

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122730.8 + phase: 0 /pseudo
(739 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
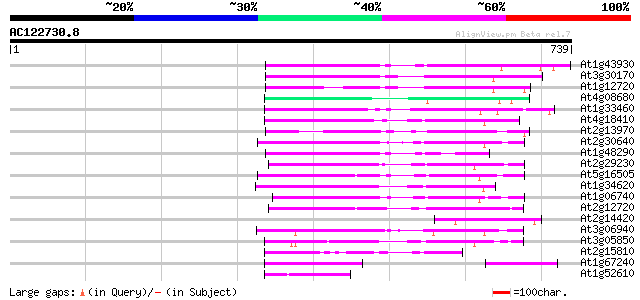
Score E
Sequences producing significant alignments: (bits) Value
At1g43930 hypothetical protein 167 1e-41
At3g30170 unknown protein 164 2e-40
At1g12720 hypothetical protein 140 3e-33
At4g08680 putative MuDR-A-like transposon protein 136 4e-32
At1g33460 mutator transposase MUDRA, putative 130 2e-30
At4g18410 MuDR transposable element - like protein 125 1e-28
At2g13970 Mutator-like transposase 124 2e-28
At2g30640 Mutator-like transposase 122 7e-28
At1g48290 hypothetical protein 118 1e-26
At2g29230 Mutator-like transposase 117 3e-26
At5g16505 mutator-like transposase-like protein (MQK4.25) 111 1e-24
At1g34620 Mutator-like protein 107 2e-23
At1g06740 mudrA-like protein 105 9e-23
At2g12720 putative protein 103 5e-22
At2g14420 Mutator-like transposase 102 8e-22
At3g06940 putative mudrA protein 100 2e-21
At3g05850 unknown protein 99 7e-21
At2g15810 Mutator-like transposase 96 9e-20
At1g67240 hypothetical protein 87 4e-17
At1g52610 hypothetical protein 73 7e-13
>At1g43930 hypothetical protein
Length = 946
Score = 167 bits (424), Expect = 1e-41
Identities = 121/424 (28%), Positives = 189/424 (44%), Gaps = 59/424 (13%)
Query: 337 GLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTAFDREMNEIKK 396
GL+ A+ ++P A++R C RH+ N+++ +L+ L W+ ++ F M +K
Sbjct: 508 GLVKAIHSVIPQAEHRQCARHIMENWKRNSHDMELQRLFWKIVRSYTEGEFGAHMRALKS 567
Query: 397 ISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDIRV 456
+ A+ LL+T WS++ F G C+ +NN++E+FN I R+KP++ MLEDIR
Sbjct: 568 YNASAFELLLKTLPVTWSRAFFKIGSCCNDNLNNLSESFNRTIREARRKPLLDMLEDIRR 627
Query: 457 YLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFLFV 516
M R EK +I K + G QVC++ +
Sbjct: 628 QCMVRNEKRYVIADRLRTRFTKRAHAEIEKMI-----AGSQVCQRWM------------- 669
Query: 517 LYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRKWLLTGLPCCHAI 576
A +EI+ + DS+++++ C C KW +TG+PC HA
Sbjct: 670 -----------------ARHNKHEIKVGPV--DSFTVDMNNNTCGCMKWQMTGIPCIHAA 710
Query: 577 SCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPVNGQSLWRRTEYTDLQPPPIKR-QPGR 635
S + + V+ YVSD + ++ Y+ I PV G+ LW + PPP +R PGR
Sbjct: 711 SVIIGKRQKVEDYVSDWYTTSMWKQTYNDGIGPVQGKLLWPTVNKVGVLPPPWRRGNPGR 770
Query: 636 PKKN-RRKDVDE-----KRDEQQLKKAKYGMKCSRCKAEGHNKSTCKLPP-PPPQPEPGT 688
PK + RRK V E +L + M CS C+ EGHNK CK PP P
Sbjct: 771 PKNHARRKGVFESSTASSSSTTELSRLHRVMTCSNCQGEGHNKQGCKNETVAPPAKRPRG 830
Query: 689 QPPATQFPT----------SQPTSDHAPTTQVSATQ----ASASQVSTTQFVAAKVSSGT 734
+P + P +P + AP++ + +Q A SQ S S G
Sbjct: 831 RPRKDKDPRMVFFGQGQTWQEPQAQGAPSSHGAPSQVFHGAPLSQPSQEAPSQGAPSQGA 890
Query: 735 QSVA 738
QS++
Sbjct: 891 QSLS 894
>At3g30170 unknown protein
Length = 995
Score = 164 bits (415), Expect = 2e-40
Identities = 108/372 (29%), Positives = 170/372 (45%), Gaps = 44/372 (11%)
Query: 337 GLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTAFDREMNEIKK 396
GL+ A+ +LP A++R C +H+ +N+++ +L+ L W+ A++ F+ M E+++
Sbjct: 480 GLVKAIHTILPHAEHRQCSKHIMDNWKRDSHDLELQRLFWKIARSYTVEEFNNHMMELQQ 539
Query: 397 ISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDIRV 456
+ +AY L T WS++ F G C+ +NN++E+FN I R+KPI+ MLEDIR
Sbjct: 540 YNRQAYESLQLTSPVTWSRAFFRIGTCCNDNLNNLSESFNRTIRQARRKPILDMLEDIRR 599
Query: 457 YLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFLFV 516
