLSQQSQQN-NRKRKGPSSSGPANSTGTGNTVGPSNSQPST 404
Query: 300 PFIHPP 305
P H P
Sbjct: 405 PSTHTP 410
>At4g08410 extensin-like protein
Length = 707
Score = 36.2 bits (82), Expect = 0.045
Identities = 24/75 (32%), Positives = 33/75 (44%), Gaps = 3/75 (4%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPI--DSLRGVPFVP 343
SP++ SLPP PPP + + P V ++ PPPP S +G P
Sbjct: 118 SPKVDYKSLPPPYVYSSPPPPYYSPSPKVNYKSPPPPYV-YSSPPPPYYSPSPKGDYKSP 176
Query: 344 PMPLYYAGPDPQLHS 358
P P Y+ P P +S
Sbjct: 177 PPPYVYSSPPPPYYS 191
Score = 34.7 bits (78), Expect = 0.13
Identities = 22/74 (29%), Positives = 31/74 (41%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP+ PPP + + L P V + PPP V + PP
Sbjct: 93 SPKVDYKSPPPSYVYSSPPPPYYSPSPKVDYKSLPPPYVYSSPPPPYYSPSPKVNYKSPP 152
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 153 PPYVYSSPPPPYYS 166
Score = 31.6 bits (70), Expect = 1.1
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 493 SPKVDYKSPPPPYVYSFPPPPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKVNYKSPP 552
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 553 PPYVYSSPPPPYYS 566
Score = 31.6 bits (70), Expect = 1.1
Identities = 23/77 (29%), Positives = 33/77 (41%), Gaps = 7/77 (9%)
Query: 286 SPRIIRPSLPPNSAPFIH---PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF- 341
SP++ S PP P+I+ PPP + + P V + PPP V +
Sbjct: 568 SPKVEYKSPPP---PYIYSSPPPPYYAPSPKVDYKSPPPPYVYSSPPPPYYSPSPKVDYK 624
Query: 342 VPPMPLYYAGPDPQLHS 358
PP P Y+ P P +S
Sbjct: 625 SPPPPYVYSSPPPPYYS 641
Score = 31.2 bits (69), Expect = 1.5
Identities = 24/84 (28%), Positives = 36/84 (42%), Gaps = 21/84 (25%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPP-IDSLRGVPFV-- 342
SP++ S PP P+++ P P+ +P V + PPPP I S +P+
Sbjct: 393 SPKVDYKSPPP---PYVYSSPPPPYYS-------PSPKVDYKSPPPPYIYSSTPLPYYSP 442
Query: 343 --------PPMPLYYAGPDPQLHS 358
PP P Y+ P P +S
Sbjct: 443 SPKVDYKSPPPPYVYSSPPPPYYS 466
Score = 31.2 bits (69), Expect = 1.5
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 243 SPKVDYKSPPPPYVYSSPPPPYYSPSPKVNYKSPPPPYVYGSPPPPYYSPSPKVDYKSPP 302
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 303 PPYVYSSPPPPYYS 316
Score = 31.2 bits (69), Expect = 1.5
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 293 SPKVDYKSPPPPYVYSSPPPPYYSPSPKVNYKSPPPPYVYGSPPPPYYSPSPKVDYKSPP 352
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 353 PPYVYSSPPPPYYS 366
Score = 31.2 bits (69), Expect = 1.5
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 343 SPKVDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSTPPPYYSPSPKVDYKSPP 402
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 403 PPYVYSSPPPPYYS 416
Score = 30.8 bits (68), Expect = 1.9
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 593 SPKVDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKVNYKSPP 652
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 653 PPYVYSSPPPPYYS 666
Score = 30.4 bits (67), Expect = 2.5
Identities = 22/77 (28%), Positives = 35/77 (44%), Gaps = 7/77 (9%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAA--PVVLFAGPPPP--IDSLRGVPF 341
SP++ S PP P+++ P P+ E + P +++ PPPP S +
Sbjct: 543 SPKVNYKSPPP---PYVYSSPPPPYYSPSPKVEYKSPPPPYIYSSPPPPYYAPSPKVDYK 599
Query: 342 VPPMPLYYAGPDPQLHS 358
PP P Y+ P P +S
Sbjct: 600 SPPPPYVYSSPPPPYYS 616
Score = 30.4 bits (67), Expect = 2.5
Identities = 20/74 (27%), Positives = 29/74 (39%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 518 SPKVDYKSPPPPYVYSSPPPPYYSPSPKVNYKSPPPPYVYSSPPPPYYSPSPKVEYKSPP 577
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P ++
Sbjct: 578 PPYIYSSPPPPYYA 591
Score = 30.4 bits (67), Expect = 2.5
Identities = 22/77 (28%), Positives = 32/77 (40%), Gaps = 7/77 (9%)
Query: 286 SPRIIRPSLPPNSAPFIH---PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF- 341
SP++ S PP P+++ PPP + + P V + PPP V +
Sbjct: 268 SPKVNYKSPPP---PYVYGSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKVNYK 324
Query: 342 VPPMPLYYAGPDPQLHS 358
