

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122725.8 - phase: 0 /pseudo
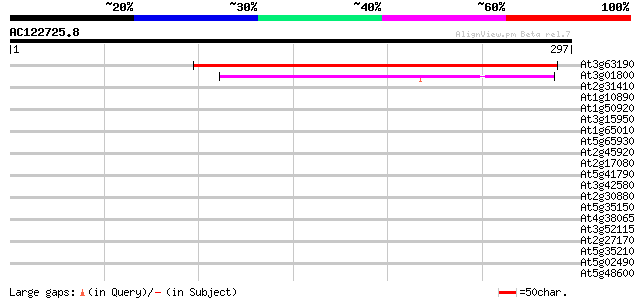
(297 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g63190 unknown protein 305 2e-83
At3g01800 putative ribosome recycling factor 102 3e-22
At2g31410 unknown protein 38 0.008
At1g10890 unknown protein 38 0.008
At1g50920 hypothetical protein 33 0.14
At3g15950 unknown protein 33 0.18
At1g65010 hypothetical protein 33 0.24
At5g65930 kinesin-like calmodulin-binding protein 32 0.32
At2g45920 unknown protein 32 0.32
At2g17080 hypothetical protein 32 0.32
At5g41790 myosin heavy chain-like protein 32 0.41
At3g42580 putative protein 32 0.41
At2g30880 unknown protein 32 0.41
At5g35150 putative protein 32 0.54
At4g38065 hypothetical protein 32 0.54
At3g52115 gamma response I protein 32 0.54
At2g27170 putative chromosome associated protein 32 0.54
At5g35210 putative protein 31 0.70
At5g02490 dnaK-type molecular chaperone hsc70.1 - like 31 0.70
At5g48600 chromosome condensation protein 31 0.92
>At3g63190 unknown protein
Length = 275
Score = 305 bits (781), Expect = 2e-83
Identities = 152/193 (78%), Positives = 181/193 (93%)
Query: 98 VRAATIEEIEAEKASIENDVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSPV 157
+R+ATIEEIEAEK++IE DVKS+ME+T++ ++T+F+SIRTGR+ +MLDKIEVEYYGSPV
Sbjct: 80 IRSATIEEIEAEKSAIETDVKSKMEKTIETLRTSFNSIRTGRSNAAMLDKIEVEYYGSPV 139
Query: 158 SLKTIAQISTPDASSLLVSPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSDR 217
SLK+IAQISTPD SSLL+ PYDKSSLKAIEKAIVNSD+G+TPNNDG+VIRLS+P LTSDR
Sbjct: 140 SLKSIAQISTPDGSSLLLQPYDKSSLKAIEKAIVNSDLGVTPNNDGDVIRLSLPPLTSDR 199
Query: 218 RKELTKIVSKQVEEGKVALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYIK 277
RKEL+K+V+KQ EEGKVALRNIRRDALK+Y+KLEK+KKLSEDNVKDLSSDLQKL D Y+K
Sbjct: 200 RKELSKVVAKQSEEGKVALRNIRRDALKSYDKLEKEKKLSEDNVKDLSSDLQKLIDVYMK 259
Query: 278 KVDALYKQKEKVL 290
K++ LYKQKEK L
Sbjct: 260 KIEELYKQKEKEL 272
>At3g01800 putative ribosome recycling factor
Length = 284
Score = 102 bits (253), Expect = 3e-22
Identities = 64/195 (32%), Positives = 100/195 (50%), Gaps = 20/195 (10%)
Query: 112 SIENDVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSPVSLKTIAQISTPDAS 171
+++ S+ME +D + + + +RTGRA P MLD I VE G + L +A +S D
Sbjct: 87 TVKAAASSQMEAAIDALSRDLTKLRTGRAAPGMLDHIVVETGGVKMPLNHLALVSVLDPK 146
Query: 172 SLLVSPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSD--------------- 216
+L V+PYD ++K +EKAIV S +G+ P DG+ + SIP S
Sbjct: 147 TLSVNPYDPDTVKELEKAIVASPLGLNPKLDGQRLVASIPAYESSSLSNFISLRLSLDPI 206
