

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122727.10 - phase: 1 /pseudo
(136 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
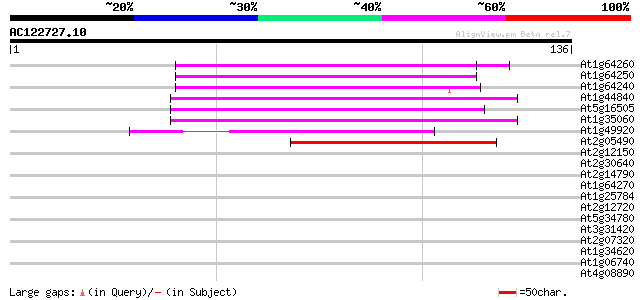
Score E
Sequences producing significant alignments: (bits) Value
At1g64260 hypothetical protein 50 3e-07
At1g64250 hypothetical protein 49 1e-06
At1g64240 hypothetical protein 47 4e-06
At1g44840 hypothetical protein 46 5e-06
At5g16505 mutator-like transposase-like protein (MQK4.25) 45 9e-06
At1g35060 hypothetical protein 45 2e-05
At1g49920 hypothetical protein 43 6e-05
At2g05490 Mutator-like transposase 40 5e-04
At2g12150 pseudogene 37 0.004
At2g30640 Mutator-like transposase 36 0.007
At2g14790 Mutator-like transposase 35 0.016
At1g64270 putative transposon 34 0.020
At1g25784 mutator-like transposase, putative 34 0.020
At2g12720 putative protein 33 0.045
At5g34780 3-methyl-2-oxobutanoate dehydrogenase-like protein 32 0.10
At3g31420 hypothetical protein 32 0.10
At2g07320 Mutator-like transposase 32 0.10
At1g34620 Mutator-like protein 32 0.10
At1g06740 mudrA-like protein 32 0.10
At4g08890 putative transposon protein 32 0.13
>At1g64260 hypothetical protein
Length = 1490
Score = 50.4 bits (119), Expect = 3e-07
Identities = 22/73 (30%), Positives = 37/73 (50%)
Query: 41 FLRTFKNTEMKKKVRNMGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGR 100
F R F + + ++R G + + Y N I++ + + KW+D P+ +W AHD GR
Sbjct: 485 FSRVFPSFCLGARIRRAGSTSQKDEFVSYMNDIKEKNPEARKWLDQFPQNRWALAHDNGR 544
Query: 101 RWGHITTNISECF 113
R+G + N F
Sbjct: 545 RYGIMEINTKALF 557
Score = 46.6 bits (109), Expect = 4e-06
Identities = 24/81 (29%), Positives = 40/81 (48%), Gaps = 1/81 (1%)
Query: 41 FLRTFKNTEMKKKVRNMGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGR 100
FL F++ ++ V G + + Y N I++ + + KW+D P+ +W AHD G
Sbjct: 1249 FLGVFRDYNLESLVEQAGSTNQKEEFDSYMNDIKEKNPEAWKWLDQIPRHKWALAHDSGL 1308
Query: 101 RWGHITTNISECFNNVLKGGP 121
R+G I + E V +G P
Sbjct: 1309 RYGIIEID-REALFAVCRGFP 1328
>At1g64250 hypothetical protein
Length = 790
Score = 48.5 bits (114), Expect = 1e-06
Identities = 23/73 (31%), Positives = 36/73 (48%)
Query: 41 FLRTFKNTEMKKKVRNMGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGR 100
F F++ +K V G + + Y N+I + + KW+D P+ QW QAHD GR
Sbjct: 506 FHYVFQDDYLKNLVYEAGSTSEKEEFDSYMNEIEKKNSEARKWLDQFPQYQWAQAHDSGR 565
Query: 101 RWGHITTNISECF 113
R+ +T + F
Sbjct: 566 RYRVMTIDAENLF 578
>At1g64240 hypothetical protein
Length = 938
Score = 46.6 bits (109), Expect = 4e-06
Identities = 24/77 (31%), Positives = 38/77 (49%), Gaps = 3/77 (3%)
Query: 41 FLRTFKNTEMKKKVRNMGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGR 100
F F++ ++ V+ G + + Y I + + KW+D P+ QW AHD GR
Sbjct: 675 FSGIFRDYYLEDLVKRAGSTSQKEEFDSYMKDIEKKNSEARKWLDQFPQNQWALAHDNGR 734
Query: 101 RWGHI---TTNISECFN 114
R+G + TT + E FN
Sbjct: 735 RYGIMEIETTTLFEDFN 751
>At1g44840 hypothetical protein
Length = 926
Score = 46.2 bits (108), Expect = 5e-06
Identities = 23/84 (27%), Positives = 41/84 (48%)
Query: 40 NFLRTFKNTEMKKKVRNMGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQG 99
N +KN + + V+N GY+ T + QI + + K++ D W + + +G
Sbjct: 668 NIQTKYKNKALTQLVKNAGYAFTGTKFKEFYGQIETTNQNCGKYLHDIGMANWTRHYFRG 727
Query: 100 RRWGHITTNISECFNNVLKGGPQS 123
+R+ +T+NI+E N L G S
Sbjct: 728 QRFNLMTSNIAETLNKALNKGRSS 751
>At5g16505 mutator-like transposase-like protein (MQK4.25)
Length = 597