PP P Y P P +S
Sbjct: 325 SPPPPYVYGSPPPPYYS 341
Score = 30.4 bits (67), Expect = 2.5
Identities = 23/82 (28%), Positives = 32/82 (38%), Gaps = 2/82 (2%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + P V + PPP V + PP
Sbjct: 143 SPKVNYKSPPPPYVYSSPPPPYYSPSPKGDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPP 202
Query: 345 MPLYYAGPDPQLHSKIVNQIDY 366
P Y+ P P +S ++DY
Sbjct: 203 PPYVYSSPPPPYYSP-TPKVDY 223
Score = 30.0 bits (66), Expect = 3.3
Identities = 23/77 (29%), Positives = 33/77 (41%), Gaps = 7/77 (9%)
Query: 286 SPRIIRPSLPPNSAPFIH---PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFV 342
SP++ S PP P+I+ P P + + P V + PPP V +
Sbjct: 418 SPKVDYKSPPP---PYIYSSTPLPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKVDYK 474
Query: 343 PPMPLY-YAGPDPQLHS 358
PP P Y Y+ P P +S
Sbjct: 475 PPPPPYVYSSPPPPYYS 491
Score = 29.6 bits (65), Expect = 4.2
Identities = 23/80 (28%), Positives = 30/80 (36%), Gaps = 3/80 (3%)
Query: 282 PPRVSP--RIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGV 339
PP SP ++ S PP PPP + + P V + PPP V
Sbjct: 212 PPYYSPTPKVDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKV 271
Query: 340 PF-VPPMPLYYAGPDPQLHS 358
+ PP P Y P P +S
Sbjct: 272 NYKSPPPPYVYGSPPPPYYS 291
Score = 29.6 bits (65), Expect = 4.2
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 443 SPKVDYKSPPPPYVYSSPPPPYYSPSPKVDYKPPPPPYVYSSPPPPYYSPSPKVDYKSPP 502
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 503 PPYVYSFPPPPYYS 516
Score = 28.9 bits (63), Expect = 7.2
Identities = 21/74 (28%), Positives = 28/74 (37%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP+ S PP PPP + + P V + PPP V + PP
Sbjct: 168 SPKGDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPTPKVDYKSPP 227
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 228 PPYVYSSPPPPYYS 241
Score = 28.5 bits (62), Expect = 9.5
Identities = 23/81 (28%), Positives = 31/81 (37%), Gaps = 5/81 (6%)
Query: 282 PPRVSPRIIRPSLPPNSAPFIH---PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRG 338
PP SP PP P+++ PPP + + P V PPP
Sbjct: 462 PPYYSPSPKVDYKPP-PPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSFPPPPYYSPSPK 520
Query: 339 VPF-VPPMPLYYAGPDPQLHS 358
V + PP P Y+ P P +S
Sbjct: 521 VDYKSPPPPYVYSSPPPPYYS 541
Score = 28.5 bits (62), Expect = 9.5
Identities = 21/84 (25%), Positives = 33/84 (39%), Gaps = 6/84 (7%)
Query: 286 SPRIIRPSLPPNSAPFIH---PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPFV 342
SP++ S PP P+++ PPP + + P V + PPP V +
Sbjct: 318 SPKVNYKSPPP---PYVYGSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKVDYK 374
Query: 343 PPMPLYYAGPDPQLHSKIVNQIDY 366
P P Y P + ++DY
Sbjct: 375 SPPPPYVYSSTPPPYYSPSPKVDY 398
>At4g08400 extensin-like protein
Length = 513
Score = 36.2 bits (82), Expect = 0.045
Identities = 24/75 (32%), Positives = 33/75 (44%), Gaps = 3/75 (4%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPI--DSLRGVPFVP 343
SP++ SLPP PPP + + P V ++ PPPP S +G P
Sbjct: 100 SPKVDYKSLPPPYVYSSPPPPYYSPSPKVNYKSPPPPYV-YSSPPPPYYSPSPKGDYKSP 158
Query: 344 PMPLYYAGPDPQLHS 358
P P Y+ P P +S
Sbjct: 159 PPPYVYSSPPPPYYS 173
Score = 34.7 bits (78), Expect = 0.13
Identities = 22/74 (29%), Positives = 31/74 (41%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP+ PPP + + L P V + PPP V + PP
Sbjct: 75 SPKVDYKSPPPSYVYSSPPPPYYSPSPKVDYKSLPPPYVYSSPPPPYYSPSPKVNYKSPP 134
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 135 PPYVYSSPPPPYYS 148
Score = 33.1 bits (74), Expect = 0.38
Identities = 23/79 (29%), Positives = 30/79 (37%), Gaps = 2/79 (2%)
Query: 282 PPRVSP--RIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGV 339
PP SP ++ S PP PPP + + P V + PPP V
Sbjct: 194 PPYYSPTPKVDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKV 253
Query: 340 PFVPPMPLYYAGPDPQLHS 358
+ P P YY P P +S
Sbjct: 254 NYKSPPPPYYRSPPPPYYS 272
Score = 33.1 bits (74), Expect = 0.38
Identities = 26/85 (30%), Positives = 37/85 (42%), Gaps = 8/85 (9%)