M R K ++I K + +G + C + + N + IY
Sbjct: 600 QCMVRSAKRFLIAEKLQTRFTKRAHEEIEKMI-----VGLRQCERYMARENLHEIY---- 650
Query: 517 LYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRKWLLTGLPCCHAI 576
+N SY +++ +K C CRK + G+PC HA
Sbjct: 651 -----------------------------VNNVSYFVDMEQKTCDCRKCQMVGIPCIHAT 681
Query: 577 SCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPVNGQSLWRRTEYTDLQPPPIKR-QPGR 635
+ + V+ YVSD + K R+ Y I PV G LW RT + PPP +R GR
Sbjct: 682 CVIIGKKEKVEDYVSDYYTKVRWRETYLKGIRPVQGMPLWCRTNRLPVLPPPWRRGNAGR 741
Query: 636 P----KKNRRKDVDEKRDEQQLKKAKYGMKCSRCKAEGHNKSTCKLPPP-PPQPEPGTQP 690
P ++ R + + ++ + K M CS C EGHNK C +P P P +P
Sbjct: 742 PSNYARRKGRNEAAAPSNPNKMSREKRIMTCSNCHQEGHNKQRCNIPTVLNPTKRPRGRP 801
Query: 691 PATQFPTSQPTS 702
Q P S S
Sbjct: 802 RKNQQPQSHQGS 813
>At1g12720 hypothetical protein
Length = 585
Score = 140 bits (353), Expect = 3e-33
Identities = 99/360 (27%), Positives = 151/360 (41%), Gaps = 72/360 (20%)
Query: 337 GLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTAFDREMNEIKK 396
GL+ A+ +LP A++R C +H+ +N+++ +L+ L W+ +++ F+ M +K
Sbjct: 190 GLVKAIHNVLPQAEHRQCSKHIMDNWKRDSHDMELQRLFWKISRSYTIEEFNTHMANLKS 249
Query: 397 ISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDIRV 456
+ +AY L T W+ I R+KP++ MLEDIR
Sbjct: 250 YNPQAYASLQLTSPMTWT------------------------IRQARRKPLLDMLEDIRR 285
Query: 457 YLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFLFV 516
M R K P +I K + G C + L N + IY
Sbjct: 286 QCMVRTAKRFIIAERLKSRFTPRAHAEIEKMI-----AGSAGCERHLARNNLHEIY---- 336
Query: 517 LYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRKWLLTGLPCCHAI 576
+N Y +++ +K C CRKW + G+PC H
Sbjct: 337 -----------------------------VNDVGYFVDMDKKTCGCRKWEMVGIPCVHTP 367
Query: 577 SCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPVNGQSLWRRTEYTDLQPPPIKR-QPGR 635
+ + V+ YVSD + K R+ Y I PV G LW R + PPP +R PGR
Sbjct: 368 CVIIGRKEKVEDYVSDYYTKVRWRETYRDGIRPVQGMPLWPRMSRLPVLPPPWRRGNPGR 427
Query: 636 ----PKKNRRKDVDEKRDEQQLKKAKYGMKCSRCKAEGHNKSTCK-----LPPPPPQPEP 686
+K R + ++ ++ +A M CS CK EGHNKS+CK LPPP P+ P
Sbjct: 428 QSNYARKKGRYETASSSNKNKMSRANRIMTCSNCKQEGHNKSSCKNATVLLPPPRPRGRP 487
>At4g08680 putative MuDR-A-like transposon protein
Length = 761
Score = 136 bits (343), Expect = 4e-32
Identities = 92/373 (24%), Positives = 151/373 (39%), Gaps = 70/373 (18%)
Query: 336 QGLLPALAELLPDADNRFCVRHLYNNFRKKYPGKK-LKELMWRAAKATYTTAFDREMNEI 394
+GLL A+ + LP+A++R CV+H+ N +K + K LK L+W+ A + + + +N +
Sbjct: 376 KGLLSAVKQELPNAEHRMCVKHIVENLKKNHAKKDMLKTLVWKLAWSYNEKEYGKNLNNL 435
Query: 395 KKISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDI 454
+ E YN +L WS+ + G C+ + NN TE+FNS I R K ++ MLE I
Sbjct: 436 RCYDEALYNDVLNEEPHTWSRCFYKLGSCCEDVDNNATESFNSTITKARAKSLIPMLETI 495
Query: 455 RVYLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFL 514
R M R K +K +
Sbjct: 496 RRQGMTRIVKRNKKSLRHEGRFTK------------------------------------ 519
Query: 515 FVLYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNG--------DSYSINLLRKECSCRKWL 566
YA +L + D +R +G + + +++ + +CSC KW
Sbjct: 520 ----------YALKMLALEKTDADRSKVYRCTHGVFEVYIDENGHRVDIPKTQCSCGKWQ 569
Query: 567 LTGLPCCHAISCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPVNGQSLWRRTEYTDLQP 626
++G+PC H+ M +D + Y+S+ F + + Y P+ G W + Y +
Sbjct: 570 ISGIPCEHSYGAMIEAGLDAENYISEFFSTDLWRDSYETATMPLRGPKYWLNSSYGLVTA 629
Query: 627 PPIKRQPGRPKKNRRKD----VDEKRDEQQLKKAKYG-----------MKCSRCKAEGHN 671
PP PGR K+ +K+ + K + + KK K + C C GHN
