

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122729.3 + phase: 0
(404 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
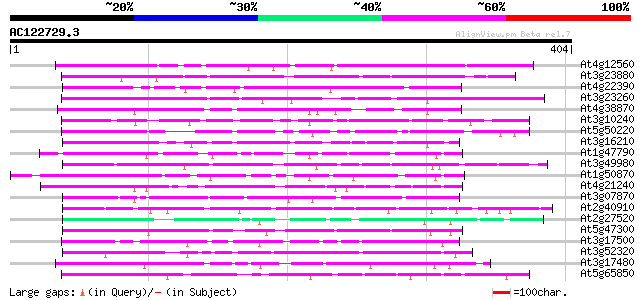
Score E
Sequences producing significant alignments: (bits) Value
At4g12560 putative protein 123 2e-28
At3g23880 unknown protein 118 5e-27
At4g22390 unknown protein 98 7e-21
At3g23260 unknown protein 90 3e-18
At4g38870 putative protein 80 3e-15
At3g10240 hypothetical protein 79 3e-15
At5g50220 putative protein 79 6e-15
At3g16210 hypothetical protein 78 8e-15
At1g47790 hypothetical protein 77 2e-14
At3g49980 putative protein 76 3e-14
At1g50870 hypothetical protein 76 4e-14
At4g21240 putative protein 75 8e-14
At3g07870 unknown protein 74 1e-13
At2g40910 unknown protein 73 2e-13
At2g27520 hypothetical protein 72 4e-13
At5g47300 putative protein 72 5e-13
At3g17500 hypothetical protein 71 9e-13
At3g52320 putative protein 70 2e-12
At3g17480 hypothetical protein 70 3e-12
At5g65850 unknown protein 69 4e-12
>At4g12560 putative protein
Length = 408
Score = 123 bits (309), Expect = 2e-28
Identities = 102/368 (27%), Positives = 184/368 (49%), Gaps = 37/368 (10%)
Query: 34 ALLWLDLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLH-LSQTRPYHLLIRNS 92
A + +D+ +I RLP K+L+ + +SK LI+DP F++ HLH + QT + +++
Sbjct: 2 ATIPMDIVNDIFLRLPAKTLVRCRALSKPCYHLINDPDFIESHLHRVLQTGDHLMILLRG 61
Query: 93 ELLLVDSRLPSVTAIIPDTTHNF-RLNPSDNHPIMIDSCDGIICFENRNDNHVDLVVWNP 151
L L L S+ + + D H R P++ + S +G+I N + DL V+NP
Sbjct: 62 ALRLYSVDLDSLDS-VSDVEHPMKRGGPTE----VFGSSNGLIGLSN---SPTDLAVFNP 113
Query: 152 CTGKFKILPPLE-NIPNGKT---HTLYSIGYDRFVDNYKVV-----AFSCHRQINKSYKY 202
T + LPP ++P+G + + Y +GYD D+YKVV ++ S+ Y
Sbjct: 114 STRQIHRLPPSSIDLPDGSSTRGYVFYGLGYDSVSDDYKVVRMVQFKIDSEDELGCSFPY 173
Query: 203 CNSQVRVHTLGTNFWRRIPNFPSNIMGL---------PNGYVGKFVSGTINWAIENQKNY 253
+V+V +L N W+RI + S+I L GY G +++W + +
Sbjct: 174 ---EVKVFSLKKNSWKRIESVASSIQLLFYFYYHLLYRRGY-GVLAGNSLHWVLPRRPGL 229
Query: 254 DSW-VILSLDLGNESYQEISRPDFGLDDPVHI-FTLGVSKDCLCVLV-YTETLLGIWVMK 310
++ +I+ DL E ++ + P+ + V I +GV CLC++ Y ++ + +W+MK
Sbjct: 230 IAFNLIVRFDLALEEFEIVRFPEAVANGNVDIQMDIGVLDGCLCLMCNYDQSYVDVWMMK 289
Query: 311 DYGNKNSWTKLFAVPYAKVGYHGFGFVDLHYISEEDDQVFLHF-CSKVYVYNYKNSTVKT 369
+Y ++SWTK+F V K F ++ S++ +V L +K+ ++ ++ + T
Sbjct: 290 EYNVRDSWTKVFTVQKPK-SVKSFSYMRPLVYSKDKKKVLLELNNTKLVWFDLESKKMST 348
Query: 370 LDIQGQPS 377
L I+ PS
Sbjct: 349 LRIKDCPS 356
>At3g23880 unknown protein
Length = 364
Score = 118 bits (296), Expect = 5e-27
Identities = 99/338 (29%), Positives = 159/338 (46%), Gaps = 30/338 (8%)
Query: 38 LDLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKF-VKHHLHL------SQTRPYHLLIR 90
L++ EIL RLPVKSL KCV S SLIS+ F +KH L L + T+ + +I
Sbjct: 16 LEMMEEILLRLPVKSLTRFKCVCSSWRSLISETLFALKHALILETSKATTSTKSPYGVIT 75
Query: 91 NSELLLVDSRLPSV--TAIIPDTTHNFRLNPSDNHPIMIDSCDGIICFENRNDNHVDLVV 148
S L + S+ + + + H+ L D + + + +C G++CF D L +
Sbjct: 76 TSRYHLKSCCIHSLYNASTVYVSEHDGELLGRDYYQV-VGTCHGLVCFHVDYDK--SLYL 132
Query: 149 WNPCTGKFKILPPLE-NIPNGKTHTLYSIGYDRFVDNYKVVAFSCHRQINKSYKYCNSQV 207
WNP + L + + + Y GYD D+YKVVA R K +
Sbjct: 133 WNPTIKLQQRLSSSDLETSDDECVVTYGFGYDESEDDYKVVALLQQRHQVK------IET 186
Query: 208 RVHTLGTNFWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIENQKNYDSWVILSLDLGNES 267
++++ WR +FPS ++ G +++GT+NWA + + SW I+S D+ +
Sbjct: 187 KIYSTRQKLWRSNTSFPSGVVVADKSRSGIYINGTLNWAATSSSS--SWTIISYDMSRDE 244