Query: 286 SPRIIRPSLPPNSAPFIH---PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF- 341
SP++ S PP P+I+ PPP + + P V + PPP V +
Sbjct: 399 SPKVDYKSPPP---PYIYNSPPPPYYSPSPKVNYKTPPPPYVYSSPPPPYYSPSPKVNYK 455
Query: 342 VPPMPLYYAGPDPQLHSKIVNQIDY 366
PP P Y+ P P +S N +DY
Sbjct: 456 SPPPPYVYSSPPPPYYSPSPN-VDY 479
Score = 31.6 bits (70), Expect = 1.1
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 274 SPKVDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPTPKVDYKSPP 333
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 334 PPYVYSSPPPPYYS 347
Score = 30.8 bits (68), Expect = 1.9
Identities = 21/77 (27%), Positives = 33/77 (42%), Gaps = 7/77 (9%)
Query: 286 SPRIIRPSLPPNSAPFIH---PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF- 341
SP++ S PP P+++ PPP + + P + + PPP V +
Sbjct: 374 SPKVDYKSPPP---PYVYNSPPPPYYSPSPKVDYKSPPPPYIYNSPPPPYYSPSPKVNYK 430
Query: 342 VPPMPLYYAGPDPQLHS 358
PP P Y+ P P +S
Sbjct: 431 TPPPPYVYSSPPPPYYS 447
Score = 30.4 bits (67), Expect = 2.5
Identities = 21/74 (28%), Positives = 29/74 (38%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 299 SPKVDYKSPPPPYVYSSPPPPYYSPTPKVDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPP 358
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 359 PPYVYSSPPPPYYS 372
Score = 30.4 bits (67), Expect = 2.5
Identities = 23/82 (28%), Positives = 32/82 (38%), Gaps = 2/82 (2%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + P V + PPP V + PP
Sbjct: 125 SPKVNYKSPPPPYVYSSPPPPYYSPSPKGDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPP 184
Query: 345 MPLYYAGPDPQLHSKIVNQIDY 366
P Y+ P P +S ++DY
Sbjct: 185 PPYVYSSPPPPYYSP-TPKVDY 205
Score = 30.0 bits (66), Expect = 3.3
Identities = 24/84 (28%), Positives = 35/84 (41%), Gaps = 7/84 (8%)
Query: 286 SPRIIRPSLPPNSAPFIH--PPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-V 342
SP++ S PP P+ PPP + + P V + PPP V +
Sbjct: 250 SPKVNYKSPPP---PYYRSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKVDYKS 306
Query: 343 PPMPLYYAGPDPQLHSKIVNQIDY 366
PP P Y+ P P +S ++DY
Sbjct: 307 PPPPYVYSSPPPPYYSP-TPKVDY 329
Score = 29.6 bits (65), Expect = 4.2
Identities = 21/74 (28%), Positives = 28/74 (37%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP++ S PP PPP + + P V + PPP V + PP
Sbjct: 349 SPKVDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYNSPPPPYYSPSPKVDYKSPP 408
Query: 345 MPLYYAGPDPQLHS 358
P Y P P +S
Sbjct: 409 PPYIYNSPPPPYYS 422
Score = 29.3 bits (64), Expect = 5.5
Identities = 23/80 (28%), Positives = 30/80 (36%), Gaps = 3/80 (3%)
Query: 282 PPRVSP--RIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGV 339
PP SP ++ S PP PPP + + P V + PPP V
Sbjct: 318 PPYYSPTPKVDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPSPKV 377
Query: 340 PF-VPPMPLYYAGPDPQLHS 358
+ PP P Y P P +S
Sbjct: 378 DYKSPPPPYVYNSPPPPYYS 397
Score = 28.9 bits (63), Expect = 7.2
Identities = 21/74 (28%), Positives = 28/74 (37%), Gaps = 1/74 (1%)
Query: 286 SPRIIRPSLPPNSAPFIHPPPLRPFGGHMGFHELAAPVVLFAGPPPPIDSLRGVPF-VPP 344
SP+ S PP PPP + + P V + PPP V + PP
Sbjct: 150 SPKGDYKSPPPPYVYSSPPPPYYSPSPKVDYKSPPPPYVYSSPPPPYYSPTPKVDYKSPP 209
Query: 345 MPLYYAGPDPQLHS 358
P Y+ P P +S
Sbjct: 210 PPYVYSSPPPPYYS 223
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.130 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,369,997
Number of Sequences: 26719
Number of extensions: 679384
Number of successful extensions: 3223
Number of sequences better than 10.0: 194
Number of HSP's better than 10.0 without gapping: 39
Number of HSP's successfully gapped in prelim test: 160
Number of HSP's that attempted gapping in prelim test: 2280
Number of HSP's gapped (non-prelim): 668
length of query: 522
length of database: 11,318,596
effective HSP length: 104
effective length of query: 418
effective length of database: 8,539,820
effective search space: 3569644760
effective search space used: 3569644760
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC123571.8


