

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122725.14 + phase: 0
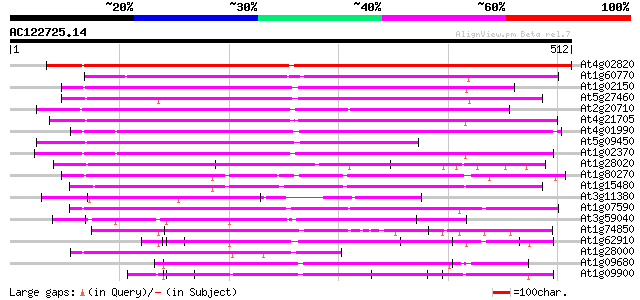
(512 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g02820 Unknown protein (At4g02820) 584 e-167
At1g60770 unknown protein (At1g60770) 273 2e-73
At1g02150 hypothetical protein 256 3e-68
At5g27460 putative protein 237 1e-62
At2g20710 unknown protein 237 1e-62
At4g21705 unknown protein 221 7e-58
At4g01990 unknown protein 219 3e-57
At5g09450 unknown protein 209 2e-54
At1g02370 unknown protein 206 2e-53
At1g28020 hypothetical protein 176 2e-44
At1g80270 unknown protein (At1g80270) 172 4e-43
At1g15480 hypothetical protein 159 3e-39
At3g11380 hypothetical protein 145 4e-35
At1g07590 DNA-binding protein like protein 133 3e-31
At3g59040 unknown protein (At3g59040) 100 3e-21
At1g74850 hypothetical protein 97 2e-20
At1g62910 unknown protein 92 5e-19
At1g28000 hypothetical protein 92 7e-19
At1g09680 hypothetical protein 91 1e-18
At1g09900 hypothetical protein 91 2e-18
>At4g02820 Unknown protein (At4g02820)
Length = 532
Score = 584 bits (1506), Expect = e-167
Identities = 289/479 (60%), Positives = 376/479 (78%), Gaps = 5/479 (1%)
Query: 34 GGDTLGRRLLSLVYPKRSAVIAINKWKEEGHTLPRKYQLNRMIRELRKNKRYKHALEVCE 93
G DTLG RLLSLVY KRSAV+ I KWKEEGH++ RKY+LNR++RELRK KRYKHALE+CE
Sbjct: 59 GRDTLGGRLLSLVYTKRSAVVTIRKWKEEGHSV-RKYELNRIVRELRKIKRYKHALEICE 117
Query: 94 WMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTCTALLHAYVQNNL 153
WM +Q DIKL GDYAV LDLI+K+RGLNSAEKFFED+PD+MRG CT+LLH+YVQN L
Sbjct: 118 WMVVQEDIKLQAGDYAVHLDLISKIRGLNSAEKFFEDMPDQMRGHAACTSLLHSYVQNKL 177
Query: 154 TNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKVNTSPDVVTFNLL 213
++KAEAL KM ECGFL+S +PYN M+S+YIS G+ EKVP L +ELK+ TSPD+VT+NL
Sbjct: 178 SDKAEALFEKMGECGFLKSCLPYNHMLSMYISRGQFEKVPVLIKELKIRTSPDIVTYNLW 237
Query: 214 LTACASENDVETAERVLLQLKKAKVDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEME 273
LTA AS NDVE AE+V L+ K+ K++PDWVTYS LTNLY + D +EKA LKEME
Sbjct: 238 LTAFASGNDVEGAEKVYLKAKEEKLNPDWVTYSVLTNLYAKT----DNVEKARLALKEME 293
Query: 274 KRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCMISSLVKLGDFA 333
K S++ RVAY+SL+SLHAN+G+ D VN W K+K+ F KM+D EY+ MIS++VKLG+F
Sbjct: 294 KLVSKKNRVAYASLISLHANLGDKDGVNLTWKKVKSSFKKMNDAEYLSMISAVVKLGEFE 353
Query: 334 GVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKGVCLSYSSWELLT 393
+ LY EWESVSGT D R+ NL+L Y+++ ++ + E F ++VEKG+ SYS+WE+LT
Sbjct: 354 QAKGLYDEWESVSGTGDARIPNLILAEYMNRDEVLLGEKFYERIVEKGINPSYSTWEILT 413
Query: 394 RGYLKKKDVKKFLHYFGKAISSVKQWIPDPRLVQEAFTVIQEQAHIEGAEQLLVILRNAG 453
YLK+KD++K L FGKAI SVK+W + RLV+ A ++EQ +++GAE+L+ +L+ AG
Sbjct: 414 WAYLKRKDMEKVLDCFGKAIDSVKKWTVNVRLVKGACKELEEQGNVKGAEKLMTLLQKAG 473
Query: 454 HVNTNIYNLFLKTYAAAGKMPLVVAERMKKDNVQLDKETHRLLDLTSKMCVSDVSGILS 512
+VNT +YN L+TYA AG+M L+V ERM KDNV+LD+ET L+ LTS+M V+++S +S
Sbjct: 474 YVNTQLYNSLLRTYAKAGEMALIVEERMAKDNVELDEETKELIRLTSQMRVTEISSTIS 532
>At1g60770 unknown protein (At1g60770)
Length = 491
Score = 273 bits (698), Expect = 2e-73
Identities = 152/440 (34%), Positives = 253/440 (56%), Gaps = 12/440 (2%)
Query: 69 KYQLNRMIRELRKNKRYKHALEVCEWMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFF 128
K+++ I++LR Y AL++ E M+ + K V D A+ LDL+ K R + + E +F
Sbjct: 55 KWEVGDTIKKLRNRGLYYPALKLSEVMEERGMNKTVS-DQAIHLDLVAKAREITAGENYF 113
Query: 129 EDLPDKMRGQPTCTALLHAYVQNNLTNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGK 188
DLP+ + + T +LL+ Y + LT KAE L++KM E S + YN +M+LY G+
Sbjct: 114 VDLPETSKTELTYGSLLNCYCKELLTEKAEGLLNKMKELNITPSSMSYNSLMTLYTKTGE 173
Query: 189 LEKVPKLFEELKV-NTSPDVVTFNLLLTACASENDVETAERVLLQLKK-AKVDPDWVTYS 246
EKVP + +ELK N PD T+N+ + A A+ ND+ ERV+ ++ + +V PDW TYS
Sbjct: 174 TEKVPAMIQELKAENVMPDSYTYNVWMRALAATNDISGVERVIEEMNRDGRVAPDWTTYS 233
Query: 247 TLTNLYIRNASVDDCLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGK 306
+ ++Y+ +A + EKA L+E+E + ++ AY L++L+ +G + EV RIW
Sbjct: 234 NMASIYV-DAGLSQKAEKA---LQELEMKNTQRDFTAYQFLITLYGRLGKLTEVYRIWRS 289
Query: 307 MKACFCKMSDDEYVCMISSLVKLGDFAGVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQ 366
++ K S+ Y+ MI LVKL D G E L+KEW++ T D+R+ N+L+ +Y +G
Sbjct: 290 LRLAIPKTSNVAYLNMIQVLVKLNDLPGAETLFKEWQANCSTYDIRIVNVLIGAYAQEGL 349
Query: 367 MEMAEIFCNQLVEKGVCLSYSSWELLTRGYLKKKDVKKFLHYFGKAISSVK----QWIPD 422
++ A + +G L+ +WE+ Y+K D+ + L KA+S K +W+P
Sbjct: 350 IQKANELKEKAPRRGGKLNAKTWEIFMDYYVKSGDMARALECMSKAVSIGKGDGGKWLPS 409
Query: 423 PRLVQEAFTVIQEQAHIEGAEQLLVILRN-AGHVNTNIYNLFLKTYAAAGKMPLVVAERM 481
P V+ + +++ + GAE LL IL+N ++ I+ ++TYAAAGK + R+
Sbjct: 410 PETVRALMSYFEQKKDVNGAENLLEILKNGTDNIGAEIFEPLIRTYAAAGKSHPAMRRRL 469
Query: 482 KKDNVQLDKETHRLLDLTSK 501
K +NV++++ T +LLD S+
Sbjct: 470 KMENVEVNEATKKLLDEVSQ 489
>At1g02150 hypothetical protein
Length = 524
Score = 256 bits (653), Expect = 3e-68
Identities = 145/418 (34%), Positives = 240/418 (56%), Gaps = 10/418 (2%)
Query: 48 PKRSAVIAINKWKEEGHTLPRKYQLNRMIRELRKNKRYKHALEVCEWMKLQND-IKLVEG 106
P+ A +N+W++ G L K++L R+++ELRK KR ALEV +WM + + +L
Sbjct: 79 PELGAASVLNQWEKAGRKLT-KWELCRVVKELRKYKRANQALEVYDWMNNRGERFRLSAS 137
Query: 107 DYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTCTALLHAYVQNNLTNKAEALMSKMSE 166
D A+QLDLI KVRG+ AE+FF LP+ + + +LL+AYV+ KAEAL++ M +
Sbjct: 138 DAAIQLDLIGKVRGIPDAEEFFLQLPENFKDRRVYGSLLNAYVRAKSREKAEALLNTMRD 197
Query: 167 CGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKV-NTSPDVVTFNLLLTACASENDVET 225
G+ P+P+N MM+LY++ + +KV + E+K + D+ ++N+ L++C S VE
Sbjct: 198 KGYALHPLPFNVMMTLYMNLREYDKVDAMVFEMKQKDIRLDIYSYNIWLSSCGSLGSVEK 257
Query: 226 AERVLLQLKK-AKVDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEMEKRTSRETRVAY 284
E V Q+K + P+W T+ST+ +YI+ EKA L+++E R + R+ Y
Sbjct: 258 MELVYQQMKSDVSIYPNWTTFSTMATMYIKMGET----EKAEDALRKVEARITGRNRIPY 313
Query: 285 SSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCMISSLVKLGDFAGVENLYKEWES 344
LLSL+ ++GN E+ R+W K+ + + Y ++SSLV++GD G E +Y+EW
Sbjct: 314 HYLLSLYGSLGNKKELYRVWHVYKSVVPSIPNLGYHALVSSLVRMGDIEGAEKVYEEWLP 373
Query: 345 VSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKGVCLSYSSWELLTRGYLKKKDVKK 404
V + D R+ NLL+ +YV Q+E AE + +VE G S S+WE+L G+ +K+ + +
Sbjct: 374 VKSSYDPRIPNLLMNAYVKNDQLETAEGLFDHMVEMGGKPSSSTWEILAVGHTRKRCISE 433
Query: 405 FLHYFGKAISS--VKQWIPDPRLVQEAFTVIQEQAHIEGAEQLLVILRNAGHVNTNIY 460
L A S+ W P ++ F + +E++ + E +L +LR +G + Y
Sbjct: 434 ALTCLRNAFSAEGSSNWRPKVLMLSGFFKLCEEESDVTSKEAVLELLRQSGDLEDKSY 491
>At5g27460 putative protein
Length = 491
Score = 237 bits (605), Expect = 1e-62
Identities = 147/447 (32%), Positives = 244/447 (53%), Gaps = 13/447 (2%)
Query: 48 PKRSAVIAINKWKEEGHTLPRKYQLNRMIRELRKNKRYKHALEVCEWMKLQNDIKLVEGD 107
P+RS + + + GH + +L + + L ++ RY AL++ EWM+ Q DI+ D
Sbjct: 50 PRRSVTSLLQERIDSGHAVSLS-ELRLISKRLIRSNRYDLALQMMEWMENQKDIEFSVYD 108
Query: 108 YAVQLDLITKVRGLNSAEKFFEDLPDK---MR-GQPTCTALLHAYVQNNLTNKAEALMSK 163
A++LDLI K GL E++FE L MR + LL AYV+N + +AEALM K
Sbjct: 109 IALRLDLIIKTHGLKQGEEYFEKLLHSSVSMRVAKSAYLPLLRAYVKNKMVKEAEALMEK 168
Query: 164 MSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKVNTSP-DVVTFNLLLTACASEND 222
++ GFL +P P+N MM LY ++G+ EKV + +K N P +V+++NL + AC +
Sbjct: 169 LNGLGFLVTPHPFNEMMKLYEASGQYEKVVMVVSMMKGNKIPRNVLSYNLWMNACCEVSG 228
Query: 223 VETAERVLLQLKKAK-VDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEMEKRTSRETR 281
V E V ++ K V+ W + TL N+YI++ EKA L++ EK +R R
Sbjct: 229 VAAVETVYKEMVGDKSVEVGWSSLCTLANVYIKSGFD----EKARLVLEDAEKMLNRSNR 284
Query: 282 VAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCMISSLVKLGDFAGVENLYKE 341
+ Y L++L+A++GN + V R+W K+ ++S Y+C++SSLVK GD E ++ E