Query: 268 YQEISRPDFGLDDPVHIFTLGVSKDCLCVLVYTE-TLLGIWVMKDYGNKNSWTKLFAVPY 326
++E+ P TLG + CL ++ Y + +WVMK++G SW+KL ++P
Sbjct: 245 FKELPGP-VCCGRGCFTMTLGDLRGCLSMVCYCKGANADVWVMKEFGEVYSWSKLLSIPG 303
Query: 327 AKVGYHGFGFVDLHYISEEDDQVFLHFCSKVYVYNYKN 364
FV +IS + V L F S + +YN N
Sbjct: 304 LT------DFVRPLWIS-DGLVVLLEFRSGLALYNCSN 334
>At4g22390 unknown protein
Length = 401
Score = 98.2 bits (243), Expect = 7e-21
Identities = 91/312 (29%), Positives = 147/312 (46%), Gaps = 43/312 (13%)
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIRNSELLLVD 98
DL E+ RL +L+ + +SK SLI P+FV HL HL+I LL
Sbjct: 7 DLINEMFLRLRATTLVKCRVLSKPCFSLIDSPEFVSSHLRRRLETGEHLMI-----LLRG 61
Query: 99 SRLPSVTAIIPDTTHNFRLNPSDNHPI-------MIDSCDGIICFENRNDNHVDLVVWNP 151
RL + + D+ N P HP+ + S +G+I N + VDL ++NP
Sbjct: 62 PRL--LRTVELDSPENVSDIP---HPLQAGGFTEVFGSFNGVIGLCN---SPVDLAIFNP 113
Query: 152 CTGKFKILP------PLENIPNGKTHTLYSIGYDRFVDNYKVVAF-SCHRQINKSYKYCN 204
T K LP P +I + + Y +GYD D++KVV C + K C
Sbjct: 114 STRKIHRLPIEPIDFPERDIT--REYVFYGLGYDSVGDDFKVVRIVQCKLKEGKKKFPCP 171
Query: 205 SQVRVHTLGTNFWRRI-PNFPSNIMG-------LPNGYVGKFVSGTINWAIENQKNYDSW 256
+V+V +L N W+R+ F I+ LP G V+ ++W + ++ ++
Sbjct: 172 VEVKVFSLKKNSWKRVCLMFEFQILWISYYYHLLPRRGYGVVVNNHLHWILPRRQGVIAF 231
Query: 257 -VILSLDLGNESYQEISRP-DFGLDDPVHIFTLGVSKDCLCVLVYTE-TLLGIWVMKDYG 313
I+ DL ++ +S P + ++D + I GV C+C++ Y E + + +WV+K+Y
Sbjct: 232 NAIIKYDLASDDIGVLSFPQELYIEDNMDI---GVLDGCVCLMCYDEYSHVDVWVLKEYE 288
Query: 314 NKNSWTKLFAVP 325
+ SWTKL+ VP
Sbjct: 289 DYKSWTKLYRVP 300
>At3g23260 unknown protein
Length = 362
Score = 89.7 bits (221), Expect = 3e-18
Identities = 91/364 (25%), Positives = 146/364 (40%), Gaps = 37/364 (10%)
Query: 38 LDLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIRNSELLLV 97
++L EIL R+P K L L+ SK N+L F K H + P +++++S + L
Sbjct: 8 VELQEEILSRVPAKYLARLRSTSKQWNALSKTGSFAKKHSANATKEPLIIMLKDSRVYLA 67
Query: 98 DSRLPSVTAIIPDTTH-NFRLNPSDNHPIMIDSCDGIICFENRNDNHVDLVVWNPCTGKF 156
L V + + RL D H + CDG++ + +N L VWNPC+G+
Sbjct: 68 SVNLHGVHNNVAQSFELGSRLYLKDPHISNVFHCDGLLLLCSIKEN--TLEVWNPCSGEA 125
Query: 157 KILPPLENIPNGKTHTLYSIGYDR--FVDNYKVVAFSCHRQINKSYK-------YCNSQV 207
K++ P + K Y++GYD YKV+ + +K + N
Sbjct: 126 KLIKPRHSY--YKESDFYALGYDNKSSCKKYKVLRVISQVHVQGDFKIEYEIYDFTNDSW 183
Query: 208 RVHTLGTNFWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIENQKNYDSWVILSLDLGNES 267
RVH T R + S V G+ W + N + + LS D E
Sbjct: 184 RVHGATTELSIRQKHPVS-------------VKGSTYWVVRN-RYFPYKYFLSFDFSTER 229
Query: 268 YQEISRPDFGLDDPVHIFTLG-VSKDCLCVLVY-----TETLLGIWVMKDYGNKNSWTKL 321
+Q +S P P + L V ++ LC+ Y T L +WV G+ SW+K
Sbjct: 230 FQSLSLPQ---PFPYLVTDLSVVREEQLCLFGYYNWSTTSEDLNVWVTTSLGSVVSWSKF 286
Query: 322 FAVPYAKVGYHGFGFVDLHYISEEDDQVFLHFCSKVYVYNYKNSTVKTLDIQGQPSILYN 381
+ K F + + E++ + KV + ++ LD G+ S L +
Sbjct: 287 LTIQIIKPRVDMFDYGMSFLVDEQNKSLVCWISQKVLHIVGEIYHIQDLDDHGRDSALRS 346
Query: 382 SSRV 385
S V
Sbjct: 347 SCSV 350
>At4g38870 putative protein
Length = 426
Score = 79.7 bits (195), Expect = 3e-15
Identities = 88/315 (27%), Positives = 141/315 (43%), Gaps = 46/315 (14%)
Query: 35 LLWLDLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLL------ 88
LL +DL +EIL +L +K L+ CVSK S+I DP F+K L+ S RP L+
Sbjct: 52 LLPVDLIMEILKKLSLKPLIRFLCVSKLWASIIRDPYFMKLFLNESLKRPKSLVFVFRAQ 111
Query: 89 ----IRNSELLLVDSRLPSVTAIIPDTTHNFRLNPSDNHPIMID-SCDGIICFENRNDNH 143
I +S L + S ++ ++ + + + I S G+IC+ +
Sbjct: 112 SLGSIFSSVHLKSTREISSSSSSSSASSITYHVTCYTQQRMTISPSVHGLICYGPPS--- 168
