

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122729.4 - phase: 0
(113 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
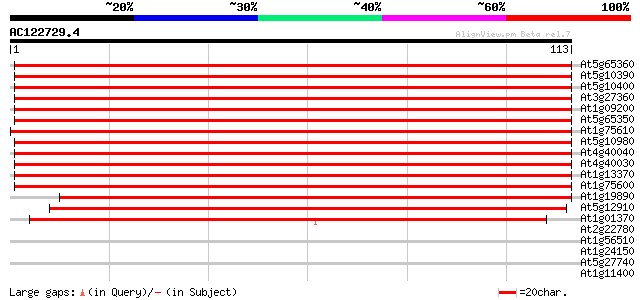
Sequences producing significant alignments: (bits) Value
At5g65360 histone H3 (sp|P05203) 184 6e-48
At5g10390 histone H3 - like protein 184 6e-48
At5g10400 histone H3 - like protein 184 6e-48
At3g27360 histone H3 like protein 184 6e-48
At1g09200 histone H3 like protein 184 6e-48
At5g65350 histone H3 179 2e-46
At1g75610 histone H3, putative 179 3e-46
At5g10980 histon H3 protein 178 3e-46
At4g40040 Histon H3 178 3e-46
At4g40030 histone H3.3 178 3e-46
At1g13370 unknown protein 177 6e-46
At1g75600 histone H3, putative 177 1e-45
At1g19890 hypothetical protein 160 1e-40
At5g12910 histone H3 -like protein 148 4e-37
At1g01370 centromeric histone H3 HTR12 91 1e-19
At2g22780 putative glyoxysomal malate dehydrogenase precursor 27 1.3
At1g56510 hypothetical protein 27 1.7
At1g24150 unknown protein 27 1.7
At5g27740 replication factor C - like 27 2.2
At1g11400 unknown protein 27 2.2
>At5g65360 histone H3 (sp|P05203)
Length = 136
Score = 184 bits (467), Expect = 6e-48
Identities = 96/112 (85%), Positives = 102/112 (90%)
Query: 2 ATRKSALATGGMKKPHRFRPGTVALREIRKYQKITELLIRKLPFKRLVHEIAQDFKTDLR 61
A RKSA ATGG+KKPHRFRPGTVALREIRKYQK TELLIRKLPF+RLV EIAQDFKTDLR
Sbjct: 25 AARKSAPATGGVKKPHRFRPGTVALREIRKYQKSTELLIRKLPFQRLVREIAQDFKTDLR 84
Query: 62 FQISAVSALQEIAEACIVGLFEDMNLCAIHAKRVTIVPKDIKLACRIRCERA 113
FQ SAV+ALQE AEA +VGLFED NLCAIHAKRVTI+PKDI+LA RIR ERA
Sbjct: 85 FQSSAVAALQEAAEAYLVGLFEDTNLCAIHAKRVTIMPKDIQLARRIRGERA 136
>At5g10390 histone H3 - like protein
Length = 136
Score = 184 bits (467), Expect = 6e-48
Identities = 96/112 (85%), Positives = 102/112 (90%)
Query: 2 ATRKSALATGGMKKPHRFRPGTVALREIRKYQKITELLIRKLPFKRLVHEIAQDFKTDLR 61
A RKSA ATGG+KKPHRFRPGTVALREIRKYQK TELLIRKLPF+RLV EIAQDFKTDLR
Sbjct: 25 AARKSAPATGGVKKPHRFRPGTVALREIRKYQKSTELLIRKLPFQRLVREIAQDFKTDLR 84
Query: 62 FQISAVSALQEIAEACIVGLFEDMNLCAIHAKRVTIVPKDIKLACRIRCERA 113
FQ SAV+ALQE AEA +VGLFED NLCAIHAKRVTI+PKDI+LA RIR ERA
Sbjct: 85 FQSSAVAALQEAAEAYLVGLFEDTNLCAIHAKRVTIMPKDIQLARRIRGERA 136
>At5g10400 histone H3 - like protein
Length = 136
Score = 184 bits (467), Expect = 6e-48
Identities = 96/112 (85%), Positives = 102/112 (90%)