Sbjct: 630 PPEPILPGRKKEKSKKEKFARIKGKNESPKKKKRKKNEVEKLGKKGKIIHCKSCGEAGHN 689
Query: 672 KSTCKLPPPPPQP 684
CK P P
Sbjct: 690 ALRCKKFPKEKVP 702
>At1g33460 mutator transposase MUDRA, putative
Length = 826
Score = 130 bits (328), Expect = 2e-30
Identities = 109/402 (27%), Positives = 179/402 (44%), Gaps = 62/402 (15%)
Query: 336 QGLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKL-KELMWRAAKATYTTAFDREMNEI 394
+G++ A+ LP+A++R CV+H+ N +K++ L K+ +W A + T + +NE+
Sbjct: 442 KGIISAVKNALPNAEHRPCVKHIVENLKKRHGSLDLLKKFVWNLAWSYSDTQYKANLNEM 501
Query: 395 KKISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDI 454
+ Y +++ W +S F G C+ + NN TE+FN+ IV R K +V M+E I
Sbjct: 502 RAYIMSLYEDVMKEEPNTWCRSWFRIGSYCEDVDNNATESFNATIVKARAKALVPMMETI 561
Query: 455 RVYLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFL 514
R M R K + K+ + +KKI + V ++ +++
Sbjct: 562 RRQAMARISKRKAKIGKW--------KKKISEYV-------SEILKEE------------ 594
Query: 515 FVLYPLV*HLYAWNVLVR-RAIEYDYEIRHRSMNGDSYSINLLRKE--CSCRKWLLTGLP 571
W + V+ + +E +G+S +++L +E CSC KW +TG+P
Sbjct: 595 ------------WELAVKCEVTKGTHEKFEVWFDGNSNAVSLKTEEWDCSCCKWQVTGIP 642
Query: 572 CCHAISCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPVNGQSLWRR---TEYTDLQPP- 627
C HA + + DV+ +V F + Y PV GQ W LQ P
Sbjct: 643 CEHAYAAINDVGKDVEDFVIPMFYTIAWREQYDTGPDPVRGQMYWPTGLGLITAPLQDPV 702
Query: 628 PIKRQPGRPKKNRR----KDVDEKRDEQQLKKAKYG--MKCSRCKAEGHNKSTCKLPPPP 681
P R+ G K R + +K+ Q+LK K G M C C GHN + CK
Sbjct: 703 PPGRKKGEKKNYHRIKGPNESPKKKGPQRLKVGKKGVVMHCKSCGEAGHNAAGCK---KF 759
Query: 682 PQPEPGTQPPATQFP--TSQPTSDHAPTTQ----VSATQASA 717
P+ + G + + TSQPT + TT ++ TQ S+
Sbjct: 760 PKEKKGRKKKNQEIKAGTSQPTMELQETTHGADTITLTQRSS 801
>At4g18410 MuDR transposable element - like protein
Length = 633
Score = 125 bits (313), Expect = 1e-28
Identities = 87/342 (25%), Positives = 149/342 (43%), Gaps = 44/342 (12%)
Query: 336 QGLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKL-KELMWRAAKATYTTAFDREMNEI 394
+GL+ A+ LP ++R CV+H+Y N +K Y K + K L+W A + T + + + +I
Sbjct: 303 KGLIKAVQLELPKIEHRMCVQHIYGNLKKTYGSKTMIKPLLWNLAWSYNETEYKQHLEKI 362
Query: 395 KKISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDI 454
+ + Y +++T + W ++ G C+ + NN E+FN + R+K V MLE I
Sbjct: 363 RCYDTKVYESVMKTNPRSWVRAFQKIGSFCEDVDNNSVESFNGSLNKAREKQFVAMLETI 422
Query: 455 RVYLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFL 514
R M R K + + P + K L G+
Sbjct: 423 RRMAMVRIAKRSVESHTHTGVCTPYV----------MKFLAGE----------------- 455
Query: 515 FVLYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRKWLLTGLPCCH 574
H A V YE+RH GD++ ++L C+C KW + G+PC H
Sbjct: 456 --------HKVASTAKVAPGTNGMYEVRH---GGDTHRVDLAAYTCTCIKWQICGIPCEH 504
Query: 575 AISCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPVNGQSLWRRTEYTDL---QPPPIK- 630
A + + + + +V FR ++ Y+ ++P G W + + +PP +
Sbjct: 505 AYGVILHKKLQPEDFVCQWFRTAMWKKNYTDGLFPQRGPKFWPESNGPRVFAAEPPEGEE 564
Query: 631 -RQPGRPKKNRRKDVDEKRDEQQLKKAKYGMKCSRCKAEGHN 671
++ + +K R+K V+E ++Q K K M C C A HN
Sbjct: 565 DKKMTKAEKKRKKGVNESPTKKQPKAKKRIMHCGVCGAADHN 606
>At2g13970 Mutator-like transposase
Length = 597
Score = 124 bits (311), Expect = 2e-28
Identities = 95/352 (26%), Positives = 146/352 (40%), Gaps = 83/352 (23%)
Query: 337 GLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTAFDREMNEIKK 396
GL A+ ++P A++R RH+ +N++K +L+ L W+ T
Sbjct: 190 GLGKAIHTVIPQAEHRQYARHIMDNWKKNSHDMELQRLFWKTNPLT-------------- 235