Query: 144 VDLVVWNPCTGKFKILPPLENIPNGKTHTLYSIGYDRFVDNYKVVAFSCHRQINKSYKYC 203
LV++NPCT + LP I G+ IGYD NYKVV + + ++ +
Sbjct: 169 -SLVIYNPCTRRSITLP---KIKAGRRAINQYIGYDPLDGNYKVVCITRGMPMLRNRRGL 224
Query: 204 NSQVRVHTLGT--NFWRRI-----PNFPSNIMGLPNG--YVGKFVSGTINWAIENQKNYD 254
+++V TLGT + WR I P+ P + NG Y F+ +N +
Sbjct: 225 AEEIQVLTLGTRDSSWRMIHDIIPPHSPVSEELCINGVLYYRAFIGTKLNES-------- 276
Query: 255 SWVILSLDLGNESYQEISRP-DFGLDDPVHIFTLGVSKDCLCVLVY---TETLLGIWVMK 310
I+S D+ +E + I P +F L + L V+ Y T ++G+W+++
Sbjct: 277 --AIMSFDVRSEKFDLIKVPCNFR-----SFSKLAKYEGKLAVIFYEKKTSGIIGLWILE 329
Query: 311 DYGNKNSWTKLFAVP 325
D N K FA+P
Sbjct: 330 DASNGEWSKKTFALP 344
>At3g10240 hypothetical protein
Length = 389
Score = 79.3 bits (194), Expect = 3e-15
Identities = 94/371 (25%), Positives = 161/371 (43%), Gaps = 74/371 (19%)
Query: 38 LDLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLI--RNSELL 95
LDL EIL RLP KS+ +CVSK +S+ ++P F+ +L TR LL+ + +E
Sbjct: 29 LDLVSEILLRLPEKSVARFRCVSKPWSSITTEPYFI----NLLTTRSPRLLLCFKANEKF 84
Query: 96 LVDSRLPSVTAIIPDTTHNFRL-NPSDNHPIMID-----------------SCDGIICFE 137
V S IP F N S ++ +ID S +G+ICF+
Sbjct: 85 FVSS--------IPQHRQTFETWNKSHSYSQLIDRYHMEFSEEMNYFPPTESVNGLICFQ 136
Query: 138 NRNDNHVDLVVWNPCTGKFKILPPLENIPNGKTHTL-YSIGYDRFVDNYKVVAFSCHRQI 196
L+VWNP T + ILP PNG ++ L +GYD +KV+ +
Sbjct: 137 ----ESARLIVWNPSTRQLLILPK----PNGNSNDLTIFLGYDPVEGKHKVMCM----EF 184
Query: 197 NKSYKYCNSQVRVHTLGT--NFWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIENQKNYD 254
+ +Y C RV TLG+ WR + + + G+ ++G + + I K+
Sbjct: 185 SATYDTC----RVLTLGSAQKLWRTVKTHNKHRSDYYDS--GRCINGVV-YHIAYVKDMC 237
Query: 255 SWVILSLDLGNESYQEISRPDFGLDDPVHIFTLGVSKDCLCVLVYTETLLGIWVMKDYGN 314
WV++S D+ +E + I P + V I G C+ + + + +W+++ +
Sbjct: 238 VWVLMSFDVRSEIFDMIELPSSDVHKDVLIDYNG-RLACVGREIIEKNGIRLWILEKH-- 294
Query: 315 KNSW-TKLFAVPYAKVG---------YHGFGFV-DLHYISEEDDQVFLHFCSKVYVYNYK 363
N W +K F P + GF ++ Y+ + H +K++ Y+
Sbjct: 295 -NKWSSKDFLAPLVHIDKSLSTNKFLLKGFTHAGEIIYV-----ESMFHKSAKIFFYDPV 348
Query: 364 NSTVKTLDIQG 374
+T + +++G
Sbjct: 349 RNTSRRFELKG 359
>At5g50220 putative protein
Length = 357
Score = 78.6 bits (192), Expect = 6e-15
Identities = 90/349 (25%), Positives = 156/349 (43%), Gaps = 62/349 (17%)
Query: 38 LDLFLEILYRLPVKSLLVLKCVSKSLNSLI-SDPKFVKHHLHLSQTRPYHLLIRNSELLL 96
+DL +EIL +LP KSL+ +CVSK +S+I S F+ + S ++P ++ + L
Sbjct: 35 IDLMVEILKKLPAKSLIKFQCVSKQWSSIIGSSRDFIDSIVTRSLSQPSRDILISFSTTL 94
Query: 97 VDSRLPSVTAIIPDTTHNFRLNPSDNHPIMIDSCDGIICFENRNDNHVDLVVWNPCTGKF 156
+S L +++ P T + +N ++ + ++NP T +
Sbjct: 95 TNS-LKQISSSFPLRT---------------------LDILTKNQSYTEAAIYNPTTRQS 132
Query: 157 KILPPLENIPNGKTHTLYS-IGYDRFVDNYKVVAFSCHRQINKSYKYCNSQVRVHTLGTN 215
LP G +H S +GYD F + YKV+ N + C+ V TLG
Sbjct: 133 LSLP---ETTAGHSHVSTSFLGYDPFKNQYKVICLD-----NYKRRCCH----VFTLGDA 180
Query: 216 F--WRRIP-NFPSNIMGLPNGYVGKFVSGTINWAIENQKNYDS-WVILSLDLGNESYQEI 271
WR+I NF LP + GTI + K Y S +V+L D+ +E + ++
Sbjct: 181 IRKWRKIQYNFGLYFPLLP----PVCIKGTIYY---QAKQYGSTYVLLCFDVISEKFDQV 233
Query: 272 SRPDFGLDDPVHIFTLGVSKDCLCVLVYTETLLGIWVMKDYGNKNSWTKLFAVPYAKVGY 331
P +D H +TL ++ + + + IWVMK+ K W+K+F A
Sbjct: 234 EAPKTMMD---HRYTL-INYQGKLGFMCCQNRVEIWVMKNDEKKQEWSKIFFYEMA---- 285
Query: 332 HGFGFVDLHYISEEDDQVFLH----FCSKVYVYNY--KNSTVKTLDIQG 374
GF + + + VF++ C +YVY Y K ++++ ++++G
Sbjct: 286 -GFEKWHIARATPSGEIVFVNRLLLSCQTLYVYYYGPKRNSMRRVEVEG 333
>At3g16210 hypothetical protein
Length = 360