Sbjct: 285 LGYFFLITLYASLGNKEGVVRLWEVSKSVCGRISCVNYICVLSSLVKTGDLEEAERVFSE 344
Query: 342 WESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKGVCLSYSSWELLTRGYLKKKD 401
WE+ DVRVSN+LL +YV G++ AE ++E+G +Y +WE+L G++K ++
Sbjct: 345 WEAQCFNYDVRVSNVLLGAYVRNGEIRKAESLHGCVLERGGTPNYKTWEILMEGWVKCEN 404
Query: 402 VKKFLHYFGKAISSVKQ--WIPDPRLVQEAFTVIQEQAHIEGAEQLLVILRNAGHVNTNI 459
++K + + +++ W P +V +++ IE A + L G + +
Sbjct: 405 MEKAIDAMHQVFVLMRRCHWRPSHNIVMAIAEYFEKEEKIEEATAYVRDLHRLGLASLPL 464
Query: 460 YNLFLKTYAAAGKMPLVVAERMKKDNV 486
Y L L+ + A + + E MK D +
Sbjct: 465 YRLLLRMHEHAKRPAYDIYEMMKLDKL 491
>At2g20710 unknown protein
Length = 490
Score = 237 bits (604), Expect = 1e-62
Identities = 148/436 (33%), Positives = 233/436 (52%), Gaps = 10/436 (2%)
Query: 25 GVTAASSSSGGDTLGRRLLSLVYPKRSAVIAINKWKEEGHTLPRKYQLNRMIRELRKNKR 84
G T S DTL RR+ P S + ++ W ++G+ L + +L+ +I+ LRK R
Sbjct: 27 GKTTPSPLDPYDTLQRRVARSGDPSASIIKVLDGWLDQGN-LVKTSELHSIIKMLRKFSR 85
Query: 85 YKHALEVCEWMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTCTAL 144
+ HAL++ +WM ++ EGD A++LDLI KV GL AEKFFE +P + R AL
Sbjct: 86 FSHALQISDWMSEHRVHEISEGDVAIRLDLIAKVGGLGEAEKFFETIPMERRNYHLYGAL 145
Query: 145 LHAYVQNNLTNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKVNT- 203
L+ Y + +KAE + +M E GFL+ +PYN M++LY+ GK V KL E++ T
Sbjct: 146 LNCYASKKVLHKAEQVFQEMKELGFLKGCLPYNVMLNLYVRTGKYTMVEKLLREMEDETV 205
Query: 204 SPDVVTFNLLLTACASENDVETAERVLLQLKKAK-VDPDWVTYSTLTNLYIRNASVDDCL 262
PD+ T N L A + +DVE E+ L++ + + + DW TY+ N YI+
Sbjct: 206 KPDIFTVNTRLHAYSVVSDVEGMEKFLMRCEADQGLHLDWRTYADTANGYIKAG----LT 261
Query: 263 EKAASTLKEMEKRTSRETRV-AYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVC 321
EKA L++ E+ + + R AY L+S + G +EV R+W K + Y+
Sbjct: 262 EKALEMLRKSEQMVNAQKRKHAYEVLMSFYGAAGKKEEVYRLWSLYKE-LDGFYNTGYIS 320
Query: 322 MISSLVKLGDFAGVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKG 381
+IS+L+K+ D VE + +EWE+ D+R+ +LL+T Y +G ME AE N LV+K
Sbjct: 321 VISALLKMDDIEEVEKIMEEWEAGHSLFDIRIPHLLITGYCKKGMMEKAEEVVNILVQKW 380
Query: 382 VCLSYSSWELLTRGYLKKKDVKKFLHYFGKAISSVKQ-WIPDPRLVQEAFTVIQEQAHIE 440
S+WE L GY ++K + + +AI K W P ++ ++ Q +E
Sbjct: 381 RVEDTSTWERLALGYKMAGKMEKAVEKWKRAIEVSKPGWRPHQVVLMSCVDYLEGQRDME 440
Query: 441 GAEQLLVILRNAGHVN 456
G ++L +L GH++
Sbjct: 441 GLRKILRLLSERGHIS 456
>At4g21705 unknown protein
Length = 492
Score = 221 bits (563), Expect = 7e-58
Identities = 139/470 (29%), Positives = 234/470 (49%), Gaps = 11/470 (2%)
Query: 37 TLGRRLLSLVYPKRSAVIAINKWKEEGHTLPRKYQLNRMIRELRKNKRYKHALEVCEWMK 96
TL ++ L PK S + W + G + +L R++ +LR+ KR+ HALEV +WM
Sbjct: 26 TLYSKISPLGDPKSSVYPELQNWVQCGKKVSVA-ELIRIVHDLRRRKRFLHALEVSKWMN 84
Query: 97 LQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTCTALLHAYVQNNLTNK 156
++AV LDLI +V G +AE++FE+L ++ + T ALL+ YV+ K
Sbjct: 85 ETGVCVFSPTEHAVHLDLIGRVYGFVTAEEYFENLKEQYKNDKTYGALLNCYVRQQNVEK 144
Query: 157 AEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELK-VNTSPDVVTFNLLLT 215
+ KM E GF+ S + YN +M LY + G+ EKVPK+ EE+K N +PD ++ + +
Sbjct: 145 SLLHFEKMKEMGFVTSSLTYNNIMCLYTNIGQHEKVPKVLEEMKEENVAPDNYSYRICIN 204
Query: 216 ACASENDVETAERVLLQLKKAK-VDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEMEK 274
A + D+E L +++ + + DW TY+ YI DC ++A LK E
Sbjct: 205 AFGAMYDLERIGGTLRDMERRQDITMDWNTYAVAAKFYIDGG---DC-DRAVELLKMSEN 260
Query: 275 RTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCMISSLVKLGDFAG 334
R ++ Y+ L++L+A +G EV R+W K + + +Y+ ++ SLVK+
Sbjct: 261 RLEKKDGEGYNHLITLYARLGKKIEVLRLWDLEKDVCKRRINQDYLTVLQSLVKIDALVE 320
Query: 335 VENLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKGVCLSYSSWELLTR 394
E + EW+S D RV N ++ Y+ + E AE L +G + SWEL+
Sbjct: 321 AEEVLTEWKSSGNCYDFRVPNTVIRGYIGKSMEEKAEAMLEDLARRGKATTPESWELVAT 380
Query: 395 GYLKKKDVKKFLHYFGKAIS---SVKQWIPDPRLVQEAFTVIQEQAHIEGAEQLLVILRN 451
