

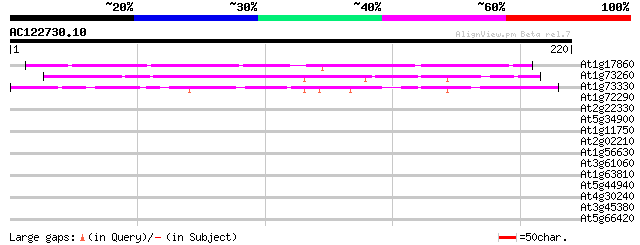
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122730.10 - phase: 0
(220 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g17860 Unknown protein (F2H15.9) 82 2e-16
At1g73260 putative trypsin inhibitor (At1g73260) 71 4e-13
At1g73330 unknown protein 58 3e-09
At1g72290 drought induced protein, putative 34 0.069
At2g22330 putative cytochrome P450 32 0.26
At5g34900 putative protein 30 1.0
At1g11750 putative ATP-dependent Clp protease proteolytic subunit 30 1.0
At2g02210 putative protein on transposon FARE2.10 (cds3) 29 1.7
At1g56630 lipase precursor, putative 29 1.7
At3g61060 unknown protein 28 5.0
At1g63810 hypothetical protein 28 5.0
At5g44940 putative protein 27 6.5
At4g30240 unknown protein 27 6.5
At3g45380 putative protein 27 6.5
At5g66420 unknown protein 27 8.5
>At1g17860 Unknown protein (F2H15.9)
Length = 196
Score = 82.4 bits (202), Expect = 2e-16
Identities = 68/200 (34%), Positives = 95/200 (47%), Gaps = 13/200 (6%)
Query: 7 LTLSFLLFVFTTNLSLAFSNDAVEQVLDINGNPIFPGGKYYILPAIRGPLGGGLRLGKSS 66
L + LL VF ++ + + AVE V DING + G YYILP IRG GGGL +
Sbjct: 5 LYIFLLLAVFISHRGVT-TEAAVEPVKDINGKSLLTGVNYYILPVIRG-RGGGLTMSNLK 62
Query: 67 NSDCEVTVVQDYNEVINGVPVKFSIPEISPGIIFTGTPIDIEFTKKPNCVESSKW-LIFV 125
C +V+QD EV G+PVKFS P I T ++I+F+ +S W L
Sbjct: 63 TETCPTSVIQDQFEVSQGLPVKFS-PYDKSRTIPVSTDVNIKFS------PTSIWELANF 115
Query: 126 DSVIQKACVGIGGPENYPGFRTLSGTFNIEKHESGFGYRLGYCVKDSPTCLDIGRAHEEV 185
D ++ + G E PG +T+ F I+K E Y++ +C C I R
Sbjct: 116 DETTKQWFISTCGVEGNPGQKTVDNWFKIDKFEK--DYKIRFCPTVCNFCKVICRDVGVF 173
Query: 186 EDEGGSRLHLTHQVAFAVVF 205
+G RL L+ V V+F
Sbjct: 174 VQDGKRRLALS-DVPLKVMF 192
>At1g73260 putative trypsin inhibitor (At1g73260)
Length = 215
Score = 71.2 bits (173), Expect = 4e-13
Identities = 62/202 (30%), Positives = 96/202 (46%), Gaps = 15/202 (7%)
Query: 14 FVFTTNLSLAFSNDAVEQVLDINGNPIFPGGKYYILPAIRGPLGGGLRLGKSSNSDCEVT 73
F L+ +++A V+DI+GN +F YY+LP IRG GGGL L C
Sbjct: 12 FYLVLALTAVLASNAYGAVVDIDGNAMFHES-YYVLPVIRGR-GGGLTLAGRGGQPCPYD 69
Query: 74 VVQDYNEVINGVPVKFSIPEISPGIIFTGTPIDIEFTKKPN-CVESSKWLIFVDSVIQKA 132
+VQ+ +EV G+PVKFS + + ++IE C++S+ W + +K
Sbjct: 70 IVQESSEVDEGIPVKFSNWRLKVAFVPESQNLNIETDVGATICIQSTYWRVGEFDHERKQ 129
Query: 133 CVGIGG--PENYPGFRTLSGTFNIEKHESGFGYRLGYCVK----DSPTCLDIGRAHEEVE 186
+ G PE + G +L F IEK Y+ +C + +P C D+G +E+
Sbjct: 130 YFVVAGPKPEGF-GQDSLKSFFKIEKSGED-AYKFVFCPRTCDSGNPKCSDVGIFIDEL- 186
Query: 187 DEGGSRLHLTHQVAFAVVFVDA 208
G RL L+ + F V+F A
Sbjct: 187 --GVRRLALSDK-PFLVMFKKA 205
>At1g73330 unknown protein
Length = 209
Score = 58.2 bits (139), Expect = 3e-09
Identities = 68/223 (30%), Positives = 103/223 (45%), Gaps = 32/223 (14%)
Query: 1 MKHVLSLTLSFLLFVFTTNLSLAFSNDAVEQVLDINGNPIFPGGKYYILPAIRGPLGGGL 60
MK +S+T FL+ SLA ++ VE D G + PG Y+I+PA P GG
Sbjct: 1 MKATISITTIFLVVALAAP-SLARPDNHVE---DSVGRLLRPGQTYHIVPA--NPETGG- 53