Score = 78.2 bits (191), Expect = 8e-15
Identities = 79/294 (26%), Positives = 136/294 (45%), Gaps = 40/294 (13%)
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIRNSELLLVD 98
+L +EIL RL +K L +CV K+ LI+DP F + + +S + +N +L V+
Sbjct: 8 ELAIEILVRLSMKDLARFRCVCKTWRDLINDPGFTETYRDMSPAKFVSFYDKNFYMLDVE 67
Query: 99 SRLPSVTAIIPDTTHNFRLNPSDNHPIMIDS------CDGIICFENRNDNHVDLVVWNPC 152
+ P +T + +F L+ S MID CDG +C +N L+VWNP
Sbjct: 68 GKHPVITNKL-----DFPLDQS-----MIDESTCVLHCDGTLCVTLKNHT---LMVWNPF 114
Query: 153 TGKFKILPPLENIPNGKTHTLYSIGYDRFVDNYKVVAFSCHRQINKSYKYCNSQVRVHTL 212
+ +FKI+P N + + GYD D+YKVV F ++ ++ + + R +
Sbjct: 115 SKQFKIVP---NPGIYQDSNILGFGYDPVHDDYKVVTFIDRLDVSTAHVF---EFRTGSW 168
Query: 213 GTNFWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIENQKNYDSWVILSLDLGNESYQEIS 272
G + P++ G F+ + W I + + D + IL +L Y+++
Sbjct: 169 GESLRISYPDWHY------RDRRGTFLDQYLYW-IAYRSSADRF-ILCFNLSTHEYRKLP 220
Query: 273 RPDFGLDDPVHIFTLGVSKDCLCVLVY--TETLLGIWVMKDYGNKNSWTKLFAV 324
P + + V LGV+ LC+ Y + + I VM+ G SW+K+ ++
Sbjct: 221 LPVY--NQGVTSSWLGVTSQKLCITEYEMCKKEIRISVMEKTG---SWSKIISL 269
>At1g47790 hypothetical protein
Length = 389
Score = 77.0 bits (188), Expect = 2e-14
Identities = 93/322 (28%), Positives = 143/322 (43%), Gaps = 47/322 (14%)
Query: 22 RKQNSSDPFGLFALLWLDLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQ 81
R++ S P F L DL EIL RLPVKS++ +CVSK +S+I+DP F+K + S
Sbjct: 14 RRRLQSKPTSSFPL---DLASEILLRLPVKSVVRFRCVSKLWSSIITDPYFIKTYETQSS 70
Query: 82 TRPYHLL-IRNSELLLV--------DSRLPSVTAIIPDTTHNFRLNPSDNHPIMIDSCDG 132
TR L + S+ L V DS S AI +L ++P +S G
Sbjct: 71 TRQSLLFCFKQSDKLFVFSIPKHHYDSNSSSQAAI---DRFQVKLPQEFSYPSPTESVHG 127
Query: 133 IICFENRNDNHV--DLVVWNPCTGKFKILPPLENIPNGKTHTLYSIGYDRFVDNYKVVAF 190
+ICF HV ++VWNP +F LP + K T++ +GYD +KVV
Sbjct: 128 LICF------HVLATVIVWNPSMRQFLTLPKPRK--SWKELTVF-LGYDPIEGKHKVVCL 178
Query: 191 SCHRQINKSYKYCNSQVRVHTLGT--NFWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIE 248
+R + + +V TLG+ WR + + N G+ + G + + I
Sbjct: 179 PRNRTCD--------ECQVLTLGSAQKSWRTVKT--KHKHRSTNDTWGRCIKGVV-YYIA 227
Query: 249 NQKNYDSWVILSLDLGNESYQEISRPDFGLDDPVHIFTLGVSKDCLCV-LVYTETLLGI- 306
+ W I+S + +E + I P L++ + CV +YT GI
Sbjct: 228 YVYHTRVWCIMSFHVKSEKFDMIKLP---LENIYRDVMINYEGRLACVDKLYTLNNDGIR 284
Query: 307 -WVMKDYGNKNSW-TKLFAVPY 326
W+++D K+ W +K F Y
Sbjct: 285 LWILED-AEKHKWSSKQFLARY 305
>At3g49980 putative protein
Length = 382
Score = 76.3 bits (186), Expect = 3e-14
Identities = 90/366 (24%), Positives = 157/366 (42%), Gaps = 42/366 (11%)
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIRNSELLLVD 98
+L EIL R+P SL L+ K N L +D F + H + L+ E +
Sbjct: 8 ELLEEILCRVPATSLKQLRLTCKEWNRLFNDRTFSRK--HFDKAPKQFLITVLEERCRLS 65
Query: 99 SRLPSVTAIIPDTTHNFRLNPSDNHP-------IMIDSCDGIICFENRNDNHVDLVVWNP 151
S ++ + P L+P D H + I CDG+ D + VVWNP
Sbjct: 66 SLSINLHSGFPSEEFTGELSPIDYHSNSSQVIIMKIFHCDGLFVCTILKDTRI--VVWNP 123
Query: 152 CTGKFKILPPLENI-PNGKTHTLYSIGYDRFVD-NYKVVAFSCHRQINKSYKYCNSQVRV 209
CTG+ K + EN+ NG+ L ++ D +YK++++ K Y Y + + ++
Sbjct: 124 CTGQKKWIQTGENLDENGQDFVLGYYQDNKSSDKSYKILSY-------KGYNYGDQEFKI 176
Query: 210 HTLGTNFWRR--IPNFPSNIMGLPNGYVGKFVSGTINWAIENQKNYDSWVILSLDLGNES 267
+ + +N WR + P N + V + G W + K+ + ++S D E
Sbjct: 177 YDIKSNTWRNLDVTPIPGNYFTCSDYRVS--LKGNTYWFAYDLKD-EQLGLISFDYTTER 233
Query: 268 YQEISRPDFGLDDPVHIFTLG-VSKDCLCVLVYTETL--LGIWV---MKDYGNKNSWTKL 321
++ + P F D H ++L V ++ L V++ + IW+ M D + SW KL