Y +K ++ A+ ++W P LV + + ++ ++ E + LRN
Sbjct: 381 AYAEKGTLENAFKCMKTALGVEVGSRKWRPGLTLVTSVLSWVGDEGSLKEVESFVASLRN 440
Query: 452 AGHVNTNIYNLFLKTYAAAGKMPL-VVAERMKKDNVQLDKETHRLLDLTS 500
VN +Y+ +K G + + +RMK D +++D+ET +L S
Sbjct: 441 CIGVNKQMYHALVKADIREGGRNIDTLLQRMKDDKIEIDEETTVILSTRS 490
>At4g01990 unknown protein
Length = 502
Score = 219 bits (558), Expect = 3e-57
Identities = 135/451 (29%), Positives = 242/451 (52%), Gaps = 12/451 (2%)
Query: 56 INKWKEEGHTLPRKYQLNRMIRELRKNKRYKHALEVCEWMKLQNDIKLVEGDYAVQLDLI 115
+N++ EG + +K+ L R ++LRK ++ + ALE+ EWM+ + +I D+A++L+LI
Sbjct: 60 LNQFVMEGVPV-KKHDLIRYAKDLRKFRQPQRALEIFEWME-RKEIAFTGSDHAIRLNLI 117
Query: 116 TKVRGLNSAEKFFEDLPDKMRGQPTCTALLHAYVQNNLTNKAEALMSKMSECGFLRSPVP 175
K +GL +AE +F L D ++ Q T +LL+ Y KA+A M + + + +P
Sbjct: 118 AKSKGLEAAETYFNSLDDSIKNQSTYGSLLNCYCVEKEEVKAKAHFENMVDLNHVSNSLP 177
Query: 176 YNRMMSLYISNGKLEKVPKLFEELKVNT-SPDVVTFNLLLTACASENDVETAERVLLQLK 234
+N +M++Y+ G+ EKVP L +K + +P +T+++ + +C S D++ E+VL ++K
Sbjct: 178 FNNLMAMYMGLGQPEKVPALVVAMKEKSITPCDITYSMWIQSCGSLKDLDGVEKVLDEMK 237
Query: 235 -KAKVDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEMEKRTSRETRVAYSSLLSLHAN 293
+ + W T++ L +YI+ KA LK +E + + R Y L++L+
Sbjct: 238 AEGEGIFSWNTFANLAAIYIKVGLYG----KAEEALKSLENNMNPDVRDCYHFLINLYTG 293
Query: 294 MGNVDEVNRIWGKMKACFCKMSDDEYVCMISSLVKLGDFAGVENLYKEWESVSGTNDVRV 353
+ N EV R+W +K + +++ Y+ M+ +L KL D GV+ ++ EWES T D+R+
Sbjct: 294 IANASEVYRVWDLLKKRYPNVNNSSYLTMLRALSKLDDIDGVKKVFAEWESTCWTYDMRM 353
Query: 354 SNLLLTSYVDQGQMEMAEIFCNQLVEKGVCLSYSSWELLTRGYLKKKDVKKFLHYFGKAI 413
+N+ ++SY+ Q E AE N ++K + +LL LK L +F A+
Sbjct: 354 ANVAISSYLKQNMYEEAEAVFNGAMKKCKGQFSKARQLLMMHLLKNDQADLALKHFEAAV 413
Query: 414 -SSVKQWIPDPRLVQEAFTVIQEQAHIEGAEQLLVILRNAGHVNTNIYNLFLKTYAAAGK 472
K W L+ F +E ++GAE+ L +++ Y L +KTY AAGK
Sbjct: 414 LDQDKNWTWSSELISSFFLHFEEAKDVDGAEEFCKTLTKWSPLSSETYTLLMKTYLAAGK 473
Query: 473 MPLVVAERMKKDNVQLDKETHRLLDLTSKMC 503
+ +R+++ + +D+E LL SK+C
Sbjct: 474 ACPDMKKRLEEQGILVDEEQECLL---SKIC 501
>At5g09450 unknown protein
Length = 409
Score = 209 bits (533), Expect = 2e-54
Identities = 126/354 (35%), Positives = 200/354 (55%), Gaps = 10/354 (2%)
Query: 25 GVTAASSSSGGDTLGRRLLSLVYPKRSAVIAINKWKEEGHTLPRKYQLNRMIRELRKNKR 84
G + S D L R+ L PKRSA + KW EG+ + +L + +ELR+ +R
Sbjct: 44 GNSLVEESEEKDDLKSRIFRLRLPKRSATTVLEKWIGEGNQMTIN-ELREISKELRRTRR 102
Query: 85 YKHALEVCEWMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTCTAL 144
YKHALEV EWM + K+ + DYA ++DLI+KV G+++AE++FE L + T T+L
Sbjct: 103 YKHALEVTEWMVQHEESKISDADYASRIDLISKVFGIDAAERYFEGLDIDSKTAETYTSL 162
Query: 145 LHAYVQNNLTNKAEALMSKMSECGFLR-SPVPYNRMMSLYISNGKLEKVPKLFEELK-VN 202
LHAY + T +AEAL ++ E L + YN MM+LY+S G++EKVP++ E LK
Sbjct: 163 LHAYAASKQTERAEALFKRIIESDSLTFGAITYNEMMTLYMSVGQVEKVPEVIEVLKQKK 222
Query: 203 TSPDVVTFNLLLTACASENDVETAERVLLQLK-KAKVDPDWVTYSTLTNLYIRNASVDDC 261
SPD+ T+NL L++CA+ +++ ++L +++ A + WV Y LT++YI ++ V +
Sbjct: 223 VSPDIFTYNLWLSSCAATFNIDELRKILEEMRHDASSNEGWVRYIDLTSIYINSSRVTN- 281
Query: 262 LEKAASTLK-EMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYV 320
A STL E EK S+ + Y L+ LH +GN +++IW ++ +S Y+
Sbjct: 282 ---AESTLPVEAEKSISQREWITYDFLMILHTGLGNKVMIDQIWKSLRNTNQILSSRSYI 338
Query: 321 CMISSLVKLGDFAGVENLYKEW-ESVSGTNDVRVSNLLLTSYVDQGQMEMAEIF 373
C++SS + LG E + +W ES + D +L ++ D G +A F
Sbjct: 339 CVLSSYLMLGHLREAEEIIHQWKESKTTEFDASACLRILNAFRDVGLEGIASGF 392
>At1g02370 unknown protein
Length = 537
Score = 206 bits (525), Expect = 2e-53
Identities = 140/481 (29%), Positives = 247/481 (51%), Gaps = 13/481 (2%)
Query: 23 RRGVTAASSSSGGDTLGRRLLSLVYPKRSAVIAINKWKEEGHTLPRKYQLNRMIRELRKN 82
R V A + +S L ++L L + +N++ EG T+ RK L R + LRK
Sbjct: 58 RTSVAAPTVASRQRELYKKLSMLSVTGGTVAETLNQFIMEGITV-RKDDLFRCAKTLRKF 116
Query: 83 KRYKHALEVCEWMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDL-PDKMRGQPTC 141