Query: 217 ---RRKELTKIVSKQVEEGKVALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTD 273
+ + KIV+K E K ++R R+ AL +K L +D VK L ++ +LT
Sbjct: 207 YLYELQAMCKIVTKSSEVVKQSIRRARQKALDTIKK--AGSSLPKDEVKRLEKEVDELTK 264
Query: 274 DYIKKVDALYKQKEK 288
++K + + K KEK
Sbjct: 265 KFVKSAEDMCKSKEK 279
>At2g31410 unknown protein
Length = 199
Score = 37.7 bits (86), Expect = 0.008
Identities = 34/155 (21%), Positives = 73/155 (46%), Gaps = 8/155 (5%)
Query: 140 ATPSMLDKIEVEYYGSPVSLKTIAQISTPDASSLLVSPYDKSSLKAIEKAIV---NSDVG 196
A ++ D+ ++ G+P + K A S+ + + ++ + + V N
Sbjct: 30 AAEAIADEASMDIDGAPPAAKRSAVASSENPDKPIALAVERPTYDGVIAGKVSGRNWKQP 89
Query: 197 MTPNNDGEVIRLSIPQLTS-DRRKELTKIVSKQVEEGKVALRNIRRDALKAYEKLEKDKK 255
T + G ++ P L R++E+ K ++V E K IR + ++ +K E+ +K
Sbjct: 90 RTHRSSGRFVKNRKPDLEEMKRQREIKKAYKERVNELK---EEIRSNKVEKRKKKEEREK 146
Query: 256 LSEDNVKDLSSDLQKLTD-DYIKKVDALYKQKEKV 289
++NV + LQK+T+ +KK+ KQ++++
Sbjct: 147 RKKENVLRTGTKLQKITNPKTLKKISMSKKQRKQL 181
>At1g10890 unknown protein
Length = 592
Score = 37.7 bits (86), Expect = 0.008
Identities = 27/120 (22%), Positives = 65/120 (53%), Gaps = 11/120 (9%)
Query: 179 DKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSDRRKELTKIVSKQVEEGKVA-LR 237
++ ++K +E+AI + + E I++ I L + RK L + V+ Q+EE K A L
Sbjct: 83 EEETVKRVEEAIRKK---VEESLQSEKIKMEILTLLEEGRKRLNEEVAAQLEEEKEASLI 139
Query: 238 NIRRDALKAYEKLEKDKKLSEDNVKDL-------SSDLQKLTDDYIKKVDALYKQKEKVL 290
+ + ++ E+ ++++E+N+K + + + Q+ ++ ++++ L +QKE+ +
Sbjct: 140 EAKEKEEREQQEKEERERIAEENLKRVEEAQRKEAMERQRKEEERYRELEELQRQKEEAM 199
>At1g50920 hypothetical protein
Length = 671
Score = 33.5 bits (75), Expect = 0.14
Identities = 40/183 (21%), Positives = 75/183 (40%), Gaps = 20/183 (10%)
Query: 106 IEAEKASIENDVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSPVSLKTIAQI 165
+ + A+ E + R+E + + K N R A P D IE L I Q+
Sbjct: 339 MSVKNAACERLLDQRVEAKMKSKKINDHLNRFHVAIPKPRDSIE--------RLPCIPQV 390
Query: 166 STPDASSLLVSPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSDRRKELTKIV 225
V A+EK D+ N V S+ + + E + +
Sbjct: 391 ---------VLEAKAKEAAAMEKRKTEKDLE-EENGGAGVYSASLKKNYILQHDEWKEDI 440
Query: 226 SKQVEEGKVALRNIRRDALKAYEKLEKDKKLSEDNVK--DLSSDLQKLTDDYIKKVDALY 283
++ +G I D L+ +LE+++ + E V+ D+ D++KL+D+ +K++ +
Sbjct: 441 MPEILDGHNVADFIDPDILQRLAELEREEGIREAGVEEADMEMDIEKLSDEQLKQLSEIR 500
Query: 284 KQK 286
K+K
Sbjct: 501 KKK 503
>At3g15950 unknown protein
Length = 622
Score = 33.1 bits (74), Expect = 0.18
Identities = 45/208 (21%), Positives = 81/208 (38%), Gaps = 21/208 (10%)
Query: 99 RAATIEEIEAEKASIENDVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSPVS 158