Sbjct: 234 FERLWLP-FQCDISDHDYSLSVVGEEKLSVVLQLKDAPRREIWITNKMDDETKEMSWRKL 292
Query: 322 FAVPYAKVGYHGFGFVDLHYISEEDDQVFLHFCSKVYVYNYKNSTVKTLDIQGQPSILYN 381
F V +VG + + ++ +E+ ++ + C K TV ++ N
Sbjct: 293 FEV---EVGTRYYMWSGRPFLVDEEKKIVV-CCEKCQKKCQPVITVSMVEADS------N 342
Query: 382 SSRVYF 387
++RVYF
Sbjct: 343 ATRVYF 348
>At1g50870 hypothetical protein
Length = 396
Score = 75.9 bits (185), Expect = 4e-14
Identities = 94/343 (27%), Positives = 145/343 (41%), Gaps = 44/343 (12%)
Query: 1 MRQPLTHSKRKHRHNVVSTTVRKQNSSDPFGLFALLWLDLFLEILYRLPVKSLLVLKCVS 60
M Q +++R HR ++Q+SS P L LDL LEIL RLPVKS+L +CVS
Sbjct: 1 MEQREENTERIHRERR-----KRQSSSIPKTTTLLCPLDLILEILLRLPVKSVLRFRCVS 55
Query: 61 KSLNSLISDPKFVKHHLHLSQTRPYHLLIRNSELLLVDSRLPSVTAIIPDTTHNFRLNPS 120
K S +DP F + S TRP L+ ++ L P + TH++ +
Sbjct: 56 KLWLSTTTDPYFTNSYEARSSTRPSLLMFFKNKDKLFVFTFPHHNQNSKE-THSYSQHVD 114
Query: 121 DNHPIMIDSCDGIICFENRNDNH--------VDLVVWNPCTGKFKILPPLENIPNGKTHT 172
H C CF H ++WNP +F ILP E G +
Sbjct: 115 SYHIKYPKYC----CFPFTESVHGLISFRISTKPIIWNPTMRQFLILPKPEKSWKGLS-- 168
Query: 173 LYSIGYDRFVDNYKVVAFSCHRQINKSYKYCNSQVRVHTLGT--NFWRRIPNFPSNIMGL 230
+GYD +K++ C + N S + RV TLG+ WRRI + + L
Sbjct: 169 -VFLGYDPVEGKHKLM---CMNRDNTS-----DECRVLTLGSAQEKWRRIKSNLKHRSIL 219
Query: 231 PNGYVGKFVSGTINWA--IENQKNYDSWVILSLDLGNESYQEISRPDFGLDDPVHIFTLG 288
Y G+ ++G I + I+ + I+S ++ +E + I+ P + +
Sbjct: 220 --RYYGQCINGVIYYQAYIDQMGFISNPTIMSFEVRSEKFDTITLPSGSFAN----MLIP 273
Query: 289 VSKDCLCVLVYTETLLG---IWVMKDYGNKNSWT-KLFAVPYA 327
CV + + G +W ++D K+ W+ KLF P A
Sbjct: 274 YQGRLACVNNTMDDVNGGITLWTLED-AEKHIWSCKLFLAPLA 315
>At4g21240 putative protein
Length = 417
Score = 74.7 bits (182), Expect = 8e-14
Identities = 88/323 (27%), Positives = 143/323 (44%), Gaps = 44/323 (13%)
Query: 23 KQNSSDPFGLFALLWLDLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQT 82
K++ S +F L+ LD+ +IL RLP KS + + VSK S+ + P F++ S T
Sbjct: 23 KEDESRLGKIFELIPLDMIPDILLRLPAKSAVRFRIVSKLWLSITTRPYFIRSFAFPSST 82
Query: 83 RPYHLL---IRNSELLLV-----DSRLPSVTAI-IPDTTHNFRLNPSDNHPIMIDSCDGI 133
R + R+ L + D V I H++ NPS +S +G+
Sbjct: 83 RLCLMACVKARDMRLFISLHQHDDGSYAHVDRCEIKSPKHDY-YNPSS------ESVNGL 135
Query: 134 ICFENRNDNHVDLVVWNPCTGKFKILPPLENIPNGKTHTLYSIGYDRFVDNYKVVAFSCH 193
+CF + ++VVWNP + LP + + +GYD D YKV++ S
Sbjct: 136 VCF----GDFYNIVVWNPSMRQHVTLPEPKPHSTVRYFIRSCLGYDPVEDKYKVLSIS-- 189
Query: 194 RQINKSYKYCNSQVRVHTLG-TNFWRRIPNFPSNIMGLPNG--YVGKFVSG------TIN 244
Y N V TLG WR I N P +I LP G VG ++G I
Sbjct: 190 -----GYHNGNHDPLVFTLGPQESWRVIQNSPLDI-PLPTGGSRVGTCINGHVYYEAQIR 243
Query: 245 WAIENQKNYDSWVILSLDLGNESYQEISRPDFGLDDPVHIFTLGVSKDCLCVLVYTETLL 304
+ +++ N+++ +++S D+ E + I +P D + F L + L + +
Sbjct: 244 FKVDDIFNFEN-ILMSFDVRYEKFNTIKKP---ADPTLRNFMLNY-QGKLAWFCSDYSSI 298
Query: 305 GIWVMKDYGNKNSWT-KLFAVPY 326
WV+ D G+K W+ K F +P+
Sbjct: 299 RFWVLDD-GDKQEWSLKNFILPF 320
>At3g07870 unknown protein
Length = 417
Score = 73.9 bits (180), Expect = 1e-13
Identities = 81/309 (26%), Positives = 134/309 (43%), Gaps = 34/309 (11%)
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLL-----IRNSE 93
D+ +I RLP+ S+ L V +S S+++ + S T+P LL IRN
Sbjct: 31 DIIADIFSRLPISSIARLMFVCRSWRSVLTQHGRLSSSSS-SPTKPCLLLHCDSPIRNG- 88
Query: 94 LLLVDSRLPSVTAIIPDTTHNFRLNPSDNHPIMIDSCDGIICFENRNDNHVDLVVWNPCT 153
L +D L I R S ++ SC+G++C + N L ++NP T
Sbjct: 89 LHFLD--LSEEEKRIKTKKFTLRFASSMPEFDVVGSCNGLLCLSDSLYND-SLYLYNPFT 145
Query: 154 GKFKILPPLENIPNGKTHTLYSIGYDRFVDNYKVVAFSCHRQINKS----------YKYC 203
LP N + + ++ G+ YKV+ R + + +Y