Query: 2 ATRKSALATGGMKKPHRFRPGTVALREIRKYQKITELLIRKLPFKRLVHEIAQDFKTDLR 61
A RKSA ATGG+KKPHRFRPGTVALREIRKYQK TELLIRKLPF+RLV EIAQDFKTDLR
Sbjct: 25 AARKSAPATGGVKKPHRFRPGTVALREIRKYQKSTELLIRKLPFQRLVREIAQDFKTDLR 84
Query: 62 FQISAVSALQEIAEACIVGLFEDMNLCAIHAKRVTIVPKDIKLACRIRCERA 113
FQ SAV+ALQE AEA +VGLFED NLCAIHAKRVTI+PKDI+LA RIR ERA
Sbjct: 85 FQSSAVAALQEAAEAYLVGLFEDTNLCAIHAKRVTIMPKDIQLARRIRGERA 136
>At3g27360 histone H3 like protein
Length = 136
Score = 184 bits (467), Expect = 6e-48
Identities = 96/112 (85%), Positives = 102/112 (90%)
Query: 2 ATRKSALATGGMKKPHRFRPGTVALREIRKYQKITELLIRKLPFKRLVHEIAQDFKTDLR 61
A RKSA ATGG+KKPHRFRPGTVALREIRKYQK TELLIRKLPF+RLV EIAQDFKTDLR
Sbjct: 25 AARKSAPATGGVKKPHRFRPGTVALREIRKYQKSTELLIRKLPFQRLVREIAQDFKTDLR 84
Query: 62 FQISAVSALQEIAEACIVGLFEDMNLCAIHAKRVTIVPKDIKLACRIRCERA 113
FQ SAV+ALQE AEA +VGLFED NLCAIHAKRVTI+PKDI+LA RIR ERA
Sbjct: 85 FQSSAVAALQEAAEAYLVGLFEDTNLCAIHAKRVTIMPKDIQLARRIRGERA 136
>At1g09200 histone H3 like protein
Length = 136
Score = 184 bits (467), Expect = 6e-48
Identities = 96/112 (85%), Positives = 102/112 (90%)
Query: 2 ATRKSALATGGMKKPHRFRPGTVALREIRKYQKITELLIRKLPFKRLVHEIAQDFKTDLR 61
A RKSA ATGG+KKPHRFRPGTVALREIRKYQK TELLIRKLPF+RLV EIAQDFKTDLR
Sbjct: 25 AARKSAPATGGVKKPHRFRPGTVALREIRKYQKSTELLIRKLPFQRLVREIAQDFKTDLR 84
Query: 62 FQISAVSALQEIAEACIVGLFEDMNLCAIHAKRVTIVPKDIKLACRIRCERA 113
FQ SAV+ALQE AEA +VGLFED NLCAIHAKRVTI+PKDI+LA RIR ERA
Sbjct: 85 FQSSAVAALQEAAEAYLVGLFEDTNLCAIHAKRVTIMPKDIQLARRIRGERA 136
>At5g65350 histone H3
Length = 139
Score = 179 bits (454), Expect = 2e-46
Identities = 92/112 (82%), Positives = 102/112 (90%)
Query: 2 ATRKSALATGGMKKPHRFRPGTVALREIRKYQKITELLIRKLPFKRLVHEIAQDFKTDLR 61
A R+SA ATGG+KKPHRFRPGTVALR+IRKYQK TE+LIRKLPF+RLV EIAQDFKTDLR
Sbjct: 25 AARQSAPATGGVKKPHRFRPGTVALRDIRKYQKSTEILIRKLPFQRLVREIAQDFKTDLR 84
Query: 62 FQISAVSALQEIAEACIVGLFEDMNLCAIHAKRVTIVPKDIKLACRIRCERA 113
FQ SAV+ALQE AEA +VGLFED NLCAIHAKRVTI+PK+I+LA RIR ERA
Sbjct: 85 FQSSAVAALQEAAEAYLVGLFEDTNLCAIHAKRVTIMPKEIQLARRIRGERA 136
>At1g75610 histone H3, putative
Length = 115
Score = 179 bits (453), Expect = 3e-46
Identities = 92/113 (81%), Positives = 100/113 (88%)
Query: 1 MATRKSALATGGMKKPHRFRPGTVALREIRKYQKITELLIRKLPFKRLVHEIAQDFKTDL 60
+A RKSA TGG+KKPHR+RPGTVALREIRKYQK TELLIRKLPF+RLV EIAQDFKTDL