+R +HA E+ +WM+ + + D+A+ LDLI K +GL +AE +F +L P Q T
Sbjct: 117 RRPQHAFEIFDWME-KRKMTFSVSDHAICLDLIGKTKGLEAAENYFNNLDPSAKNHQSTY 175
Query: 142 TALLHAYVQNNLTNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKV 201
AL++ Y KA+A M E F+ + +P+N MMS+Y+ + EKVP L + +K
Sbjct: 176 GALMNCYCVELEEEKAKAHFEIMDELNFVNNSLPFNNMMSMYMRLSQPEKVPVLVDAMKQ 235
Query: 202 N-TSPDVVTFNLLLTACASENDVETAERVLLQL-KKAKVDPDWVTYSTLTNLYIRNASVD 259
SP VT+++ + +C S ND++ E+++ ++ K ++ W T+S L +Y +
Sbjct: 236 RGISPCGVTYSIWMQSCGSLNDLDGLEKIIDEMGKDSEAKTTWNTFSNLAAIYTKAG--- 292
Query: 260 DCLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEY 319
EKA S LK ME++ + R ++ L+SL+A + EV R+W +K ++++ Y
Sbjct: 293 -LYEKADSALKSMEEKMNPNNRDSHHFLMSLYAGISKGPEVYRVWESLKKARPEVNNLSY 351
Query: 320 VCMISSLVKLGDFAGVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVE 379
+ M+ ++ KLGD G++ ++ EWES D+R++N+ + +Y+ E AE + ++
Sbjct: 352 LVMLQAMSKLGDLDGIKKIFTEWESKCWAYDMRLANIAINTYLKGNMYEEAEKILDGAMK 411
Query: 380 KGVCLSYSSWELLTRGYLKKKDVKKFLHYFGKAIS----SVKQWIPDPRLVQEAFTVIQE 435
K + +LL L+ + + A+S + +W LV F ++
Sbjct: 412 KSKGPFSKARQLLMIHLLENDKADLAMKHLEAAVSDSAENKDEWGWSSELVSLFFLHFEK 471
Query: 436 QAHIEGAEQLLVILRNAGHVNTNIYNLFLKTYAAAGKMPLVVAERMKKDNVQLDKETHRL 495
++GAE IL N +++ +KTYAAA K + ER+ + +++ +E L
Sbjct: 472 AKDVDGAEDFCKILSNWKPLDSETMTFLIKTYAAAEKTSPDMRERLSQQQIEVSEEIQDL 531
Query: 496 L 496
L
Sbjct: 532 L 532
>At1g28020 hypothetical protein
Length = 612
Score = 176 bits (447), Expect = 2e-44
Identities = 133/470 (28%), Positives = 231/470 (48%), Gaps = 26/470 (5%)
Query: 41 RLLSLVYPKRSAVIAINKWKEEGHTLPRKYQLNRMIRELRKNKRYKHALEVCEWMKLQND 100
R+ ++ + + +W+++G+ + + + +I++LR + + AL+V EWM +
Sbjct: 41 RITDALHRNAQIIPVLEQWRQQGNQVNPSH-VRVIIKKLRDSDQSLQALQVSEWMSKEKI 99
Query: 101 IKLVEGDYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTCTALLHAYVQNNLTN-KAEA 159
L+ D+A +L LI V GL AEKFFE +P RG T+LL++Y +++ T KAEA
Sbjct: 100 CNLIPEDFAARLHLIENVVGLEEAEKFFESIPKNARGDSVYTSLLNSYARSDKTLCKAEA 159
Query: 160 LMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKVN-TSPDVVTFNLLLTACA 218
KM + G L PVPYN MMSLY + EKV +L E+K N D VT N +L +
Sbjct: 160 TFQKMRDLGLLLRPVPYNAMMSLYSALKNREKVEELLLEMKDNDVEADNVTVNNVLKLYS 219
Query: 219 SENDVETAERVLLQLKKAK-VDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEMEKRTS 277
+ DV E+ L + + + +W T + Y+R S ++ T + +++++
Sbjct: 220 AVCDVTEMEKFLNKWEGIHGIKLEWHTTLDMAKAYLRARSSGKAMKMLRLTEQLVDQKSL 279
Query: 278 RETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCMISSLVKLGDFAGVEN 337
+ AY L+ L+ GN +EV R+W K+ + ++ Y +I SL+K+ D G E
Sbjct: 280 KS---AYDHLMKLYGEAGNREEVLRVWKLYKSKIGERDNNGYRTVIRSLLKVDDIVGAEE 336
Query: 338 LYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKGVCLSYSSWELLTR--G 395
+YK WES+ D R+ +L + Y D+G E AE N K ++ LL +
Sbjct: 337 IYKVWESLPLEFDHRIPTMLASGYRDRGMTEKAEKLMNSKTIKDRRMNKPVTPLLEQWGD 396
Query: 396 YLKKKDVKKFL------HYFGKAISSVKQWIPDPRL----VQEAFTVIQEQAHIEGAEQL 445
+K D+K + F KA+ V +W+ + ++ +++ + ++ G E+
Sbjct: 397 QMKPSDLKCLIKNLRDSKQFSKAL-QVSEWMGEKQVCNLYLEDYAARLYLTENVLGLEEA 455
Query: 446 LVILRN--AGHVNTNIYNLFLKTYAAA----GKMPLVVAERMKKDNVQLD 489
N + ++Y L +YA + G M + M+++NV D
Sbjct: 456 EKYFENIPENMKDYSVYVALLSSYAKSDKNLGNMVDEILREMEENNVDPD 505
Score = 54.7 bits (130), Expect = 1e-07
Identities = 46/177 (25%), Positives = 73/177 (40%), Gaps = 35/177 (19%)
Query: 189 LEKVPKLFEELKVNTSPDVVTFNLLLTACASENDV-ETAERVLLQLKKAKVDPDWVTYST 247
LE+ K FE + N V LL + S+ ++ + +L ++++ VDPD +T +
Sbjct: 452 LEEAEKYFENIPENMKDYSVYVALLSSYAKSDKNLGNMVDEILREMEENNVDPDLITVNH 511
Query: 248 LTNLYIRNASVDDCLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKM 307
+ +Y A S ++ ME R Y + EV+ +W +
Sbjct: 512 VLKVYA-----------AESKIQAMEMFMRRWAVEVYGDVARCKR------EVHNLWDEC 554
Query: 308 K-----------------ACFCKMSDDEYVCMISSLVKLGDFAGVENLYKEWESVSG 347