Sbjct: 146 TNSLELPECSNKYHDQ-ELVFGFGFHEMTKEYKVLKIVYFRGSSSNNNGIYRGRGRIQYK 204
Query: 204 NSQVRVHTLGTNF------WRRIPNFPSNIMGLPNGYVGKFVSGTINWAIENQKNYDSWV 257
S+V++ TL + WR + P + + + V+G +++ +++
Sbjct: 205 QSEVQILTLSSKTTDQSLSWRSLGKAPYKFVKRSSEAL---VNGRLHFVTRPRRHVPDRK 261
Query: 258 ILSLDLGNESYQEISRPDFG-LDDPVHIFTLGVSKDCLCVLVYTET-LLGIWVMKDYGNK 315
+S DL +E ++EI +PD G L+ H L K CLC +VY L IWVMK YG K
Sbjct: 262 FVSFDLEDEEFKEIPKPDCGGLNRTNH--RLVNLKGCLCAVVYGNYGKLDIWVMKTYGVK 319
Query: 316 NSWTKLFAV 324
SW K +++
Sbjct: 320 ESWGKEYSI 328
>At2g40910 unknown protein
Length = 449
Score = 73.2 bits (178), Expect = 2e-13
Identities = 100/386 (25%), Positives = 179/386 (45%), Gaps = 54/386 (13%)
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIRNSELLLVD 98
DL +EI+ RLP KS++ KC+SK +SLIS ++ + L S TR I + LVD
Sbjct: 72 DLLMEIVMRLPAKSMVRFKCISKQWSSLIS-CRYFCNSLFTSVTRKKQPHI---HMCLVD 127
Query: 99 SR----LPSVTAIIPDTT---HNFRLNPSDNHPIMIDSCDGIICFENRNDNHVDLVVWNP 151
L S+++ PD T + L+ + ++ G++CF R ++NP
Sbjct: 128 HGGQRVLLSLSSTSPDNTCYVVDQDLSLTGMGGFFLNLVRGLLCFSVRE----KACIYNP 183
Query: 152 CTGKFKILPPLEN----IPNGKTHTLYSIGYDRFVDNYKVVAFSCHRQINKS-YKYCNSQ 206
T + LP +++ + + + Y IG+D D YK+V C I+ + + S+
Sbjct: 184 TTRQRLTLPAIKSDIIALKDERKDIRYYIGHDPVNDQYKLV---CTIGISSAFFTNLKSE 240
Query: 207 VRVHTL-GTNFWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIENQKNYDSWVILSLDLGN 265
V L WR++ S P+ VG+F++G++ + + + D+ ++S D+ +
Sbjct: 241 HWVFVLEAGGSWRKVRTLESYHPHAPS-TVGQFINGSVVYYMA-WLDMDTCAVVSFDITS 298
Query: 266 ESYQEI----SRPDFGLDDPVHIFTLGVSKDCLCVLVYTET------LLGIWVMKDYGNK 315
E I D L P G+ + + V+ T L+ +WV+KD G K
Sbjct: 299 EELTTIIVTLEAGDVAL--PAGRMKAGLIQYHGEIAVFDHTHLKEKFLVDLWVLKDAGMK 356
Query: 316 NSWTK--LFAVPYAKVGYHGFGFVDLHYI---SEEDDQVFL---HFCSKVYV--YNYKNS 365
W+K L P K H D+ + + +D +V L + CS++Y+ Y+ ++
Sbjct: 357 -KWSKKRLVLQPCQKHLVHD----DIELVVKGTTQDGKVILAPVNMCSQLYILCYDMGSN 411
Query: 366 TVKTLDIQGQPSILYNSSRVYFESLY 391
++ ++I+G P I ++ YF+ Y
Sbjct: 412 DLRKVEIKGVPDIWFD-KECYFDLKY 436
>At2g27520 hypothetical protein
Length = 347
Score = 72.4 bits (176), Expect = 4e-13
Identities = 80/355 (22%), Positives = 141/355 (39%), Gaps = 48/355 (13%)
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIRNSELLLVD 98
DL EIL RLP SL L+ K N+L DP+F+ H + + L++ N + +
Sbjct: 9 DLVDEILSRLPATSLGRLRFTCKRWNALFKDPEFITKQFHKAAKQDLVLMLSNFGVYSMS 68
Query: 99 SRLPSVTAIIPDTTHNFRLNPSDNHPIMIDSCDGIICFENRNDNHVDLVVWNPCTGKFKI 158
+ L + P++ + C+G++ N LVV NPCTG+ +
Sbjct: 69 TNLKEI--------------PNNIEIAQVFHCNGLLLCSTEEGNKTKLVVVNPCTGQTRW 114
Query: 159 LPPLENIPNGKTHTLYSIGY-----DRFVDNYKVVAFSCHRQINKSYKYCNSQVRVHTLG 213
+ P + H + ++GY + D+YK++ + Y V + L
Sbjct: 115 IEPRTDY--NYNHDI-ALGYGNNSTKKSYDSYKILRIT----------YGCKLVEIFELK 161
Query: 214 TNFWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIENQKNYDSWVILSLDLGNESYQEISR 273
+N WR + N+ + Y G G W +Y + ILS D E+++ +
Sbjct: 162 SNSWRVLSKVHPNVE--KHYYGGVSFKGNTYWL-----SYTKFNILSFDFTTETFRSVPL 214
Query: 274 PDFGLDDPVHIFTLGVSKDCLCVL--VYTETLLGIWVMKDYGNKN--SWTKLFAVPYAKV 329
P D V + V ++ L +L + +GIW+ + SW+K F + + +
Sbjct: 215 PFLYQDGFVTLALSVVREEQLLLLRSRFDMGQVGIWMCNKIDTETVLSWSKSFTLEFDSL 274
Query: 330 GYHGFGFVDLHYISEEDDQVFLHFCSKVYVYNYKNSTVKTLDIQGQPSILYNSSR 384
+ L EED +V + C +K + + + G + Y R
Sbjct: 275 RDLPVMSI-LRIFIEEDKKVIVVDCDD----RWKENMIYIVGKNGFKKLSYEKDR 324
>At5g47300 putative protein
Length = 416
Score = 72.0 bits (175), Expect = 5e-13
