

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122729.1 - phase: 0 /pseudo
(119 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
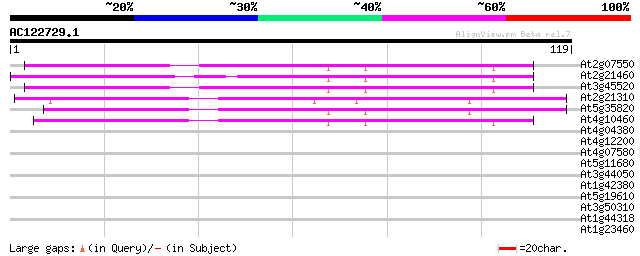
Score E
Sequences producing significant alignments: (bits) Value
At2g07550 putative retroelement pol polyprotein 50 2e-07
At2g21460 putative retroelement pol polyprotein 47 1e-06
At3g45520 copia-like polyprotein 46 3e-06
At2g21310 putative retroelement pol polyprotein 45 6e-06
At5g35820 copia-like retrotransposable element 44 1e-05
At4g10460 putative retrotransposon 42 6e-05
At4g04380 putative polyprotein 37 0.002
At4g12200 putative protein 32 0.051
At4g07580 31 0.086
At5g11680 unknown protein 29 0.33
At3g44050 kinesin -like protein 26 2.8
At1g42380 hypothetical protein 26 3.6
At5g19610 GNOM-like protein 25 6.2
At3g50310 protein kinase -like protein 25 6.2
At1g44318 delta-aminolevulinic acid dehydratase (Alad), putative 25 6.2
At1g23460 putative polygalacturonase precursor 25 6.2
>At2g07550 putative retroelement pol polyprotein
Length = 1356
Score = 49.7 bits (117), Expect = 2e-07
Identities = 38/117 (32%), Positives = 62/117 (52%), Gaps = 15/117 (12%)
Query: 4 EKMIEMADKATSLIVLCLVDKVSIDVKKETTMHGGHVGKVGVIVYDQVFDS*TIPKTKFL 63
E + E KA S IVL + D+V +KKE+T ++ D+++ S +P +
Sbjct: 65 EALEEKKKKARSAIVLSVTDRVLRKIKKEST------AAAMLLALDKLYMSKALPNRIYP 118
Query: 64 LIK-YEVK*SHH-------EEFNKILDNLENIEVHLEDEDKVILLL-CIPKSFESFR 111
K Y K S + +EF +I+ +LEN+ V + DED+ ILLL +PK+F+ +
Sbjct: 119 KQKLYSFKMSENLSVEGNIDEFLQIITDLENMNVIISDEDQAILLLTALPKAFDQLK 175
>At2g21460 putative retroelement pol polyprotein
Length = 1333
Score = 47.4 bits (111), Expect = 1e-06
Identities = 40/120 (33%), Positives = 63/120 (52%), Gaps = 15/120 (12%)
Query: 1 MSQEKMIEMADKATSLIVLCLVDKVSIDVKKETTMHGGHVGKVGVIVYDQVFDS*TIPKT 60
+ +E + E KA S IVL + D+V +KKE + +GV+ D+++ S +P
Sbjct: 62 LRRELLEEKRRKARSAIVLSVTDRVLRKIKKEQSA----AAMLGVL--DKLYMSKALPNR 115
Query: 61 KFLLIK-YEVK*SHH-------EEFNKILDNLENIEVHLEDEDKVILLL-CIPKSFESFR 111
+ K Y K S + +EF +I+ +LEN V + DED+ ILLL +PK F+ R
Sbjct: 116 IYQKQKLYSFKMSENLSIEGNIDEFLRIIADLENTNVLVSDEDQAILLLMSLPKPFDQLR 175
>At3g45520 copia-like polyprotein
Length = 1363
Score = 45.8 bits (107), Expect = 3e-06
Identities = 39/117 (33%), Positives = 59/117 (50%), Gaps = 15/117 (12%)
Query: 4 EKMIEMADKATSLIVLCLVDKVSIDVKKETTMHGGHVGKVGVIVYDQVFDS*TIPKTKFL 63
E E KA S IVL + D+V +KKET+ + D+++ S +P +L
Sbjct: 65 EAFEEKKRKARSTIVLSVSDRVLRKIKKETS------AAAMLEALDRLYMSKALPNRIYL 118
Query: 64 LIK-YEVK*SHH-------EEFNKILDNLENIEVHLEDEDKVILLL-CIPKSFESFR 111
K Y K S + +EF I+ +LEN+ V + DED+ ILLL +PK F+ +
Sbjct: 119 KQKLYSFKMSENLSIEGNIDEFLHIVADLENLNVLVSDEDQAILLLMSLPKPFDQLK 175