Query: 397 ISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDIRV 456
WS++ F G C+ +NN++E+FN I R+KP++ MLEDIR
Sbjct: 236 ----------------WSRAFFRIGSCCNDNLNNLSESFNRTIRNARRKPLLDMLEDIRR 279
Query: 457 YLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFLFV 516
M R EK +I K + G VC + + N
Sbjct: 280 QCMVRNEKRFVIAYRLKSRFTKRAHMEIEKMID-----GAAVCDRWMARHN--------- 325
Query: 517 LYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRKWLLTGLPCCHAI 576
+ VR ++ DS+S+++ + C CRKW +TG+PC HA
Sbjct: 326 -----------RIEVRVGLD------------DSFSVDMNDRTCGCRKWQMTGIPCIHAA 362
Query: 577 SCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPVNGQSLWRRTEYTDLQPPPIKR-QPGR 635
S + V+ +VSD + ++ Y I PV G+ LW + PPP +R PGR
Sbjct: 363 SVIIGNRQKVEDFVSDWYTTYMWKQTYYDGIMPVQGKMLWPIVNRVGVLPPPWRRGNPGR 422
Query: 636 PKKNRRKDVDEKRDEQQLKKAKYGMKCSRCKAEGHNKSTCK---LPPPPPQP 684
PK + R+ K Y + EGHNK+ CK + PPP +P
Sbjct: 423 PKNHDRR------------KGIYESATASTSREGHNKAGCKNETVAPPPKRP 462
>At2g30640 Mutator-like transposase
Length = 754
Score = 122 bits (306), Expect = 7e-28
Identities = 95/362 (26%), Positives = 155/362 (42%), Gaps = 56/362 (15%)
Query: 327 KLMTYPS*MQGLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTA 386
KL S Q ++ + P A + CV L + R ++ L L+W AAK
Sbjct: 438 KLTILSSRDQSIVDGVDTNFPTAFHGLCVHCLTESVRTQFNNSILVNLVWEAAKCLTDFE 497
Query: 387 FDREMNEIKKISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKP 446
F+ +M EI +IS EA + + W+ F G L N++E+ NS + P
Sbjct: 498 FEGKMGEIAQISPEAASWIRNIQHSQWATYCFE-GTRFGHLTANVSESLNSWVQDASGLP 556
Query: 447 IVTMLEDIRVYLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TL 506
I+ MLE IR LM + + R+ ++ ++P+ + + + + + CR
Sbjct: 557 IIQMLESIRRQLMTLFNERRETSMQWSGMLVPSAERHVLEAI--------EECR------ 602
Query: 507 N*Y*IYFLFVLYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRKWL 566
LYP V +A E +E+ + + +++ + C CR W
Sbjct: 603 ----------LYP-----------VHKANEAQFEVM---TSEGKWIVDIRCRTCYCRGWE 638
Query: 567 LTGLPCCHAISCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPVNGQSLWRRTEYTD--- 623
L GLPC HA++ + + +V ++ F Y Y+ I+PV ++ W+ TE
Sbjct: 639 LYGLPCSHAVAALLACRQNVYRFTESYFTVANYRRTYAETIHPVPDKTEWKTTEPAGESG 698
Query: 624 -------LQPPPIKRQPGRPKKNRRKDVDEKRDEQQLKKAKYGMKCSRCKAEGHNKSTCK 676
++PP RQP RPKK R + D R K ++CSRC GH ++TC
Sbjct: 699 EDGEEIVIRPPRDLRQPPRPKKRRSQGEDRGRQ-------KRVVRCSRCNQAGHFRTTCT 751
Query: 677 LP 678
P
Sbjct: 752 AP 753
>At1g48290 hypothetical protein
Length = 444
Score = 118 bits (296), Expect = 1e-26
Identities = 78/295 (26%), Positives = 128/295 (42%), Gaps = 55/295 (18%)
Query: 337 GLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTAFDREMNEIKK 396
GL+ A+ +P A++R C +H+ +N+++ +L+ L W+ A++ + + E+K
Sbjct: 164 GLVKAIHNRIPAAEHRQCAKHIMDNWKRNSHDMELQRLFWKIARSYTIGEYTANLEELKT 223
Query: 397 ISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDIRV 456
+ A L+ T WS++ F G C+ +NN++E+FN I R+KP++ MLEDIR
Sbjct: 224 YNPGAAASLMNTKPMEWSRAFFRIGSCCNDNLNNLSESFNRTIRQARRKPLLDMLEDIRS 283
Query: 457 YLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFLFV 516
M R EK + ++I K + G Q C + +
Sbjct: 284 QCMVRNEKRYIIAGRWKSRFTKRAHEEIEKMI-----AGSQFCERSM------------- 325
Query: 517 LYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRKWLLTGLPCCHAI 576
A +EI H G YS+++ C CRKW +T
Sbjct: 326 -----------------ARHNKHEISH---FGRKYSVDMNDNTCGCRKWQMT-------- 357
Query: 577 SCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPVNGQSLWRRTEYTDLQPPPIKR 631