K KM DD Y +ISSL+KL D GVE +Y EW+ +G
Sbjct: 555 KNNKEEEVEDGKKRLPVVTTLLKMDDDGYRTVISSLLKLDDVQGVEKVYGEWKPKAG 611
>At1g80270 unknown protein (At1g80270)
Length = 596
Score = 172 bits (436), Expect = 4e-43
Identities = 126/470 (26%), Positives = 223/470 (46%), Gaps = 25/470 (5%)
Query: 48 PKRSAVIAINKWKEEGHTLPRKYQLNRMIRELRKNKRYKHALEVCEWMKLQNDIKLVEGD 107
P S A++KW EEG+ + R ++ + + +LR+ + Y AL++ EW++ I++ E D
Sbjct: 140 PGLSIGSALDKWVEEGNEITR-VEIAKAMLQLRRRRMYGRALQMSEWLEANKKIEMTERD 198
Query: 108 YAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTCTALLHAYVQNNLTNKAEALMSKMSEC 167
YA +LDL K+RGL E + +P +G+ LL V K+E + +KM +
Sbjct: 199 YASRLDLTVKIRGLEKGEACMQKIPKSFKGEVLYRTLLANCVAAGNVKKSELVFNKMKDL 258
Query: 168 GFLRSPVPYNRMMSLY--ISNGKLEKVPKLFEELKVNTSPDVVTFNLLLTACASENDVET 225
GF S ++M+ L+ I K+ V L E K N P ++T+ +L+ + ND+
Sbjct: 259 GFPLSGFTCDQMLLLHKRIDRKKIADVLLLME--KENIKPSLLTYKILIDVKGATNDISG 316
Query: 226 AERVLLQLKKAKVDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEMEKRTSRETRVAYS 285
E++L +K V+ D+ T LT + A + D KA LKEME + R A+
Sbjct: 317 MEQILETMKDEGVELDFQT-QALTARHYSGAGLKD---KAEKVLKEMEGESLEANRRAFK 372
Query: 286 SLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCMISSLVKLGDFAGVENLYKEWESV 345
LLS++A++G DEV RIW K C K +E + I + KL E ++++ +
Sbjct: 373 DLLSIYASLGREDEVKRIW---KICESKPYFEESLAAIQAFGKLNKVQEAEAIFEKIVKM 429
Query: 346 SGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKGVCLSYSSWELLTRGYLKKKDVKKF 405
++LL YVD + + ++ E G + ++W+ L + Y++ +V+K
Sbjct: 430 DRRASSSTYSVLLRVYVDHKMLSKGKDLVKRMAESGCRIEATTWDALIKLYVEAGEVEKA 489
Query: 406 LHYFGKAISSVKQWIPDPRLVQEAFTVIQEQ----AHIEGAEQLLVILRNAGHVN-TNIY 460
KA +L+ +F I ++ + E++ + +R AG+ + +
Sbjct: 490 DSLLDKASKQ-----SHTKLMMNSFMYIMDEYSKRGDVHNTEKIFLKMREAGYTSRLRQF 544
Query: 461 NLFLKTYAAAGKMPLVVAERMKKDNVQLDKETHRLL---DLTSKMCVSDV 507
++ Y A + +R+K DN+ +K L D K +SD+
Sbjct: 545 QALMQAYINAKSPAYGMRDRLKADNIFPNKSMAAQLAQGDPFKKTAISDI 594
>At1g15480 hypothetical protein
Length = 623
Score = 159 bits (402), Expect = 3e-39
Identities = 120/435 (27%), Positives = 204/435 (46%), Gaps = 14/435 (3%)
Query: 55 AINKWKEEGHTLPRKYQLNRMIRELRKNKRYKHALEVCEWMKLQNDIKLVEGDYAVQLDL 114
A++KW E+G RK + M+ +LRK + + AL++ EW+ ++ E DYA +LDL
Sbjct: 174 ALDKWVEQGKDTNRKEFESAML-QLRKRRMFGRALQMTEWLDENKQFEMEERDYACRLDL 232
Query: 115 ITKVRGLNSAEKFFEDLPDKMRGQPTCTALLHAYVQNNLTNKAEALMSKMSECGFLRSPV 174
I+KVRG E + + +P+ RG+ LL +V + AEA+ +KM + GF S
Sbjct: 233 ISKVRGWYKGEAYIKTIPESFRGELVYRTLLANHVATSNVRTAEAVFNKMKDLGFPLSTF 292
Query: 175 PYNRMMSLY--ISNGKLEKVPKLFEELKVNTSPDVVTFNLLLTACASENDVETAERVLLQ 232
N+M+ LY + K+ V L E K N P++ T+ +L+ S ND+ E+++
Sbjct: 293 TCNQMLILYKRVDKKKIADVLLLLE--KENLKPNLNTYKILIDTKGSSNDITGMEQIVET 350
Query: 233 LKKAKVDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEMEKRTSRETRVAYSSLLSLHA 292
+K V+ D + L R+ + EKA LKEME + E R LLS++
Sbjct: 351 MKSEGVELDLRARA----LIARHYASAGLKEKAEKVLKEMEGESLEENRHMCKDLLSVYG 406
Query: 293 NMGNVDEVNRIWGKMKACFCKMSDDEYVCMISSLVKLGDFAGVENLYKEWESVSGTNDVR 352
+ DEV R+W K C +E + I + K+ E ++++ +S
Sbjct: 407 YLQREDEVRRVW---KICEENPRYNEVLAAILAFGKIDKVKDAEAVFEKVLKMSHRVSSN 463
Query: 353 VSNLLLTSYVDQGQMEMAEIFCNQLVEKGVCLSYSSWELLTRGYLKKKDVKKFLHYFGKA 412
V ++LL YVD + + Q+ + G + +W+ + + Y++ +V+K KA
Sbjct: 464 VYSVLLRVYVDHKMVSEGKDLVKQMSDSGCNIGALTWDAVIKLYVEAGEVEKAESSLSKA 523
Query: 413 ISSVKQWIPDPRLVQEAFTVIQEQAHIEGAEQLLVILRNAGHVNTN-IYNLFLKTYAAAG 471
I S KQ P + + E++ ++ AG+ + Y ++ Y A
Sbjct: 524 IQS-KQIKPLMSSFMYLMHEYVRRGDVHNTEKIFQRMKQAGYQSRFWAYQTLIQAYVNAK 582
Query: 472 KMPLVVAERMKKDNV 486
+ ERMK DN+
Sbjct: 583 APAYGMKERMKADNI 597
>At3g11380 hypothetical protein
Length = 541
Score = 145 bits (367), Expect = 4e-35
Identities = 105/355 (29%), Positives = 168/355 (46%), Gaps = 44/355 (12%)
Query: 30 SSSSGGDTLGRRLLSLVYPKRSAVIAINKWKEEGHTLPRKYQ---LNRMIRELRKNKRYK 86