>At2g21310 putative retroelement pol polyprotein
Length = 838
Score = 45.1 bits (105), Expect = 6e-06
Identities = 36/127 (28%), Positives = 68/127 (53%), Gaps = 16/127 (12%)
Query: 2 SQEKMI-EMADKATSLIVLCLVDKVSIDVKKETTMHGGHVGKVGVIVYDQVFDS*TIPKT 60
+++K++ E KA S ++L L + V V KE T G + V D++F + ++P
Sbjct: 59 TEDKVLKEKRGKARSTVILSLGNHVLRKVIKEKTAAGM------IRVLDKLFMAKSLPNR 112
Query: 61 KFL---LIKYEVK*S-----HHEEFNKILDNLENIEVHLEDEDK-VILLLCIPKSFESFR 111
+L L Y++ S + +F K++ +LEN++V + DED+ ++LL+ +PK F+ +
Sbjct: 113 IYLKQRLYGYKMSDSMTIEENVNDFFKLISDLENVKVSVPDEDQAIVLLMSLPKQFDQLK 172
Query: 112 RPFSMAK 118
K
Sbjct: 173 DTLKYGK 179
>At5g35820 copia-like retrotransposable element
Length = 1342
Score = 44.3 bits (103), Expect = 1e-05
Identities = 35/120 (29%), Positives = 61/120 (50%), Gaps = 15/120 (12%)
Query: 8 EMADKATSLIVLCLVDKVSIDVKKETTMHGGHVGKVGVIVYDQVFDS*TIPKTKFLLIK- 66
E KA S I+L L + V V K+ T G + V DQ+F + ++P +L +
Sbjct: 74 EKRGKARSTIILSLGNNVLRKVIKQKTAAGM------IKVLDQLFMAKSLPNRIYLKQRL 127
Query: 67 YEVK*SHH-------EEFNKILDNLENIEVHLEDEDK-VILLLCIPKSFESFRRPFSMAK 118
Y K S + +F K++ +LEN++V + DED+ ++LL+ +P+ F+ + K
Sbjct: 128 YGYKMSENMTMEENVNDFFKLISDLENVKVVVPDEDQAIVLLMSLPRQFDQLKETLKYCK 187
>At4g10460 putative retrotransposon
Length = 1230
Score = 41.6 bits (96), Expect = 6e-05
Identities = 37/115 (32%), Positives = 56/115 (48%), Gaps = 15/115 (13%)
Query: 6 MIEMADKATSLIVLCLVDKVSIDVKKETTMHGGHVGKVGVIVYDQVFDS*TIPKTKFLLI 65
M E KA S IVL + D+V KKE T + D+++ S +P +L
Sbjct: 65 MEEKRQKARSTIVLSVSDQVLRKSKKEKTAPSM------LEALDKLYMSKALPNRIYLKQ 118
Query: 66 K-YEVK*SHH-------EEFNKILDNLENIEVHLEDEDKVILLL-CIPKSFESFR 111
K Y K + +EF +++ +LEN V + DED+ ILLL +PK F+ +
Sbjct: 119 KLYSYKMQENLSVEGNIDEFLRLIADLENTNVLVSDEDQAILLLMSLPKQFDQLK 173
>At4g04380 putative polyprotein
Length = 778
Score = 36.6 bits (83), Expect = 0.002
Identities = 26/72 (36%), Positives = 41/72 (56%), Gaps = 9/72 (12%)
Query: 49 DQVFDS*TIPKTKFLLIK-YEVK*SHH-------EEFNKILDNLENIEVHLEDEDKVILL 100
D+++ S +P +L K Y K S + +EF I+ +LEN+ V + DED+ ILL
Sbjct: 73 DRLYMSKALPNQIYLKQKLYRFKMSENLSMEGNIDEFLHIVADLENLNVLVSDEDQTILL 132
Query: 101 L-CIPKSFESFR 111
L +PKSF+ +
Sbjct: 133 LMSLPKSFDQLK 144
>At4g12200 putative protein
Length = 200
Score = 32.0 bits (71), Expect = 0.051
Identities = 14/37 (37%), Positives = 24/37 (64%), Gaps = 1/37 (2%)
Query: 75 EEFNKILDNLENIEVHLEDEDKVI-LLLCIPKSFESF 110
+EF ++D L N +V + DED+ I L++ +P+ E F
Sbjct: 12 DEFQWLIDELSNAKVEIFDEDQAIYLMISLPEQIEKF 48
>At4g07580
Length = 323
Score = 31.2 bits (69), Expect = 0.086
Identities = 17/48 (35%), Positives = 24/48 (49%)
Query: 29 VKKETTMHGGHVGKVGVIVYDQVFDS*TIPKTKFLLIKYEVK*SHHEE 76
VKKE+ GGHV K + +D TI K +++ + K S H E