V+ YVSD + +++ Y+ I PV GQ LW R + PPP +R
Sbjct: 358 ---------VEDYVSDWYTTRMWQLTYNDGIAPVQGQLLWPRVNRLGVLPPPWRR 403
>At2g29230 Mutator-like transposase
Length = 915
Score = 117 bits (292), Expect = 3e-26
Identities = 90/341 (26%), Positives = 145/341 (42%), Gaps = 47/341 (13%)
Query: 342 LAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTAFDREMNEIKKISEEA 401
L + P A + C+ HL N Y K L + RAA+A F NE+ ++
Sbjct: 617 LRRVYPKAKHGACIVHLQRNIATSYKKKHLLFHVSRAARAFRICEFHTYFNEVIRLDPAC 676
Query: 402 YNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDIRVYLMER 461
+L HW+++ F G + + +N+ E+ N+V+ R+ PI+++LE IR L+
Sbjct: 677 ARYLESVGFCHWTRAYFL-GKRYNVMTSNVAESLNAVLKEARELPIISLLEFIRTTLISW 735
Query: 462 WEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFLFVLYPLV 521
+ R+ A + P +R+ + + F K +
Sbjct: 736 FAMRREAARTEASPLPPKMREVVHRN--FEKSV--------------------------- 766
Query: 522 *HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRKWLLTGLPCCHAISCMKS 581
V R YDYEIR + Y + L+ + CSCR + L LPC HAI+ +
Sbjct: 767 ------RFAVHRLDRYDYEIREEGAS--VYHVKLMERTCSCRAFDLLHLPCPHAIAAAVA 818
Query: 582 QDIDVDQYVSDCFRKERYEICYSHIIYPV----NGQSLWRRTEYTDLQPPPIKRQPGRPK 637
+ + + ++ + E + + Y I PV + +L L PP +R GRPK
Sbjct: 819 EGVPIQGLMAPEYSVESWRMSYLGTIKPVPEVGDVFALPEPIASLRLFPPATRRPSGRPK 878
Query: 638 KNRRKDVDEKRDEQQLKKAKYGMKCSRCKAEGHNKSTCKLP 678
K R E Q + +CSRC GHN++TCK+P
Sbjct: 879 KKRITSRGEFTCPQ-----RQVTRCSRCTGAGHNRATCKMP 914
>At5g16505 mutator-like transposase-like protein (MQK4.25)
Length = 597
Score = 111 bits (278), Expect = 1e-24
Identities = 90/361 (24%), Positives = 153/361 (41%), Gaps = 56/361 (15%)
Query: 327 KLMTYPS*MQGLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTA 386
KL ++ A+ P A + FC+R++ NFR + KL + W A A
Sbjct: 283 KLTILSERQSAVVEAVETHFPTAFHGFCLRYVSENFRDTFKNTKLVNIFWSAVYALTPAE 342
Query: 387 FDREMNEIKKISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKP 446
F+ + NE+ +IS++ W+ + F G +TE + + + P
Sbjct: 343 FETKSNEMIEISQDVVQWFELYLPHLWAVAYFQ-GVRYGHFGLGITEVLYNWALECHELP 401
Query: 447 IVTMLEDIRVYLMERWEKNRQKVANYADNIL-PNIRKKI*KRV*FYKQLGGQVCRQQL*T 505
I+ M+E IR + + W NR++++ ++IL P+ ++I + V C Q L
Sbjct: 402 IIQMMEHIR-HQISSWFDNRRELSMGWNSILVPSAERRITEAV------ADARCYQVL-- 452
Query: 506 LN*Y*IYFLFVLYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRKW 565
RA E ++EI +++ ++CSCR+W
Sbjct: 453 ---------------------------RANEVEFEIVSTERTN---IVDIRTRDCSCRRW 482
Query: 566 LLTGLPCCHAISCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPVNGQSLWR-------- 617
+ GLPC HA + + S +V + CF Y+ YS +I + +SLW+
Sbjct: 483 QIYGLPCAHAAAALISCGRNVHLFAEPCFTVSSYQQTYSQMIGEIPDRSLWKDEGEEAGG 542
Query: 618 RTEYTDLQPPPIKRQPGRPKKNRRKDVDEKRDEQQLKKAKYGMKCSRCKAEGHNKSTCKL 677
E ++PP +R PGRPKK + + LK+ K ++C RC GH++ C
Sbjct: 543 GVESLRIRPPKTRRPPGRPKKKVVR-------VENLKRPKRIVQCGRCHLLGHSQKKCTQ 595
Query: 678 P 678
P
Sbjct: 596 P 596
>At1g34620 Mutator-like protein
Length = 1034
Score = 107 bits (267), Expect = 2e-23
Identities = 75/321 (23%), Positives = 145/321 (44%), Gaps = 43/321 (13%)
Query: 325 NTKLMTYPS*MQGLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYT 384
N LM + + L+++ A++ C HL+ N R Y K L LM AA+A +
Sbjct: 621 NPDLMFISDRHESIYTGLSKVYTQANHAACTVHLWRNIRHLYKPKSLCRLMSEAAQAFHV 680
Query: 385 TAFDREMNEIKKISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRK 444
T F+R +I+K++ +L+ W++ S G + + +N+ E++N+VI R+
Sbjct: 681 TDFNRIFLKIQKLNPGCAAYLVDLGFSEWTR-VHSKGRRFNIMDSNICESWNNVIREARE 739
Query: 445 KPIVTMLEDIRVYLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL* 504
P++ MLE IR LM+ + R + + + P +++++ +
Sbjct: 740 YPLICMLEYIRTTLMDWFATRRAQAEDCPTTLAPRVQERVEEN----------------- 782
Query: 505 TLN*Y*IYFLFVLYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRK 564
+ A ++ V+ ++++++ R+ G+ + + L CSC +
Sbjct: 783 ------------------YQSAMSMSVKPICNFEFQVQERT--GECFIVKLDESTCSCLE 822
Query: 565 WLLTGLPCCHAISCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPVNGQSLWRRTEYT-- 622
+ G+PC HAI+ + D ++ + E ++ Y IYP++ E
Sbjct: 823 FQGLGIPCAHAIAAAARIGVPTDSLAANGYFNELVKLSYEEKIYPIHSVGGEVAPEIAAG 882
Query: 623 ---DLQPPPIKRQPGRPKKNR 640
+LQPP ++R PGRP+K R
Sbjct: 883 TTGELQPPFVRRPPGRPRKIR 903
>At1g06740 mudrA-like protein
Length = 726
Score = 105 bits (262), Expect = 9e-23
Identities = 77/337 (22%), Positives = 140/337 (40%), Gaps = 52/337 (15%)
Query: 347 PDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTAFDREMNEIKKISEEAYNHLL 406
P A + FC+ +L F++++ L +L W AA F ++N+I++IS EA +
Sbjct: 436 PAAFHGFCLHYLTERFQREFQSSVLVDLFWEAAHCLTVLEFKSKINKIEQISPEASLWIQ 495
Query: 407 QTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDIRVYLMERWEKNR 466
W+ S F N +TE+ ++ + PI+ +E I +L+ ++ R
Sbjct: 496 NKSPARWASSYFEGTRFGQLTANVITESLSNWVEDTSGLPIIQTMECIHRHLINMLKERR 555
Query: 467 QKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFLFVLYPLV*HLYA 526
+ ++++ ++P+ K++ + + R
Sbjct: 556 ETSLHWSNVLVPSAEKQMLAAI--------EQSRAHR----------------------- 584
Query: 527 WNVLVRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRKWLLTGLPCCHAISCMKSQDIDV 586
V RA E ++E+ N +N+ C C +W + GLPC HA+ + S + DV
Sbjct: 585 ----VYRANEAEFEVMTCEGN---VVVNIENCSCLCGRWQVYGLPCSHAVGALLSCEEDV 637
Query: 587 DQYVSDCFRKERYEICYSHIIYPVNGQSLWR-----RTEYTDLQPPPIKRQPGRPKKNRR 641
+Y CF E Y Y+ + P++ + W+ R ++ P K G P+K R
Sbjct: 638 YRYTESCFTVENYRRAYAETLEPISDKVQWKENDSERDSENVIKTP--KAMKGAPRKRRV 695
Query: 642 KDVDEKRDEQQLKKAKYGMKCSRCKAEGHNKSTCKLP 678
+ D R + + C RC GH ++TC P
Sbjct: 696 RAEDRDR-------VRRVVHCGRCNQTGHFRTTCTAP 725
>At2g12720 putative protein
Length = 819
Score = 103 bits (256), Expect = 5e-22
Identities = 85/338 (25%), Positives = 139/338 (40%), Gaps = 44/338 (13%)
Query: 341 ALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTAFDREMNEIKKISEE 400
A+ E+ P A C HLY N K+ K L L+ +AA+ F NEI+++
Sbjct: 523 AIGEVYPLAARGICTYHLYKNILVKFKRKDLFPLVKKAARCYRLNDFTNAFNEIEELDPL 582
Query: 401 AYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDIRVYLME 460
+ +L + V+ W+++ F G + + N+ E+ N + + PIV MLE IR +M
Sbjct: 583 LHAYLQRAGVEMWARAHFP-GDRYNLMTTNIAESMNRALSQAKNLPIVRMLEAIR-QMMT 640
Query: 461 RW-EKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFLFVLYP 519
RW + R + + P + K + RV + L Q
Sbjct: 641 RWFAERRDDASKQHTQLTPGVEKLLQTRVTSSRLLDVQ---------------------- 678
Query: 520 LV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLLRKECSCRKWLLTGLPCCHAISCM 579
+ ++ YE + +N+ K+C+CR + LPC HAI+
Sbjct: 679 ---------TIDASRVQVAYE-------ASLHVVNVDEKQCTCRLFNKEKLPCIHAIAAA 722
Query: 580 KSQDIDVDQYVSDCFRKERYEICYSHIIYPVNGQ-SLWRRTEYTDLQPPPIKRQPGRPKK 638
+ + S ++ Y+ I P + + + + PP + QPGRPKK
Sbjct: 723 EHMGVSRISLCSPYYKSSHLVNVYACAIMPSDTEVPVPQIVIDQPCLPPIVANQPGRPKK 782
Query: 639 NRRKDVDEKRDEQQLKKAKYGMKCSRCKAEGHNKSTCK 676
R K E E + + ++ CSRCK GHN TC+
Sbjct: 783 LRMKSALEVAVETKRPRKEHA--CSRCKETGHNVKTCR 818
>At2g14420 Mutator-like transposase
Length = 241
Score = 102 bits (254), Expect = 8e-22