Sbjct: 3 LAARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTELLIRKLPFQRLVREIAQDFKTDL 62
Query: 61 RFQISAVSALQEIAEACIVGLFEDMNLCAIHAKRVTIVPKDIKLACRIRCERA 113
RFQ AV ALQE AEA +VGLFED NLCAIHAKRVTI+PKD++LA RIR ERA
Sbjct: 63 RFQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTIMPKDVQLARRIRGERA 115
>At5g10980 histon H3 protein
Length = 136
Score = 178 bits (452), Expect = 3e-46
Identities = 93/112 (83%), Positives = 99/112 (88%)
Query: 2 ATRKSALATGGMKKPHRFRPGTVALREIRKYQKITELLIRKLPFKRLVHEIAQDFKTDLR 61
A RKSA TGG+KKPHR+RPGTVALREIRKYQK TELLIRKLPF+RLV EIAQDFKTDLR
Sbjct: 25 AARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTELLIRKLPFQRLVREIAQDFKTDLR 84
Query: 62 FQISAVSALQEIAEACIVGLFEDMNLCAIHAKRVTIVPKDIKLACRIRCERA 113
FQ AV ALQE AEA +VGLFED NLCAIHAKRVTI+PKDI+LA RIR ERA
Sbjct: 85 FQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTIMPKDIQLARRIRGERA 136
>At4g40040 Histon H3
Length = 136
Score = 178 bits (452), Expect = 3e-46
Identities = 93/112 (83%), Positives = 99/112 (88%)
Query: 2 ATRKSALATGGMKKPHRFRPGTVALREIRKYQKITELLIRKLPFKRLVHEIAQDFKTDLR 61
A RKSA TGG+KKPHR+RPGTVALREIRKYQK TELLIRKLPF+RLV EIAQDFKTDLR
Sbjct: 25 AARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTELLIRKLPFQRLVREIAQDFKTDLR 84
Query: 62 FQISAVSALQEIAEACIVGLFEDMNLCAIHAKRVTIVPKDIKLACRIRCERA 113
FQ AV ALQE AEA +VGLFED NLCAIHAKRVTI+PKDI+LA RIR ERA
Sbjct: 85 FQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTIMPKDIQLARRIRGERA 136
>At4g40030 histone H3.3
Length = 136
Score = 178 bits (452), Expect = 3e-46
Identities = 93/112 (83%), Positives = 99/112 (88%)
Query: 2 ATRKSALATGGMKKPHRFRPGTVALREIRKYQKITELLIRKLPFKRLVHEIAQDFKTDLR 61
A RKSA TGG+KKPHR+RPGTVALREIRKYQK TELLIRKLPF+RLV EIAQDFKTDLR
Sbjct: 25 AARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTELLIRKLPFQRLVREIAQDFKTDLR 84
Query: 62 FQISAVSALQEIAEACIVGLFEDMNLCAIHAKRVTIVPKDIKLACRIRCERA 113
FQ AV ALQE AEA +VGLFED NLCAIHAKRVTI+PKDI+LA RIR ERA
Sbjct: 85 FQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTIMPKDIQLARRIRGERA 136
>At1g13370 unknown protein
Length = 136
Score = 177 bits (450), Expect = 6e-46
Identities = 92/112 (82%), Positives = 98/112 (87%)
Query: 2 ATRKSALATGGMKKPHRFRPGTVALREIRKYQKITELLIRKLPFKRLVHEIAQDFKTDLR 61
A RKSA TGG+KKPHRFRPGTVALREIRKYQK TELL RKLPF+RLV EIAQDFKTDLR
Sbjct: 25 AARKSAPTTGGVKKPHRFRPGTVALREIRKYQKSTELLNRKLPFQRLVREIAQDFKTDLR 84