SS + TL R+ + + K + +W ++ ++ L ++ +LR + R +
Sbjct: 33 SSPATNQTLQSRIEAASHQKAEITTVLEQWLQQQQQQGKQLNPSLLRGIVEKLRSSNRPR 92
Query: 87 HALEVCEWMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTCTALLH 146
ALEV +WM Q L E D++ + L KV L AEKFFE +P+ MR + +LL
Sbjct: 93 QALEVSDWMVEQKMYNLPE-DFSARFHLTEKVLNLEEAEKFFESIPENMRFESMYNSLLR 151
Query: 147 AYVQNN---LTNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKV-N 202
+Y + + KAE++ KM + G L P PYN M SLY S G +KV ++ E+K N
Sbjct: 152 SYARQSGEKALKKAESVFKKMKKLGLLLRPSPYNSMTSLYSSLGNRDKVDEILREMKENN 211
Query: 203 TSPDVVTFNLLLTACASENDVETAERVLLQLKKAKVDPDWVTYSTLTNLYIRNASVDDCL 262
D VT N L A+ +DV T ++ L +K D +T + Y
Sbjct: 212 VELDNVTVNNALRVYAAVSDVATMDKFLAD-RKEITRLDGLTMLAMAKAY---------- 260
Query: 263 EKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCM 322
L+SL+ G +++V+R+W K KA K ++E+ +
Sbjct: 261 -----------------------ELMSLYGEAGEIEDVHRVWDKYKATRQK-DNEEFRTL 296
Query: 323 ISSLVKLGDFAGVENL-YKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQ 376
I SL+KLGD G E + Y EWE D R+ ++L++ Y ++G + A+ N+
Sbjct: 297 IGSLLKLGDTKGAEKIYYNEWECSGLEFDNRIPDMLVSGYREKGMVMKADKLVNK 351
Score = 92.0 bits (227), Expect = 7e-19
Identities = 60/161 (37%), Positives = 84/161 (51%), Gaps = 3/161 (1%)
Query: 72 LNRMIRELRKNKRYKHALEVCEWMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFE-D 130
L +I+ LR + + ALE WM + L DYA +L L KV GL AE FFE
Sbjct: 381 LRDLIKNLRDSNQLSKALEASTWMCQKKVFNLFSEDYATRLHLTEKVLGLEEAENFFESS 440
Query: 131 LPDKMRGQPTCTALLHAYVQ-NNLTNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKL 189
+P+ M+ LL Y + +N +KAEA+ KM E G P+N ++SLY GKL
Sbjct: 441 IPENMKDYSVYDTLLSCYARSSNTQSKAEAVFEKMRELGLQSKLSPFNSLISLYSGQGKL 500
Query: 190 EKVPKLFEELK-VNTSPDVVTFNLLLTACASENDVETAERV 229
V L ++K N PD+VT N +L A A +++ E+V
Sbjct: 501 SVVNILLCDMKHKNIEPDIVTRNNVLRANAYILAIDSMEKV 541
Score = 31.6 bits (70), Expect = 1.1
Identities = 18/68 (26%), Positives = 35/68 (51%), Gaps = 3/68 (4%)
Query: 241 DWVTYSTLTNLYIRNASVDDCLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEV 300
D+ Y TL + Y R+++ KA + ++M + + ++SL+SL++ G + V
Sbjct: 447 DYSVYDTLLSCYARSSNTQ---SKAEAVFEKMRELGLQSKLSPFNSLISLYSGQGKLSVV 503
Query: 301 NRIWGKMK 308
N + MK
Sbjct: 504 NILLCDMK 511
>At1g07590 DNA-binding protein like protein
Length = 534
Score = 133 bits (334), Expect = 3e-31
Identities = 110/457 (24%), Positives = 205/457 (44%), Gaps = 19/457 (4%)
Query: 55 AINKWKEEGHTLPRKYQLNRMIRELRKNKRYKHALEVCEWMKLQNDIKLVEGDYAVQLDL 114
A+ W +G + + I LRK R K ALE+ EW+ + +L E +Y+ L+
Sbjct: 84 ALQSWMGDGFPV-HGGDVYHAINRLRKLGRNKRALELMEWIIRERPYRLGELEYSYLLEF 142
Query: 115 ITKVRGLNSAEKFFEDLPDKMRGQPTCTALLHAYVQNNLTNKAEALMSKMSECGFLRSPV 174
K+ G++ EK F +P + + + L+ A + + A M KM E G+ S +
Sbjct: 143 TVKLHGVSQGEKLFTRVPQEFQNELLYNNLVIACLDQGVIRLALEYMKKMRELGYRTSHL 202
Query: 175 PYNRMMSLYISNGKLEKVPKLFEELKVN-TSPDVVTFNLLLTACASENDVETAERVLLQL 233
YNR++ + G+ + + K +K + +P V T+++L+ A+E++++ + +
Sbjct: 203 VYNRLIIRNSAPGRRKLIAKDLALMKADKATPHVSTYHILMKLEANEHNIDGVLKAFDGM 262
Query: 234 KKAKVDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEMEKRTSRETRVAYSSLLSLHAN 293
KKA V+P+ V+Y L + +V A + +E+EK + + L+ L+
Sbjct: 263 KKAGVEPNEVSYCILAMAH----AVARLYTVAEAYTEEIEKSITGDNWSTLDILMILYGR 318
Query: 294 MGNVDEVNRIWGKMKACFCKMSDDEYVCMISSLVKLGDFAGVENLYKEWESVSGTNDVRV 353
+G E+ R W ++ F + Y+ + ++G+ E L+ E ++V G +
Sbjct: 319 LGKEKELARTWNVIRG-FHHVRSKSYLLATEAFARVGNLDRAEELWLEMKNVKGLKETEQ 377
Query: 354 SNLLLTSYVDQGQMEMAEIFCNQLVEKGVCLSYSSWELLTRGYLKKKDVKKFLHYF---- 409
N LL+ Y G +E A ++ G + ++ L G K K +K+ L
Sbjct: 378 FNSLLSVYCKDGLIEKAIGVFREMTGNGFKPNSITYRHLALGCAKAKLMKEALKNIEMGL 437
Query: 410 ----GKAISSVKQWIPDPRLVQEAFTVIQEQAHIEGAEQLLVILRNAGHVN-TNIYNLFL 464
K+I S W+ + E F E+ +E +E+L ++NA + +YN