Identities = 56/154 (36%), Positives = 85/154 (54%), Gaps = 13/154 (8%)
Query: 560 CSCRKWLLTGLPCCHAISCMKSQDIDV---DQYVSDCFRKERYEICYSHIIYPVNGQSLW 616
C+CR + ++G+PCCH +S M + + + VSD + E++++CYS + +PVNG LW
Sbjct: 81 CTCRAYDVSGIPCCHIMSAMWEEYKETKLPETVVSDWYSIEKWKLCYSSLFFPVNGMELW 140
Query: 617 RRTEYTDLQPPPIKRQPGRPKKNRR-KDVDEKRDEQQL--KKAKYGMKCSRCKAEGHNKS 673
+ + PPP + PGRPK N R +D E R Q++ + K M CS C+ GHNK
Sbjct: 141 KTHIDVVVMPPPDRIMPGRPKNNDRIRDPTEDRPPQKVPSTREKLQMTCSNCQQIGHNKR 200
Query: 674 TCKLPP-PPPQPEPGTQP------PATQFPTSQP 700
+CK P P +P +P A + P +QP
Sbjct: 201 SCKRKAVPKPSKKPQGRPRKKQKTMAEENPITQP 234
>At3g06940 putative mudrA protein
Length = 749
Score = 100 bits (250), Expect = 2e-21
Identities = 87/367 (23%), Positives = 155/367 (41%), Gaps = 66/367 (17%)
Query: 326 TKLMTYPS*MQ-GLLPALAELLP-DADNRFCVRHLYNNFRKKYPGKKLKEL-------MW 376
++++T+ + Q GL A+A++ DA + +C+ L G+ E +
Sbjct: 431 SRIITFVADFQNGLKNAIAQVFEKDAHHAYCLGQLAEKLNVDLKGQFSHEARRYMLNDFY 490
Query: 377 RAAKATYTTAFDREMNEIKKISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFN 436
A AT + + IK IS +AYN ++++ HW+ + F G + + +N + F
Sbjct: 491 SVAYATTPVGYYLALENIKSISPDAYNWVIESEPHHWANALFQ-GERYNKMNSNFGKDFY 549
Query: 437 SVIVVLRKKPIVTMLEDIRVYLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGG 496
S + + PI M+++ R +M+ + K + + P+ +K+ K + + L
Sbjct: 550 SWVSEAHEFPITQMIDEFRAKMMQSIYTRQVKSREWVTTLTPSNEEKLQKEIELAQLL-- 607
Query: 497 QVCRQQL*TLN*Y*IYFLFVLYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGDSYSINLL 556
QV + LF +NG S++++
Sbjct: 608 QVSSPE---------GSLF-----------------------------QVNGGESSVSIV 629
Query: 557 ---RKECSCRKWLLTGLPCCHAISCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPVNGQ 613
+ +C C+ W LTGLPC HAI+ + + +Y S E + + Y+ I PV
Sbjct: 630 DINQCDCDCKTWRLTGLPCSHAIAVIGCIEKSPYEYCSTYLTVESHRLMYAESIQPVPNM 689
Query: 614 SLWRRTEYTD----LQPPPIKRQPGRPKKNRRKDVDEKRDEQQLKKAKYGMKCSRCKAEG 669
+ + + PPP +R PGRPK + + +D K ++CS+CK G
Sbjct: 690 DRMMLDDPPEGLVCVTPPPTRRTPGRPKIKKVEPLD---------MMKRQLQCSKCKGLG 740
Query: 670 HNKSTCK 676
HNK TCK
Sbjct: 741 HNKKTCK 747
>At3g05850 unknown protein
Length = 777
Score = 99.4 bits (246), Expect = 7e-21
Identities = 91/356 (25%), Positives = 155/356 (42%), Gaps = 65/356 (18%)
Query: 336 QGLLPALAELLPDADNRFCVRHLYNNFRKKYPGK---KLKELM----WRAAKATYTTAFD 388
+ L ++ ++ + + +C+R+L + K G ++K L+ + AA A +F+
Sbjct: 466 KNLQESIPKVFEKSFHAYCLRYLTDELIKDLKGPFSHEIKRLIVDDFYSAAYAPRADSFE 525
Query: 389 REMNEIKKISEEAYNHLLQ-TPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPI 447
R + IK +S EAY+ ++Q + HW+ + F G + + ++ E F S PI
Sbjct: 526 RHVENIKGLSPEAYDWIVQKSQPDHWANAYF-RGARYNHMTSHSGEPFFSWASDANDLPI 584
Query: 448 VTMLEDIRVYLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN 507
M++ IR +M R N+ P++ K+ K
Sbjct: 585 TQMVDVIRGKIMGLIHVRRISANEANGNLTPSMEVKLEKE-------------------- 624
Query: 508 *Y*IYFLFVLYPLV*HLYAWNVLVRRAIEYD-YEIRHRSMNGDSYS-INLLRKECSCRKW 565
L A V V + + + +++R G++Y +N+ +CSC+ W
Sbjct: 625 ---------------SLRAQTVHVAPSADNNLFQVR-----GETYELVNMAECDCSCKGW 664
Query: 566 LLTGLPCCHAISCMKSQDIDVDQYVSDCFRKERYEICYSHIIYPV---NGQSLWRRT--E 620
LTGLPC HA++ + + Y S F Y Y+ I PV G+ +
Sbjct: 665 QLTGLPCHHAVAVINYYGRNPYDYCSKYFTVAYYRSTYAQSINPVPLLEGEMCRESSGGS 724
Query: 621 YTDLQPPPIKRQPGRPKKNRRKDVDEKRDEQQLKKAKYGMKCSRCKAEGHNKSTCK 676
+ PPP +R PGRP K +K +++ K + ++CSRCK GHNKSTCK
Sbjct: 725 AVTVTPPPTRRPPGRPPK-------KKTPAEEVMKRQ--LQCSRCKGLGHNKSTCK 771
>At2g15810 Mutator-like transposase