Score = 45.4 bits (106), Expect = 9e-06
Identities = 25/76 (32%), Positives = 38/76 (49%)
Query: 40 NFLRTFKNTEMKKKVRNMGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQG 99
NF TFKNT++ + Y++T N++ + S DVV+W + W A+ QG
Sbjct: 317 NFRDTFKNTKLVNIFWSAVYALTPAEFETKSNEMIEISQDVVQWFELYLPHLWAVAYFQG 376
Query: 100 RRWGHITTNISECFNN 115
R+GH I+E N
Sbjct: 377 VRYGHFGLGITEVLYN 392
>At1g35060 hypothetical protein
Length = 873
Score = 44.7 bits (104), Expect = 2e-05
Identities = 25/84 (29%), Positives = 39/84 (45%)
Query: 40 NFLRTFKNTEMKKKVRNMGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQG 99
N FKN + + V+N GY T NQI + +K++ D W + + G
Sbjct: 619 NIQARFKNRGLTQLVKNAGYEFTSGKFKTLYNQINAINPLCIKYLHDVGMAHWTRLYFPG 678
Query: 100 RRWGHITTNISECFNNVLKGGPQS 123
+R+ +T+NI+E N L G S
Sbjct: 679 QRFNLMTSNIAETLNKALFKGRSS 702
>At1g49920 hypothetical protein
Length = 785
Score = 42.7 bits (99), Expect = 6e-05
Identities = 23/74 (31%), Positives = 35/74 (47%), Gaps = 11/74 (14%)
Query: 30 TMSSAFDT*LNFLRTFKNTEMKKKVRNMGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPK 89
++S FD ++FL V G S + Y +I++ + + KW+D P
Sbjct: 483 SVSPGFDYNMHFL-----------VDEAGSSSQKEEFDSYMKEIKERNPEAWKWLDQFPP 531
Query: 90 KQWLQAHDQGRRWG 103
QW AHD GRR+G
Sbjct: 532 HQWALAHDDGRRYG 545
>At2g05490 Mutator-like transposase
Length = 942
Score = 39.7 bits (91), Expect = 5e-04
Identities = 17/50 (34%), Positives = 30/50 (60%)
Query: 69 YRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGRRWGHITTNISECFNNVLK 118
Y ++R WS K+++D + W +A+ + R+ +T+N SE NNVL+
Sbjct: 697 YFEELRGWSPGCAKYLEDVGFEHWTRAYCKEERYNIMTSNNSEAMNNVLR 746
>At2g12150 pseudogene
Length = 942
Score = 36.6 bits (83), Expect = 0.004
Identities = 17/49 (34%), Positives = 29/49 (58%)
Query: 69 YRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGRRWGHITTNISECFNNVL 117
Y ++R S K+++D + W +A+ +G R+ +T+N SE NNVL
Sbjct: 697 YFEELRGRSPGCAKYLEDVGFEHWTRAYCKGERYNIMTSNNSEAMNNVL 745
>At2g30640 Mutator-like transposase
Length = 754
Score = 35.8 bits (81), Expect = 0.007
Identities = 18/71 (25%), Positives = 30/71 (41%)
Query: 45 FKNTEMKKKVRNMGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGRRWGH 104
F N+ + V +T +I S + W+ + QW +G R+GH
Sbjct: 477 FNNSILVNLVWEAAKCLTDFEFEGKMGEIAQISPEAASWIRNIQHSQWATYCFEGTRFGH 536
Query: 105 ITTNISECFNN 115
+T N+SE N+
Sbjct: 537 LTANVSESLNS 547
>At2g14790 Mutator-like transposase
Length = 895
Score = 34.7 bits (78), Expect = 0.016
Identities = 16/49 (32%), Positives = 29/49 (58%)
Query: 69 YRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGRRWGHITTNISECFNNVL 117
Y +R+ S K+++D + W +AH +G R+ +++N SE N+VL
Sbjct: 607 YFELLREKSGKCAKYLEDIGFEHWTRAHCRGERYNIMSSNNSESMNHVL 655
>At1g64270 putative transposon
Length = 162
Score = 34.3 bits (77), Expect = 0.020
Identities = 19/71 (26%), Positives = 28/71 (38%), Gaps = 1/71 (1%)
Query: 39 LNFLRTFKNTEMKKKVRNMGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQ 98
L F F + + V+ G + + Y I+ + KW+ P+ QW AHDQ
Sbjct: 85 LQFHDIFGDYNLVSLVKQAGSTSQKEEFDSYIKDIKKKDSEARKWLAQFPQNQWALAHDQ 144
Query: 99 -GRRWGHITTN 108
W H N
Sbjct: 145 WSEIWSHDDRN 155
>At1g25784 mutator-like transposase, putative
Length = 1127
Score = 34.3 bits (77), Expect = 0.020
Identities = 16/49 (32%), Positives = 29/49 (58%)
Query: 69 YRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGRRWGHITTNISECFNNVL 117
Y +R+ S K+++D + W +AH +G R+ +++N SE N+VL