Identities = 72/302 (23%), Positives = 133/302 (43%), Gaps = 33/302 (10%)
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIRNSELLLVD 98
DL EIL R+P SL L+ K N+L ++ +F + HL + +L++ +S + +
Sbjct: 49 DLLEEILCRVPATSLKQLRSTCKQWNNLFNNGRFTRKHLDKAPKDFQNLMLSDSRVFSMS 108
Query: 99 ---SRLPSVTAIIPDTTHNFRLNPSDNHPI-MIDSCDGIICFENRNDNHVDLVVWNPCTG 154
+PSV A + + + D I + CDG++ + ++ + VVWNPCTG
Sbjct: 109 VSFHGIPSVEATCELSLIDSFSSFEDKFEISQVFHCDGLLLCTDADNTRI--VVWNPCTG 166
Query: 155 KFKILPPLENIPNGKTHTLYSIGYDRFVD-----NYKVVAFSCHRQINKSYKYCNSQVRV 209
K + + P + Y + ++D +YK++++S Y Y N ++ +
Sbjct: 167 KTRWIEP--------NNRCYYYAFGSYLDKSYGNSYKILSYS-------GYGYENQELAI 211
Query: 210 HTLGTNFWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIENQKNYD-SWVILSLDLGNESY 268
+ + + WR + I+ Y G + G W ++K + S ++S D E +
Sbjct: 212 YEINSQSWRFLDVTRDCILERYTDY-GVSLKGHTYWFASDEKEKNLSVFLVSFDYTTERF 270
Query: 269 QEISRPDFGLDDPVHIFTLGVSKDCLCVLVYTE--TLLGIWVMKDYGNKN--SWTKLFAV 324
+ + P + D V ++ L VL+ E + IWV G SW+ + AV
Sbjct: 271 RRLRLP-YQCPDYNTASLSVVREEKLAVLLQRENTSRTEIWVTSRIGETKVVSWSMVLAV 329
Query: 325 PY 326
+
Sbjct: 330 DF 331
>At3g17500 hypothetical protein
Length = 438
Score = 71.2 bits (173), Expect = 9e-13
Identities = 80/303 (26%), Positives = 131/303 (42%), Gaps = 38/303 (12%)
Query: 38 LDLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIRNSELLLV 97
LDL EIL R+P SL L+ KS N+ D +F + H S L+ L+L
Sbjct: 6 LDLVEEILSRVPATSLKRLRSTCKSWNNCYKDQRFTEKH---SVIAAKQFLV----LMLK 58
Query: 98 DSRLPSVTAIIPDTTHNFRLNPSDNHPIM--------IDSCDGIICFENRNDNHVDLVVW 149
D R+ SV+ + + +N + ++ I CDG++ ++D L VW
Sbjct: 59 DCRVSSVSVNLNEIHNNIAAPSIELKGVVGPQMQISGIFHCDGLLLCTTKDDR---LEVW 115
Query: 150 NPCTGKFKILPPLENIPNGKTHTLYSIGY--DRFVDNYKVVAFSCHRQINKSYKYCNSQV 207
NPCTG+ + +++ + KT++ + +GY + +YK++ + N Y S+
Sbjct: 116 NPCTGQTR---RVQHSIHYKTNSEFVLGYVNNNSRHSYKILRYWNFYMSN----YRVSEF 168
Query: 208 RVHTLGTNFWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIENQKNYDSWVILSLDLGNES 267
++ ++ WR I N L G V + G W ++K D +IL D E
Sbjct: 169 EIYDFSSDSWRFIDEV--NPYCLTEGEVS--LKGNTYWLASDEKR-DIDLILRFDFSIER 223
Query: 268 YQEISRPDFGLDDPVHIFTLGVSKDCLCVLVYTETL-LGIWVMK----DYGNKN-SWTKL 321
YQ ++ P D ++ K +L +TL IWV D+ KN W K
Sbjct: 224 YQRLNLPILKSDYETEALSVVREKQLSVLLKRNDTLEREIWVTTNDKIDHTTKNLLWIKF 283
Query: 322 FAV 324
A+
Sbjct: 284 LAI 286
>At3g52320 putative protein
Length = 390
Score = 70.1 bits (170), Expect = 2e-12
Identities = 80/315 (25%), Positives = 137/315 (43%), Gaps = 43/315 (13%)
Query: 39 DLFLEILYRLPVKSLLVLKCVSKSLNSLI------------SDPKFVKHHLHLSQTRPYH 86
++ ++IL RLP KSL+ KCVSK SLI S P + +L + + +
Sbjct: 30 EMLIDILIRLPAKSLMRFKCVSKLWLSLITSRYFTNRFFKPSSPSCLFAYLVDRENQSKY 89
Query: 87 LLIRNSELLLVDSRLPSVTAIIPDTTHNFRLNPSDNHPIM----IDSCDGIICFENRNDN 142
LL+++S D SV+ I +T PIM +++ G++C+ R
Sbjct: 90 LLLQSSSSSRHDHSDTSVSVIDQHST----------IPIMGGYLVNAARGLLCY--RTGR 137
Query: 143 HVDLVVWNPCTGKFKILPPLENIPNGKTHTLYSIGYDRFVDNYKVVAFSCHRQINKSYKY 202
V V NP T + LP I KT+ G+D F D YKV+ S ++ K
Sbjct: 138 RVK--VCNPSTRQIVELP----IMRSKTNVWNWFGHDPFHDEYKVL--SLFWEVTKEQTV 189
Query: 203 CNSQVRVHTLGTN-FWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIENQKNYDSWVILSL 261
S+ +V LG WR + + G + G + ++ N V++S
Sbjct: 190 VRSEHQVLVLGVGASWRNTKSHHTPHRPFHPYSRGMTIDGVLYYSARTDAN--RCVLMSF 247
Query: 262 DLGNESYQEISRPDFGLDDPVHIFTLGVSKDCLCVLVYTETLLGIWVMKDYGNKNSWT-- 319
DL +E + I P +H+ G C + + ++ + + V++D +K+ W+
Sbjct: 248 DLSSEEFNLIELPFENWSRTIHMNYQGKVATCQYMRLASDGFVDVCVLED-ADKSQWSNK 306