Query: 61 RLGKSSNSD--CEVTVVQDYNEVINGVPVKFSIPEISPGIIFTGTPIDIEFTKKPN---C 115
G SNS+ C + + Q N + G+P+KF + + I IEF + PN C
Sbjct: 54 --GIFSNSEEICPLDIFQSNNPLDLGLPIKF---KSELWFVKEMNSITIEF-EAPNWFLC 107
Query: 116 VESSK-WLIFVDSVIQKA-CVGIGGPENYPGFRTLSGTFNIEKHESGFGYRLGYCVK-DS 172
+ SK W + +K+ + GG N GF+ I + + G Y++ YC +
Sbjct: 108 PKESKGWRVVYSEEFKKSLIISTGGSSNPSGFQ-------IHRVDGG-AYKIVYCTNIST 159
Query: 173 PTCLDIGRAHEEVEDEGGSRLHLTHQVAFAVVFVDAASYEAGI 215
TC+++G + G RL LT A V F AA+ +A +
Sbjct: 160 TTCMNVGIF---TDISGARRLALTSDEALLVKFQKAATPKADL 199
>At1g72290 drought induced protein, putative
Length = 215
Score = 33.9 bits (76), Expect = 0.069
Identities = 41/158 (25%), Positives = 64/158 (39%), Gaps = 14/158 (8%)
Query: 9 LSFLLFVFTTNLSLAFSNDAVEQVLDINGNPIFPGGKYYILPA-IRGPLGGGL-RLGKSS 66
+SFL+ + N+ V+ D GNP+ +Y+I P GGGL +
Sbjct: 7 ISFLIILLFAATICTHGNEPVK---DTAGNPLNTREQYFIQPVKTESKNGGGLVPAAITV 63
Query: 67 NSDCEVTVVQDYNEVINGVPVKFSIPEISPGIIFTGTPIDIEFTKK--PNCVESSKWLIF 124
C + + Q G+PV F + + T + ++IEF P C E SK+
Sbjct: 64 LPFCPLGITQTLLPYQPGLPVSFVLALGVGSTVMTSSAVNIEFKSNIWPFCKEFSKFWEV 123
Query: 125 VD--SVIQKACVGIGGPENYPGFRTLSGTFNIEKHESG 160
D S ++ + IGG + +F IEK G
Sbjct: 124 DDSSSAPKEPSILIGGK-----MGDRNSSFKIEKAGEG 156
>At2g22330 putative cytochrome P450
Length = 566
Score = 32.0 bits (71), Expect = 0.26
Identities = 23/97 (23%), Positives = 40/97 (40%), Gaps = 11/97 (11%)
Query: 104 PIDIEFTKKPNCVESSKWLIFVDSVIQKACVGIGGPENYPGFRTLSGTFNIEKHESGFGY 163
P+D+ F + C + K L+F + GGP TL +++ G G+
Sbjct: 220 PVDLRFVTRHYCGNAIKRLMFGTRTFSEKTEADGGP-------TLEDIEHMDAMFEGLGF 272
Query: 164 RLGYCVKD---SPTCLDIGRAHEEVEDEGGSRLHLTH 197
+C+ D T LD+ HE++ E + + H
Sbjct: 273 TFAFCISDYLPMLTGLDL-NGHEKIMRESSAIMDKYH 308
>At5g34900 putative protein
Length = 767
Score = 30.0 bits (66), Expect = 1.0
Identities = 21/59 (35%), Positives = 28/59 (46%), Gaps = 5/59 (8%)
Query: 136 IGGPENY----PGFRTLSGTFNIEKHESGFGYRLGYCVKDSPTCLDIGRAHEEVEDEGG 190
IG PE++ P F T F GYRL + + C+D+ A EE E+EGG
Sbjct: 304 IGNPEDFKSLVPDFHTNDTEFQQVLELVKMGYRLKK-TEWNTRCVDVCAALEEAEEEGG 361
>At1g11750 putative ATP-dependent Clp protease proteolytic subunit
Length = 271
Score = 30.0 bits (66), Expect = 1.0
Identities = 15/60 (25%), Positives = 27/60 (45%)
Query: 48 ILPAIRGPLGGGLRLGKSSNSDCEVTVVQDYNEVINGVPVKFSIPEISPGIIFTGTPIDI 107
I+ A++ P G L+ G SSN + + + K P + P ++ G P+D+
Sbjct: 38 IVSALQSPYGDSLKAGLSSNVSGSPIKIDNKAPRFGVIEAKKGNPPVMPSVMTPGGPLDL 97
>At2g02210 putative protein on transposon FARE2.10 (cds3)
Length = 684
Score = 29.3 bits (64), Expect = 1.7
Identities = 21/59 (35%), Positives = 28/59 (46%), Gaps = 5/59 (8%)
Query: 136 IGGPENY----PGFRTLSGTFNIEKHESGFGYRLGYCVKDSPTCLDIGRAHEEVEDEGG 190
IG PE++ P F T F GYRL + + C+D+ A EE E+EGG
Sbjct: 241 IGNPEDFKSLVPDFHTDDTEFQHILGLVNMGYRLKK-TEWNTRCVDVCAALEEAEEEGG 298
>At1g56630 lipase precursor, putative
Length = 421
Score = 29.3 bits (64), Expect = 1.7
Identities = 15/47 (31%), Positives = 23/47 (48%), Gaps = 1/47 (2%)