Sbjct: 698 YFELLREKSAKCAKYLEDIGFEHWTRAHCRGERYNIMSSNNSESMNHVL 746
>At2g12720 putative protein
Length = 819
Score = 33.1 bits (74), Expect = 0.045
Identities = 20/78 (25%), Positives = 34/78 (42%)
Query: 40 NFLRTFKNTEMKKKVRNMGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQG 99
N L FK ++ V+ T+ N+I + + ++ + W +AH G
Sbjct: 543 NILVKFKRKDLFPLVKKAARCYRLNDFTNAFNEIEELDPLLHAYLQRAGVEMWARAHFPG 602
Query: 100 RRWGHITTNISECFNNVL 117
R+ +TTNI+E N L
Sbjct: 603 DRYNLMTTNIAESMNRAL 620
>At5g34780 3-methyl-2-oxobutanoate dehydrogenase-like protein
Length = 365
Score = 32.0 bits (71), Expect = 0.10
Identities = 18/68 (26%), Positives = 34/68 (49%), Gaps = 5/68 (7%)
Query: 59 YSMTQPSITHYRNQIRD---WSDDVVKWVDDNPKKQWLQAHDQGRRWGHITTNISECFNN 115
+ M++ S+ +R + D WS++ + N +KQ LQA +W ++E FN+
Sbjct: 152 WKMSRNSVNRFRKSVEDNGWWSEEDESKLRSNARKQLLQAIQAAEKWE--KQPLTELFND 209
Query: 116 VLKGGPQS 123
V P++
Sbjct: 210 VYDVKPKN 217
>At3g31420 hypothetical protein
Length = 1491
Score = 32.0 bits (71), Expect = 0.10
Identities = 17/57 (29%), Positives = 28/57 (48%), Gaps = 3/57 (5%)
Query: 76 WSDDVVKWVDDNPKKQWLQAHDQGRRWGHITTNISECFNNVLKGGPQSSNHTNCPGN 132
++D+ + W + +WL A D+ R+ H TT C N +L Q S+ C G+
Sbjct: 512 YADEEIYW-QTKSRSRWLNAGDRNTRYFHSTTKTRRCRNRLL--SVQDSDGDICRGD 565
>At2g07320 Mutator-like transposase
Length = 534
Score = 32.0 bits (71), Expect = 0.10
Identities = 16/67 (23%), Positives = 34/67 (49%)
Query: 54 VRNMGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGRRWGHITTNISECF 113
V N+G + +I+ + + K+++ +W + + QG R+ +T+NI+E
Sbjct: 2 VLNVGCVYKMSVFRNLYKEIKSRNYECDKYLEKIGPARWSRVYFQGERYNLMTSNIAETL 61
Query: 114 NNVLKGG 120
NN ++ G
Sbjct: 62 NNAIRLG 68
>At1g34620 Mutator-like protein
Length = 1034
Score = 32.0 bits (71), Expect = 0.10
Identities = 11/28 (39%), Positives = 20/28 (71%)
Query: 91 QWLQAHDQGRRWGHITTNISECFNNVLK 118
+W + H +GRR+ + +NI E +NNV++
Sbjct: 708 EWTRVHSKGRRFNIMDSNICESWNNVIR 735
>At1g06740 mudrA-like protein
Length = 726
Score = 32.0 bits (71), Expect = 0.10
Identities = 17/76 (22%), Positives = 35/76 (45%), Gaps = 1/76 (1%)
Query: 41 FLRTFKNTEMKKKVRNMGYSMTQPSITHYRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGR 100
F R F+++ + + +T N+I S + W+ + +W ++ +G
Sbjct: 451 FQREFQSSVLVDLFWEAAHCLTVLEFKSKINKIEQISPEASLWIQNKSPARWASSYFEGT 510
Query: 101 RWGHITTN-ISECFNN 115
R+G +T N I+E +N
Sbjct: 511 RFGQLTANVITESLSN 526
>At4g08890 putative transposon protein
Length = 1028
Score = 31.6 bits (70), Expect = 0.13
Identities = 14/49 (28%), Positives = 28/49 (56%)
Query: 69 YRNQIRDWSDDVVKWVDDNPKKQWLQAHDQGRRWGHITTNISECFNNVL 117
Y +R+ S K+++D + W + H +G+ + +++N SE N+VL
Sbjct: 616 YFGLLREKSAKCAKYLEDIGFEHWTRVHCRGKHYNIMSSNNSESMNHVL 664
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.329 0.139 0.467
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,162,912
Number of Sequences: 26719
Number of extensions: 119597
Number of successful extensions: 454
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 48
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 400
Number of HSP's gapped (non-prelim): 62
length of query: 136
length of database: 11,318,596
effective HSP length: 89
effective length of query: 47
effective length of database: 8,940,605
effective search space: 420208435
effective search space used: 420208435
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 54 (25.4 bits)
Medicago: description of AC122727.10