R + ++ IE E ++ K ++E DN SMLD+IE E+ + S
Sbjct: 152 RQSMLDAIEREFEAVTESFK-QLEDIADNKAEGDDE---SAKRQSMLDEIEREFEAATNS 207
Query: 159 LKTIAQISTPDASSLLVSPYDKSSLKAIEKAIVNSDVGM----------TPNNDGEVIRL 208
LK + + S S L+AIE+ + G+ ++D V R
Sbjct: 208 LKQLNLDDFSEGDDSAESARRNSMLEAIEREFEAATKGLEELKANDSTGDKDDDEHVARR 267
Query: 209 SIPQLTSDRRKELTKIVSKQVEEGKVALRNIRRDALKAYEK-------LEKDKKLSEDNV 261
I +R E ++++ N R L+A+E+ + + + S N
Sbjct: 268 KIMLEAIEREFEAATKGLEELKNESEQAENKRNSMLEAFEREFEAATNAKANGENSAKNP 327
Query: 262 KDLSSDLQKLTDDYIKKVDALYKQKEKV 289
+S+ +QK + Y ++ L K + V
Sbjct: 328 STISTTVQKSSGGYNAGLEGLLKPADGV 355
>At1g65010 hypothetical protein
Length = 1318
Score = 32.7 bits (73), Expect = 0.24
Identities = 51/214 (23%), Positives = 96/214 (44%), Gaps = 32/214 (14%)
Query: 102 TIEEIEAEKASIENDVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEV--EYYGSPVSL 159
T+ E +AE ++ + + E L NV SS+R S+L+KIE + S V
Sbjct: 576 TLGEAKAESMKLKESLLDK-EEDLKNVTAEISSLREWEG--SVLEKIEELSKVKESLVDK 632
Query: 160 KTIAQISTPDASSLLVSPYDKSSLKAIEK----------------AIVNSDVGMTPNNDG 203
+T Q T +A L + + +K IE+ +IV + G
Sbjct: 633 ETKLQSITQEAEEL--KGREAAHMKQIEELSTANASLVDEATKLQSIVQESEDLKEKEAG 690
Query: 204 ---EVIRLSIP-QLTSDRRKELTKIV--SKQVEEGKVA-LRNIRRDALKAYEKLEKDKKL 256
++ LS+ + +D +L IV SK ++E +VA L+ I ++ ++K+ KL
Sbjct: 691 YLKKIEELSVANESLADNVTDLQSIVQESKDLKEREVAYLKKIEELSVANESLVDKETKL 750
Query: 257 SEDNVKDLSSDLQKLTDDYIKKVDALYKQKEKVL 290
++ + +L+ ++KK++ L K+ E ++
Sbjct: 751 Q--HIDQEAEELRGREASHLKKIEELSKENENLV 782
>At5g65930 kinesin-like calmodulin-binding protein
Length = 1260
Score = 32.3 bits (72), Expect = 0.32
Identities = 38/192 (19%), Positives = 81/192 (41%), Gaps = 11/192 (5%)
Query: 104 EEIEAEKASIENDVKSRMERTLDNVKTNFSSIRTGRATPSMLDKIEVEYYGSPVSLKTIA 163
EE+EA +E + + +E TLD K G S++ ++ G L
Sbjct: 657 EELEAIHNGLELERRKLLEVTLDRDKLRSLCDEKGTTIQSLMSELR----GMEARLAKSG 712
Query: 164 QISTPDASSLLVSPYDKSSLKAIEK--AIVNSDVGMTPNNDGEVIR----LSIPQLTSDR 217
+ + ++ + L I+K + N ++ + +N ++ L +
Sbjct: 713 NTKSSKETKSELAEMNNQILYKIQKELEVRNKELHVAVDNSKRLLSENKILEQNLNIEKK 772
Query: 218 RKELTKIVSKQVEEGKVALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYIK 277
+KE +I K+ E+ K L+ + E L +D +E ++ +SD+ L ++ +K
Sbjct: 773 KKEEVEIHQKRYEQEKKVLKLRVSELENKLEVLAQDLDSAESTIESKNSDMLLLQNN-LK 831
Query: 278 KVDALYKQKEKV 289
+++ L + KE +
Sbjct: 832 ELEELREMKEDI 843
>At2g45920 unknown protein
Length = 325
Score = 32.3 bits (72), Expect = 0.32
Identities = 21/62 (33%), Positives = 33/62 (52%), Gaps = 7/62 (11%)