Query: 320 KLFAVPYAKVGY-HG 333
K F +P +++ + HG
Sbjct: 307 KTFVLPISQMNFVHG 321
>At3g17480 hypothetical protein
Length = 373
Score = 69.7 bits (169), Expect = 3e-12
Identities = 80/333 (24%), Positives = 139/333 (41%), Gaps = 40/333 (12%)
Query: 34 ALLWLDLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIRNSE 93
++L DL +IL R+P SL+ L+ K N++++D +F+K H ++ +L+R+
Sbjct: 9 SVLTEDLVEDILSRVPATSLVRLRSTCKQWNAILNDRRFIKKHFDTAEKEYLDMLLRSLR 68
Query: 94 LL--------LVDSRLPSVTAIIPDTTHNFRLNPSDNHPIMIDSCDGIICFENRNDNHVD 145
+ L D+ PS+ N + C+G++ F N +
Sbjct: 69 VSSMSVNLHGLHDNIDPSIELKRELNLKGLLCNSGKVESFEVFHCNGLLLFTNTS----T 124
Query: 146 LVVWNPCTGKFKILPPLENIPNGKTHTLYSIGYD--RFVDNYKVVAFSCHRQINKSYKYC 203
+VVWNPCTG+ K + N + H Y++GY+ +YK++ F
Sbjct: 125 IVVWNPCTGQTKWIQ--TESANTRYHK-YALGYENKNLCRDYKILRFLDDG--------T 173
Query: 204 NSQVRVHTLGTNFWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIENQKNYDSWVI----L 259
N ++ ++ ++ WR + + + L G G V G W + + D ++
Sbjct: 174 NFELEIYEFNSSSWRVLDSVEID-FELDIGSQGMSVKGNTYWIVIDDHEVDGELVNCFLT 232
Query: 260 SLDLGNESYQEISRPDFGLDDPVHIFTL-GVSKDCLCVLVYTETLL--GIWVMKDYGNK- 315
S D E + F + +L V ++ L VL + E L IWV D +
Sbjct: 233 SFDFTRERFGPHVCLPFQTSWYSDVISLSSVREERLSVLFHEEDSLTMEIWVTNDITDTI 292
Query: 316 -NSWTKLFAVP-YAKVGYHGFGFVDLHYISEED 346
SW+K V Y Y+G F +I EE+
Sbjct: 293 VTSWSKFLRVDLYTHRFYNGVTF----FIDEEN 321
>At5g65850 unknown protein
Length = 392
Score = 69.3 bits (168), Expect = 4e-12
Identities = 88/359 (24%), Positives = 154/359 (42%), Gaps = 51/359 (14%)
Query: 38 LDLFLEILYRLPVKSLLVLKCVSKSLNSLISDPKFVKHHLHLSQTRPYHLLIRNSELLLV 97
+D+ +EIL RLP KS+ +CVSK S+I F + L S RP L + L
Sbjct: 37 VDIIIEILLRLPAKSIATCRCVSKLWISVICRQDFTELFLTRSLHRPQLLFCCKKDGNLF 96
Query: 98 DSRLPSVTAIIPDTT----HNFRLNPSDNHPIMIDSCDGIICFENRNDNHVDLVVWNPCT 153
P + +++ NF L + P+ +G+ICF+ + N V+ NP T
Sbjct: 97 FFSSPQLQNPYENSSAISLKNFSLCYKISRPV-----NGLICFKRKEMNETVTVICNPST 151
Query: 154 GKFKILP-PLENIPNGKTHTLYSIGYDRFVDNYKVVAFSCHRQINKSYKYCNSQVRVHTL 212
G LP P++ SIG RF V +Q Y + + +V TL
Sbjct: 152 GHTLSLPKPMKT----------SIGPSRF-----FVYEPIQKQFKVLLSYKSDEHQVLTL 196
Query: 213 GTN--FWRRIPNFPSNIMGLPNGYVGKFVSGTINWAIENQKNYDSWVILSLDLGNESYQE 270
GT WR I +I+G+ ++G + + N + D ++I+ D+ +E ++
Sbjct: 197 GTGELSWRIIECSMPHILGMSE----ICINGVLYYPAINLSSGD-YIIVCFDVRSEKFRF 251
Query: 271 ISRPDFGLDDPVHIFTL--------GVSKDCLCVLVYTETLLGIWVMKDYGNKNSWTK-L 321
I+ + + H TL + + C + + +WV++D K W+K
Sbjct: 252 ITVMEEFI-KAAHDGTLINYNGKLASLVSERYCFVDGRSKSIELWVLQD-AEKKEWSKHT 309
Query: 322 FAVPYAKVGYHGFGFVDLHYIS-EEDDQVFLHFCSK-----VYVYNYKNSTVKTLDIQG 374
+ +P H G ++L ++ +++ L C + VY +N + T+ ++ IQG
Sbjct: 310 YVLP--AWWQHRIGTLNLRFVGVTRTNEIMLSPCYQTVPFDVYYFNIERKTMMSVAIQG 366
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.140 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,116,093
Number of Sequences: 26719
Number of extensions: 469722
Number of successful extensions: 1403
Number of sequences better than 10.0: 254
Number of HSP's better than 10.0 without gapping: 190
Number of HSP's successfully gapped in prelim test: 64
Number of HSP's that attempted gapping in prelim test: 1025
Number of HSP's gapped (non-prelim): 304
length of query: 404
length of database: 11,318,596
effective HSP length: 102
effective length of query: 302
effective length of database: 8,593,258
effective search space: 2595163916
effective search space used: 2595163916
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC122729.3