Query: 115 CVESSKWLIFVDSVIQKACVGIGGPENYPGFRTLSGTFNIEKHESGF 161
C +W+IFV V+QK + + P ++ GF L N+ GF
Sbjct: 47 CEFRGRWIIFVSIVVQKLLIILRKPLSFLGF-ALGFWLNLPSSNGGF 92
>At3g61060 unknown protein
Length = 290
Score = 27.7 bits (60), Expect = 5.0
Identities = 13/31 (41%), Positives = 19/31 (60%)
Query: 88 KFSIPEISPGIIFTGTPIDIEFTKKPNCVES 118
K + P++S GI F+ T ID TK C++S
Sbjct: 244 KVTNPDVSTGIKFSMTQIDCTHTKGGLCIDS 274
>At1g63810 hypothetical protein
Length = 1026
Score = 27.7 bits (60), Expect = 5.0
Identities = 16/43 (37%), Positives = 25/43 (57%), Gaps = 3/43 (6%)
Query: 75 VQDYNEVINGVPVKFSI-PEISPGII-FTGTPIDIEFT-KKPN 114
V E I+G+P KF + E++P + G ++EF+ KKPN
Sbjct: 36 VSSIKEAIDGIPEKFQVTSELAPSFVEDIGADKEVEFSFKKPN 78
>At5g44940 putative protein
Length = 356
Score = 27.3 bits (59), Expect = 6.5
Identities = 26/83 (31%), Positives = 40/83 (47%), Gaps = 7/83 (8%)
Query: 100 FTGTPIDIEFTKKPNCVESSKWLIFVDSVIQKACVGIG----GPENYPGFR--TLSGTFN 153
F P +I+F K P C+ S+ + ++ +I K GI EN + TLS T++
Sbjct: 269 FPFEPKNIDFHKVPQCLISTLEYVQIEELILKEKSGIKLVDYFLENSAVLKKLTLSFTYH 328
Query: 154 IEKHESGFGY-RLGYCVKDSPTC 175
+K + Y +L K SPTC
Sbjct: 329 SKKKQDPESYKKLLTSTKLSPTC 351
>At4g30240 unknown protein
Length = 300
Score = 27.3 bits (59), Expect = 6.5
Identities = 18/49 (36%), Positives = 27/49 (54%), Gaps = 6/49 (12%)
Query: 149 SGTFNIEKHESGF-GYRLGYCVKDSPTCL----DIGRAHEEVEDEGGSR 192
S T ++ ++ G G R G C DSP C+ DIG + E + +GG+R
Sbjct: 157 SSTSSVAENPRGINGRREGRCYGDSPDCVIDIDDIG-SPESADKKGGTR 204
>At3g45380 putative protein
Length = 690
Score = 27.3 bits (59), Expect = 6.5
Identities = 10/24 (41%), Positives = 19/24 (78%)
Query: 70 CEVTVVQDYNEVINGVPVKFSIPE 93
CE +++++ V+NGVP+++SI E
Sbjct: 240 CEDSLLEEAWFVVNGVPIRYSIKE 263
>At5g66420 unknown protein
Length = 655
Score = 26.9 bits (58), Expect = 8.5
Identities = 31/126 (24%), Positives = 51/126 (39%), Gaps = 28/126 (22%)
Query: 40 IFPGGKYYILPAIRGPLGGGLRLGKSSNSDCEVTVVQDYNEVINGVPVKFSIPEISPGII 99
I+ G++ + A RG L G L +D V++ NEV+ PV ++P ++ G+
Sbjct: 424 IYNSGRFRM--AGRGSLAGLLPF-----ADANAVVLEMANEVL---PVVKAVPVLA-GVC 472
Query: 100 FTGTPIDIEFTKKPNCVESSKWLIFVDSVIQKACVGIGGPENYPGFRTLSGTFNIEKHES 159
T +++ K Q +G G +N+P G F E+
Sbjct: 473 ATDPFRRMDYFLK-----------------QLESIGFVGVQNFPTVGLFDGNFRQNLEET 515
Query: 160 GFGYRL 165
G GY L
Sbjct: 516 GMGYGL 521
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.141 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,252,542
Number of Sequences: 26719
Number of extensions: 240440
Number of successful extensions: 504
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 493
Number of HSP's gapped (non-prelim): 16
length of query: 220
length of database: 11,318,596
effective HSP length: 95
effective length of query: 125
effective length of database: 8,780,291
effective search space: 1097536375
effective search space used: 1097536375
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC122730.10