Query: 62 FQISAVSALQEIAEACIVGLFEDMNLCAIHAKRVTIVPKDIKLACRIRCERA 113
FQ AV ALQE AEA +VGLFED NLCAIHAKRVTI+PKD++LA RIR ERA
Sbjct: 85 FQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTIMPKDVQLARRIRAERA 136
>At1g75600 histone H3, putative
Length = 136
Score = 177 bits (448), Expect = 1e-45
Identities = 91/112 (81%), Positives = 99/112 (88%)
Query: 2 ATRKSALATGGMKKPHRFRPGTVALREIRKYQKITELLIRKLPFKRLVHEIAQDFKTDLR 61
A RKSA TGG+KKPHR+RPGTVALREIRKYQK TELLIRKLPF+RLV EIAQD+KTDLR
Sbjct: 25 AARKSAPTTGGVKKPHRYRPGTVALREIRKYQKSTELLIRKLPFQRLVREIAQDYKTDLR 84
Query: 62 FQISAVSALQEIAEACIVGLFEDMNLCAIHAKRVTIVPKDIKLACRIRCERA 113
FQ AV ALQE AEA +VGLFED NLCAIHAKRVTI+PKD++LA RIR ERA
Sbjct: 85 FQSHAVLALQEAAEAYLVGLFEDTNLCAIHAKRVTIMPKDVQLARRIRGERA 136
>At1g19890 hypothetical protein
Length = 137
Score = 160 bits (404), Expect = 1e-40
Identities = 83/103 (80%), Positives = 90/103 (86%)
Query: 11 GGMKKPHRFRPGTVALREIRKYQKITELLIRKLPFKRLVHEIAQDFKTDLRFQISAVSAL 70
GG+K+ HRFRPGTVALREIRKYQK T+LLIRKLPF+RLV EIAQDFK DLRFQ AV AL
Sbjct: 35 GGVKRAHRFRPGTVALREIRKYQKSTDLLIRKLPFQRLVREIAQDFKVDLRFQSHAVLAL 94
Query: 71 QEIAEACIVGLFEDMNLCAIHAKRVTIVPKDIKLACRIRCERA 113
QE AEA +VGLFED NLCAIHAKRVTI+ KDI+LA RIR ERA
Sbjct: 95 QEAAEAYLVGLFEDTNLCAIHAKRVTIMSKDIQLARRIRGERA 137
>At5g12910 histone H3 -like protein
Length = 131
Score = 148 bits (374), Expect = 4e-37
Identities = 74/104 (71%), Positives = 89/104 (85%)
Query: 9 ATGGMKKPHRFRPGTVALREIRKYQKITELLIRKLPFKRLVHEIAQDFKTDLRFQISAVS 68
+T +KKP+R++PGTVALREIRKYQK T+L+IRKLPF+RLV EIAQ K DLRFQ AVS
Sbjct: 27 STPPLKKPYRYKPGTVALREIRKYQKTTDLVIRKLPFQRLVKEIAQSLKADLRFQTGAVS 86
Query: 69 ALQEIAEACIVGLFEDMNLCAIHAKRVTIVPKDIKLACRIRCER 112
ALQE AEA +VG+FED NLCA+HAKR TI+PKDI+LA R+R +R
Sbjct: 87 ALQEAAEAFMVGMFEDTNLCAMHAKRSTIMPKDIQLAKRLRGDR 130
>At1g01370 centromeric histone H3 HTR12
Length = 178
Score = 90.5 bits (223), Expect = 1e-19
Identities = 52/106 (49%), Positives = 66/106 (62%), Gaps = 2/106 (1%)
Query: 5 KSALATGGMKKPHRFRPGTVALREIRKYQKITELLIRKLPFKRLVHEIAQDFKTDL--RF 62
+ A+ G KK +R+RPGTVAL+EIR +QK T LLI F R V I R+
Sbjct: 66 RQAMPRGSQKKSYRYRPGTVALKEIRHFQKQTNLLIPAASFIREVRSITHMLAPPQINRW 125
Query: 63 QISAVSALQEIAEACIVGLFEDMNLCAIHAKRVTIVPKDIKLACRI 108
A+ ALQE AE +VGLF D LCAIHA+RVT++ KD +LA R+
Sbjct: 126 TAEALVALQEAAEDYLVGLFSDSMLCAIHARRVTLMRKDFELARRL 171