Query: 227 KQVEEGKVALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYIKKVDALYKQK 286
KQ +E ++ L+ +R EKLEK + +SE+ + + +QKL D Y L K K
Sbjct: 165 KQRKETEMELKKVR-------EKLEKMRYISENRITESYMLVQKLQDKYNLATKVLRKAK 217
Query: 287 EK 288
E+
Sbjct: 218 EE 219
>At2g17080 hypothetical protein
Length = 263
Score = 32.3 bits (72), Expect = 0.32
Identities = 29/118 (24%), Positives = 52/118 (43%), Gaps = 6/118 (5%)
Query: 180 KSSLKAIEKAIVNSDVGMTPNNDGEVIRLSI-----PQLTSDRRKELTKIVSKQVEEGKV 234
+ SLK + +A N+D + + E I LS+ ++ + +VSK + + KV
Sbjct: 144 QKSLK-VTQAEDNNDDTLAVFGEAEAITLSLFDSLLSYMSGSKTCSKWSVVSKLMNKKKV 202
Query: 235 ALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYIKKVDALYKQKEKVLYI 292
+ K + + +K L D+V++L S +Q L D +L K + L I
Sbjct: 203 TCEAQENEFTKVDSEFQSEKTLKMDDVQNLESCIQDLEDGLESLSKSLIKYRVSFLNI 260
>At5g41790 myosin heavy chain-like protein
Length = 1305
Score = 32.0 bits (71), Expect = 0.41
Identities = 44/192 (22%), Positives = 88/192 (44%), Gaps = 16/192 (8%)
Query: 105 EIEAEKASIE-----NDVKSRMERTLDNVKTNFSSIR--TGRATPSMLDKIEVEYYGSPV 157
EI+ EK S E + + + E ++ VK + S + G + ++E+E G
Sbjct: 892 EIQLEKKSEEISEYLSQITNLKEEIINKVKVHESILEEINGLSEKIKGRELELETLGKQR 951
Query: 158 SLKTIAQISTPDASSLLVSPYDKSSLKAIEKAIVNSDVGMTPNN-DGEVIRLSIPQLTSD 216
S + ++ T + V +DK ++ + E + + N D ++ S + +
Sbjct: 952 S-ELDEELRTKKEEN--VQMHDKINVASSEIMALTELINNLKNELDSLQVQKSETEAELE 1008
Query: 217 RRKELTKIVSKQVEEGKVALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYI 276
R K+ +S Q+ + + AL + ++A AY LE++ K + K+ + L K+T DY
Sbjct: 1009 REKQEKSELSNQITDVQKAL--VEQEA--AYNTLEEEHKQINELFKETEATLNKVTVDY- 1063
Query: 277 KKVDALYKQKEK 288
K+ L +++ K
Sbjct: 1064 KEAQRLLEERGK 1075
>At3g42580 putative protein
Length = 903
Score = 32.0 bits (71), Expect = 0.41
Identities = 19/65 (29%), Positives = 34/65 (52%), Gaps = 1/65 (1%)
Query: 215 SDRRKELTKIVSKQVEEGKVALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDD 274
SD E ++V+K+VE+G + + A +E + S+DNV+D S+ +DD
Sbjct: 332 SDSPSEKVEMVNKKVEKGNKKVHQVSEQAETISLPIE-ELSSSDDNVEDPSNQKGSFSDD 390
Query: 275 YIKKV 279
+K +
Sbjct: 391 EMKDI 395
>At2g30880 unknown protein
Length = 504
Score = 32.0 bits (71), Expect = 0.41
Identities = 31/120 (25%), Positives = 58/120 (47%), Gaps = 10/120 (8%)
Query: 172 SLLVSPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSD--RRKELTKIVSKQV 229
S++ DK S A E A+ + T + +++ + +LT+D R++E TK+ K++
Sbjct: 229 SMIKEIADKLSETA-EAAVAAASAAHTMDEQRKIVCVEFERLTTDSQRQQEATKLKLKEL 287
Query: 230 EEGKVALRNIRRDALKAYEKLEKDKKLSEDNVKDLSSDLQKLTDDYIKKVDALYKQKEKV 289
EE L + +K E+D L E ++ S+L K + + A+ + +EKV