Length = 545
Score = 95.5 bits (236), Expect = 9e-20
Identities = 71/262 (27%), Positives = 117/262 (44%), Gaps = 58/262 (22%)
Query: 336 QGLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTAFDREMNEIK 395
+GLL A+A+LLP+A++R C RH+Y+N++K Y + W A ++ + M +K
Sbjct: 126 KGLLNAVADLLPEAEHRHCARHIYSNWKKVYGDYCHESYFWAIAYSSTDGTYTYNMRALK 185
Query: 396 KISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDIR 455
AY LL+T K++F G N + +A RKKP+++MLED+R
Sbjct: 186 SYDPLAYEDLLKTE----PKTEFQQGH------NGLRDA--------RKKPVISMLEDVR 227
Query: 456 VYLMERWEKNRQKVANYADNILPNIRKKI*KRV*FYKQLGGQVCRQQL*TLN*Y*IYFLF 515
M+R K R + A P+I V + ++C
Sbjct: 228 RQAMKRISKRRDQTAKCKSKFPPHI-----MGVLKANRKSAKLC---------------- 266
Query: 516 VLYPLV*HLYAWNVLVRRAIEYDYEIRHRSMNGD-SYSINLLRKECSCRKWLLTGLPCCH 574
V + E+ YE+ M G+ ++ N+L + C+C +W LTG+PC H
Sbjct: 267 --------------TVLKNSEHVYEV----MEGNGGFTANILHRTCACNQWNLTGIPCPH 308
Query: 575 AISCMKSQDIDVDQYVSDCFRK 596
AI + + D + +S +K
Sbjct: 309 AIYVINEHNRDPEDTISSSTQK 330
>At1g67240 hypothetical protein
Length = 851
Score = 86.7 bits (213), Expect = 4e-17
Identities = 42/129 (32%), Positives = 73/129 (56%)
Query: 336 QGLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTTAFDREMNEIK 395
+GL+ +A+LLP A++R C RH+Y N+RK Y + W A ++ + M+ ++
Sbjct: 507 KGLINVVADLLPQAEHRHCARHIYANWRKVYSDYSHESYFWAIAYSSTNGDYRWNMDALR 566
Query: 396 KISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIVTMLEDIR 455
+A++ LL+T + W ++ FS C+ + NN+ E+FN I R P++ MLE++R
Sbjct: 567 LYDPQAHDDLLKTDPRTWCRAFFSTHSRCEDVSNNLCESFNRTIRDARNLPVINMLEEVR 626
Query: 456 VYLMERWEK 464
M+R K
Sbjct: 627 RTSMKRIAK 635
Score = 45.1 bits (105), Expect = 1e-04
Identities = 32/98 (32%), Positives = 45/98 (45%), Gaps = 3/98 (3%)
Query: 627 PPIKRQPGRPKK-NRRKDVDEKR-DEQQLKKAKYGMKCSRCKAEGHNKSTCKLPPPPPQP 684
P +++ GRPKK +R K+ E + +L + + CS CK GHNK TCK P P
Sbjct: 678 PDKRKKRGRPKKFDRIKEPGESATNPNKLTREGKTVTCSNCKEIGHNKGTCKKPTVQSAP 737
Query: 685 EPGTQPPATQFPTSQPTS-DHAPTTQVSATQASASQVS 721
P P S ++AP +A + SQ S
Sbjct: 738 PRKRGRPRKSMEDDDPWSINNAPRRWRNAPNVAQSQYS 775
>At1g52610 hypothetical protein
Length = 792
Score = 72.8 bits (177), Expect = 7e-13
Identities = 40/114 (35%), Positives = 66/114 (57%), Gaps = 2/114 (1%)
Query: 336 QGLLPALAELLPDADNRFCVRHLYNNFRKKYPGKKLKELMWRAAKATYTT-AFDREMNEI 394
+GLL + +LLP A++ C R++Y N++K Y G E +RA + T +++ M +
Sbjct: 675 KGLLNLVVDLLPQAEHMHCARYIYANWKKVY-GDYCHESYFRAIAYSSTKGSYNYNMEAL 733
Query: 395 KKISEEAYNHLLQTPVKHWSKSKFSNGPLCDTLVNNMTEAFNSVIVVLRKKPIV 448
K AY +LL+T K W ++ FS C+ + NN++E+FN I RKKP++
Sbjct: 734 KSYDAFAYENLLKTEPKTWCRAFFSGHASCEDVCNNLSESFNRTIRDDRKKPVM 787
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.342 0.148 0.499
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,746,395
Number of Sequences: 26719
Number of extensions: 652810
Number of successful extensions: 7765
Number of sequences better than 10.0: 180
Number of HSP's better than 10.0 without gapping: 120
Number of HSP's successfully gapped in prelim test: 65
Number of HSP's that attempted gapping in prelim test: 5837
Number of HSP's gapped (non-prelim): 1056
length of query: 739
length of database: 11,318,596
effective HSP length: 107
effective length of query: 632
effective length of database: 8,459,663
effective search space: 5346507016
effective search space used: 5346507016
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 64 (29.3 bits)
Medicago: description of AC122730.8