Sbjct: 10 VKKESVRRGGHVEKNKEVAVVNKYDGDTIEMLKKFIVEQQKKPSKHVE 57
>At5g11680 unknown protein
Length = 207
Score = 29.3 bits (64), Expect = 0.33
Identities = 24/73 (32%), Positives = 33/73 (44%), Gaps = 4/73 (5%)
Query: 23 DKVSIDVKKETTMHGGHVGKVGVIVYD---QVFDS*TIPKTKFLLIKYEVK*SHHEEFNK 79
D V +V K HGGHV GVI VF S + P F+ + H E+FN+
Sbjct: 26 DGVEFEVDKIPGGHGGHVKAKGVIYLSNIRMVFVS-SKPVDNFVAFDMPLLYIHAEKFNQ 84
Query: 80 ILDNLENIEVHLE 92
+ + NI +E
Sbjct: 85 PIFHCNNIAGQVE 97
>At3g44050 kinesin -like protein
Length = 1229
Score = 26.2 bits (56), Expect = 2.8
Identities = 14/37 (37%), Positives = 22/37 (58%)
Query: 65 IKYEVK*SHHEEFNKILDNLENIEVHLEDEDKVILLL 101
I EV+ S EE K++ NLEN + DE++ ++ L
Sbjct: 796 IALEVEKSAAEEQKKMIGNLENQLTEMHDENEKLMSL 832
>At1g42380 hypothetical protein
Length = 143
Score = 25.8 bits (55), Expect = 3.6
Identities = 11/27 (40%), Positives = 20/27 (73%)
Query: 75 EEFNKILDNLENIEVHLEDEDKVILLL 101
++F KI+ +L +I++ + DE + ILLL
Sbjct: 12 DDFLKIVVDLNHIQIEVRDEVQAILLL 38
>At5g19610 GNOM-like protein
Length = 1375
Score = 25.0 bits (53), Expect = 6.2
Identities = 11/28 (39%), Positives = 16/28 (56%)
Query: 79 KILDNLENIEVHLEDEDKVILLLCIPKS 106
++++NLE VH DED I L + S
Sbjct: 257 EVVENLEGTNVHTADEDVQIFALVLINS 284
>At3g50310 protein kinase -like protein
Length = 342
Score = 25.0 bits (53), Expect = 6.2
Identities = 11/29 (37%), Positives = 18/29 (61%)
Query: 82 DNLENIEVHLEDEDKVILLLCIPKSFESF 110
D+ EN V ++DEDKV++ P F+ +
Sbjct: 275 DHRENFVVKVKDEDKVLMSPKCPFEFDDW 303
>At1g44318 delta-aminolevulinic acid dehydratase (Alad), putative
Length = 406
Score = 25.0 bits (53), Expect = 6.2
Identities = 11/18 (61%), Positives = 14/18 (77%)
Query: 33 TTMHGGHVGKVGVIVYDQ 50
TT HGG VG+ GVI+ D+
Sbjct: 205 TTGHGGIVGEDGVILNDE 222
>At1g23460 putative polygalacturonase precursor
Length = 457
Score = 25.0 bits (53), Expect = 6.2
Identities = 11/21 (52%), Positives = 15/21 (71%)
Query: 80 ILDNLENIEVHLEDEDKVILL 100
ILD LEN +V ++D+D LL
Sbjct: 31 ILDELENFDVLVDDDDDTKLL 51
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.330 0.144 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,482,696
Number of Sequences: 26719
Number of extensions: 92192
Number of successful extensions: 290
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 1
Number of HSP's that attempted gapping in prelim test: 267
Number of HSP's gapped (non-prelim): 17
length of query: 119
length of database: 11,318,596
effective HSP length: 95
effective length of query: 24
effective length of database: 8,780,291
effective search space: 210726984
effective search space used: 210726984
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 52 (24.6 bits)
Medicago: description of AC122729.1