>At2g22780 putative glyoxysomal malate dehydrogenase precursor
Length = 354
Score = 27.3 bits (59), Expect = 1.3
Identities = 16/38 (42%), Positives = 21/38 (55%), Gaps = 2/38 (5%)
Query: 64 ISAVSALQEIAEACIVGLFEDMNL--CAIHAKRVTIVP 99
+S A E A+AC+ GL D N+ CA A VT +P
Sbjct: 267 LSMAYAAVEFADACLRGLRGDANIVECAYVASHVTELP 304
>At1g56510 hypothetical protein
Length = 1007
Score = 26.9 bits (58), Expect = 1.7
Identities = 16/49 (32%), Positives = 25/49 (50%), Gaps = 4/49 (8%)
Query: 56 FKTDLRFQISAV----SALQEIAEACIVGLFEDMNLCAIHAKRVTIVPK 100
F T+L +S + S ++ I E CI GL +LC KR+ +P+
Sbjct: 754 FYTELPVSVSHINISNSGIEWITEDCIKGLHNLHDLCLSGCKRLVSLPE 802
>At1g24150 unknown protein
Length = 725
Score = 26.9 bits (58), Expect = 1.7
Identities = 22/82 (26%), Positives = 39/82 (46%), Gaps = 5/82 (6%)
Query: 1 MATRKSALATGGMKKPHRFRPGTVALREIRKYQKITELLIRKLPFKR--LVHEIAQ--DF 56
+A K + GG KKP P + + + RK Q T ++++ L R LV + + DF
Sbjct: 353 VAVGKKSPDDGGDKKPSSASPAQIFILDPRKSQN-TAIVLKSLGMTRDELVESLMEGHDF 411
Query: 57 KTDLRFQISAVSALQEIAEACI 78
D ++S ++ +E A +
Sbjct: 412 HPDTLERLSRIAPTKEEQSAIL 433
>At5g27740 replication factor C - like
Length = 354
Score = 26.6 bits (57), Expect = 2.2
Identities = 13/43 (30%), Positives = 25/43 (57%), Gaps = 4/43 (9%)
Query: 34 KITELLIRKLP----FKRLVHEIAQDFKTDLRFQISAVSALQE 72
K+ ELL+ +P KRL+HE+ + ++L+ ++ +A E
Sbjct: 279 KVYELLVNCIPPEVILKRLLHELLKKLDSELKLEVCHWAAYYE 321
>At1g11400 unknown protein
Length = 204
Score = 26.6 bits (57), Expect = 2.2
Identities = 12/33 (36%), Positives = 19/33 (57%)
Query: 11 GGMKKPHRFRPGTVALREIRKYQKITELLIRKL 43
G ++KP R RPG E+ KYQ L+ +++
Sbjct: 34 GTLRKPIRIRPGYTPEDEVVKYQSKGSLMKKEM 66
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.328 0.140 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,193,497
Number of Sequences: 26719
Number of extensions: 69897
Number of successful extensions: 220
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 198
Number of HSP's gapped (non-prelim): 28
length of query: 113
length of database: 11,318,596
effective HSP length: 89
effective length of query: 24
effective length of database: 8,940,605
effective search space: 214574520
effective search space used: 214574520
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 52 (24.6 bits)
Medicago: description of AC122729.4