Sbjct: 288 EEKTFTLSKEKDQLVK-----ERDAALQEAHM--WRSELGKARERVVILEGAVVRAEEKV 340
>At5g35150 putative protein
Length = 333
Score = 31.6 bits (70), Expect = 0.54
Identities = 26/91 (28%), Positives = 42/91 (45%), Gaps = 1/91 (1%)
Query: 180 KSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSDRRKELTKIVSKQVEEGKVALR-N 238
+SSL +EK N G I S P+ + + EL + + + V G+V ++ +
Sbjct: 169 ESSLHGLEKLYGKQSRLPDCNGLGPHIHFSRPKSYQEIQDELEEKLGRAVSLGEVFIQTH 228
Query: 239 IRRDALKAYEKLEKDKKLSEDNVKDLSSDLQ 269
+ D K EK + E NVKD S+L+
Sbjct: 229 TKSDGTYVDRKAEKIALIYEQNVKDRLSELE 259
>At4g38065 hypothetical protein
Length = 1050
Score = 31.6 bits (70), Expect = 0.54
Identities = 23/77 (29%), Positives = 39/77 (49%), Gaps = 9/77 (11%)
Query: 202 DGEVIRLSIPQLTSDRRKELTKIVSKQVEEGKVALRNIRRDALKAYEKLEKDKKLSE--- 258
D V+++ + + + + K K +EE L+N+ +D+ K +E+ EK K L E
Sbjct: 169 DDVVVKMEEEKSQVEEKLKWKKEQFKHLEEAYEKLKNLFKDSKKEWEE-EKSKLLDEIYS 227
Query: 259 -----DNVKDLSSDLQK 270
D+V +S DLQK
Sbjct: 228 LQTKLDSVTRISEDLQK 244
>At3g52115 gamma response I protein
Length = 588
Score = 31.6 bits (70), Expect = 0.54
Identities = 26/92 (28%), Positives = 45/92 (48%), Gaps = 13/92 (14%)
Query: 207 RLSIPQLTSDRRKELTKIVSKQVEEGKVALRNIR-------RDALKAYEKL---EKDKKL 256
RL + + R +E+ + + +VEEGK AL ++ R+ L +K+ EK+K +
Sbjct: 221 RLQVFEENVGRLEEILRQKTDEVEEGKTALEVLQGKLKLTEREMLNCKQKIADHEKEKTV 280
Query: 257 SEDNVKDLSSDLQKLTDDYIKKVDALYKQKEK 288
KD D+Q Y+ ++AL Q E+
Sbjct: 281 VMGKAKD---DMQGRHGSYLADLEALRCQSEE 309
>At2g27170 putative chromosome associated protein
Length = 1163
Score = 31.6 bits (70), Expect = 0.54
Identities = 19/66 (28%), Positives = 37/66 (55%), Gaps = 14/66 (21%)
Query: 227 KQVEEGKVALRNIRRDALKAYEKLEK---DKKLSEDNVKDLSSDLQKLTDDYIKKVDALY 283
+++E+ +VA ++ K Y+++EK D K ++++K+L+ +LQ LY
Sbjct: 237 EKLEQVEVARTKASEESTKMYDRVEKAQDDSKSLDESLKELTKELQ-----------TLY 285
Query: 284 KQKEKV 289
K+KE V
Sbjct: 286 KEKETV 291
>At5g35210 putative protein
Length = 1606
Score = 31.2 bits (69), Expect = 0.70
Identities = 27/128 (21%), Positives = 52/128 (40%), Gaps = 20/128 (15%)
Query: 148 IEVEYYGSPVSLK-TIAQISTPDASSLLVSPYDKSSLKAIEKAIVNSDVGMTPNNDGEVI 206
+E+EYY P+ +K I QI D +D + L+ A S++G P+ +
Sbjct: 305 VEIEYYSLPIGMKLKILQILCDDI-------FDVADLRDEIDAREESEIGFDPDRVATGL 357
Query: 207 RLSIPQLTSDRRKELTKIVSKQVEEGKVALRNIRRDALKAYEKLEKDKKLSEDNVKDLSS 266
++P+ R + + K+V + E + D + + ++SS
Sbjct: 358 LENVPRRVHPRFAKTSAYKEKEVTDS------------STNESKDLDSRCTNGGSNEVSS 405
Query: 267 DLQKLTDD 274
DL +D+
Sbjct: 406 DLDGNSDE 413
>At5g02490 dnaK-type molecular chaperone hsc70.1 - like
Length = 653
Score = 31.2 bits (69), Expect = 0.70
Identities = 39/138 (28%), Positives = 64/138 (46%), Gaps = 10/138 (7%)
Query: 132 FSSIRTGRATPSMLDKIEVEYYGSPVSLKTIAQISTP---DASSLL-VSPYDKSSLKAIE 187
F R ++L K E+ G P + + + QI+ DA+ +L VS DK++ K
Sbjct: 449 FEGERARTKDNNLLGKFELS--GIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGKK-N 505
Query: 188 KAIVNSDVGMTPNNDGEVIRLSIPQLTSDRRKELTKIVSKQ-VEEGKVALRNIRRDALKA 246
K + +D G D E + + S+ + K+ +K +E +RN RD K
Sbjct: 506 KITITNDKGRLSKEDIEKMVQEAEKYKSEDEEHKKKVEAKNALENYAYNMRNTIRDE-KI 564
Query: 247 YEKLEK-DKKLSEDNVKD 263
EKL DKK ED++++
Sbjct: 565 GEKLPAADKKKVEDSIEE 582
>At5g48600 chromosome condensation protein
Length = 1241
Score = 30.8 bits (68), Expect = 0.92
Identities = 34/169 (20%), Positives = 75/169 (44%), Gaps = 35/169 (20%)
Query: 111 ASIENDVKSRMERTLDNVKTNF-SSIRTGRATPSMLDKIEVEYYGSPVSLKTIAQISTPD 169
A+ EN++ S++ L+N++ +++R RA + + +E+E S ++++
Sbjct: 722 ANAENEL-SKIVDMLNNIREKVGNAVRQYRAAENEVSGLEMELAKSQREIESL------- 773
Query: 170 ASSLLVSPYDKSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSDRRKELTKIVSKQV 229
S +EK + + + P D DR KEL KI+SK+
Sbjct: 774 ----------NSEHNYLEKQLASLEAASQPKTD-----------EIDRLKELKKIISKEE 812
Query: 230 EEGKVALRNIRRDALKAYEKLEKD-KKLSEDNVKDLSSDLQKLTDDYIK 277
+E + N+ + + + +KL+ + + + +K + ++K+ D K
Sbjct: 813 KE----IENLEKGSKQLKDKLQTNIENAGGEKLKGQKAKVEKIQTDIDK 857
Score = 27.7 bits (60), Expect = 7.8
Identities = 29/123 (23%), Positives = 55/123 (44%), Gaps = 12/123 (9%)
Query: 180 KSSLKAIEKAIVNSDVGMTPNNDGEVIRLSIPQLTSDRR----------KELTKIVSKQV 229
+ SL+ +E ++ + V M +N+ S+ + R+ KE K +Q
Sbjct: 280 RDSLQNLENSLKDERVKMDESNEELKKFESVHEKHKKRQEVLDNELRACKEKFKEFERQD 339
Query: 230 EEGKVALRNIRRDALKAYEKLEKDKKLSEDNVKDL--SSDLQKLTDDYIKKVDALYKQKE 287
+ + L+++++ K +KLEKD D K+ SS+L + I K+ + +E
Sbjct: 340 VKHREDLKHVKQKIKKLEDKLEKDSSKIGDMTKESEDSSNLIPKLQENIPKLQKVLLDEE 399
Query: 288 KVL 290
K L
Sbjct: 400 KKL 402
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.133 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,870,974
Number of Sequences: 26719
Number of extensions: 231138
Number of successful extensions: 1205
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 56
Number of HSP's that attempted gapping in prelim test: 1152
Number of HSP's gapped (non-prelim): 112
length of query: 297
length of database: 11,318,596
effective HSP length: 99
effective length of query: 198
effective length of database: 8,673,415
effective search space: 1717336170
effective search space used: 1717336170
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC122725.8