Sbjct: 438 NLKTSKSIGSSTPWLETTLSIIECFA---EKGDVENSEKLFEEVKNAKYNRYAFVYNALF 494
Query: 465 KTYAAAGKMPLVVAERMKKDNVQLDKETHRLLDLTSK 501
K Y A + +RM + D E++ LL L +
Sbjct: 495 KAYVKAKVYDPNLFKRMVLGGARPDAESYSLLKLVEQ 531
>At3g59040 unknown protein (At3g59040)
Length = 526
Score = 99.8 bits (247), Expect = 3e-21
Identities = 85/387 (21%), Positives = 169/387 (42%), Gaps = 18/387 (4%)
Query: 40 RRLLSLVYPKRSAVIAINKWKEEGHT-LPRKYQLNRMIRELRKNKRYKHALEVCEWMKLQ 98
R L+ + SAV + ++K +G LPR L ++R ++ K++ E+ EW++ Q
Sbjct: 17 RGLMLEIESTGSAVPVLRQYKTDGDQGLPRDLVLGTLVR-FKQLKKWNLVSEILEWLRYQ 75
Query: 99 NDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDLPDKMRGQPTC---TALLHAYVQNNLTN 155
N E D+ + + K+ N AE+ L KM P TAL+ +Y + N
Sbjct: 76 NWWNFSEIDFLMLITAYGKLGNFNGAERVLSVL-SKMGSTPNVISYTALMESYGRGGKCN 134
Query: 156 KAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEEL----KVNTSPDVVTFN 211
AEA+ +M G S + Y ++ ++ K ++ ++FE L K PD ++
Sbjct: 135 NAEAIFRRMQSSGPEPSAITYQIILKTFVEGDKFKEAEEVFETLLDEKKSPLKPDQKMYH 194
Query: 212 LLLTACASENDVETAERVLLQLKKAKVDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKE 271
+++ + E A +V + V VTY++L S + ++ + +
Sbjct: 195 MMIYMYKKAGNYEKARKVFSSMVGKGVPQSTVTYNSLM-------SFETSYKEVSKIYDQ 247
Query: 272 MEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCMISSLVKLGD 331
M++ + V+Y+ L+ + +E ++ +M + + Y ++ + G
Sbjct: 248 MQRSDIQPDVVSYALLIKAYGRARREEEALSVFEEMLDAGVRPTHKAYNILLDAFAISGM 307
Query: 332 FAGVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKGVCLSYSSWEL 391
+ ++K D+ +L++YV+ ME AE F ++ G + ++
Sbjct: 308 VEQAKTVFKSMRRDRIFPDLWSYTTMLSAYVNASDMEGAEKFFKRIKVDGFEPNIVTYGT 367
Query: 392 LTRGYLKKKDVKKFLHYFGK-AISSVK 417
L +GY K DV+K + + K +S +K
Sbjct: 368 LIKGYAKANDVEKMMEVYEKMRLSGIK 394
Score = 73.2 bits (178), Expect = 3e-13
Identities = 69/306 (22%), Positives = 135/306 (43%), Gaps = 12/306 (3%)
Query: 70 YQLNRMIRELRKNKRYKHALEVCEWM--KLQNDIKLVEGDYAVQLDLITKVRGLNSAEKF 127
YQ+ +++ + ++K A EV E + + ++ +K + Y + + + K A K
Sbjct: 155 YQI--ILKTFVEGDKFKEAEEVFETLLDEKKSPLKPDQKMYHMMIYMYKKAGNYEKARKV 212
Query: 128 FEDLPDKMRGQPTCTALLHAYVQNNLTNK-AEALMSKMSECGFLRSPVPYNRMMSLYISN 186
F + K G P T ++ + + K + +M V Y ++ Y
Sbjct: 213 FSSMVGK--GVPQSTVTYNSLMSFETSYKEVSKIYDQMQRSDIQPDVVSYALLIKAYGRA 270
Query: 187 GKLEKVPKLFEE-LKVNTSPDVVTFNLLLTACASENDVETAERVLLQLKKAKVDPDWVTY 245
+ E+ +FEE L P +N+LL A A VE A+ V +++ ++ PD +Y
Sbjct: 271 RREEEALSVFEEMLDAGVRPTHKAYNILLDAFAISGMVEQAKTVFKSMRRDRIFPDLWSY 330
Query: 246 STLTNLYIRNASVDDCLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWG 305
+T+ + Y+ NAS +E A K ++ V Y +L+ +A +V+++ ++
Sbjct: 331 TTMLSAYV-NASD---MEGAEKFFKRIKVDGFEPNIVTYGTLIKGYAKANDVEKMMEVYE 386
Query: 306 KMKACFCKMSDDEYVCMISSLVKLGDFAGVENLYKEWESVSGTNDVRVSNLLLTSYVDQG 365
KM+ K + ++ + + +F YKE ES D + N+LL+ Q
Sbjct: 387 KMRLSGIKANQTILTTIMDASGRCKNFGSALGWYKEMESCGVPPDQKAKNVLLSLASTQD 446
Query: 366 QMEMAE 371
++E A+
Sbjct: 447 ELEEAK 452
>At1g74850 hypothetical protein
Length = 862
Score = 97.1 bits (240), Expect = 2e-20
Identities = 102/460 (22%), Positives = 189/460 (40%), Gaps = 44/460 (9%)
Query: 75 MIRELRKNKRYKHALEVCEWMKLQNDIKLVEGDYAVQLDLITKVRGLNSAEKFFEDLPDK 134
+ +E ++ +L + ++M+ Q K E Y + + L+ + L+ + F+++P +
Sbjct: 111 VFKEFAGRGDWQRSLRLFKYMQRQIWCKPNEHIYTIMISLLGREGLLDKCLEVFDEMPSQ 170
Query: 135 --MRGQPTCTALLHAYVQNNLTNKAEALMSKMSECGFLRSPVPYNRMMSLYISNG-KLEK 191
R + TAL++AY +N + L+ +M S + YN +++ G E
Sbjct: 171 GVSRSVFSYTALINAYGRNGRYETSLELLDRMKNEKISPSILTYNTVINACARGGLDWEG 230
Query: 192 VPKLFEELK-VNTSPDVVTFNLLLTACASENDVETAERVLLQLKKAKVDPDWVTYSTLTN 250
+ LF E++ PD+VT+N LL+ACA + AE V + + PD TYS L
Sbjct: 231 LLGLFAEMRHEGIQPDIVTYNTLLSACAIRGLGDEAEMVFRTMNDGGIVPDLTTYSHLVE 290
Query: 251 LYIRNASVDDCLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKAC 310
+ + LEK L EM S +Y+ LL +A G++ E ++ +M+A
Sbjct: 291 TFGKLRR----LEKVCDLLGEMASGGSLPDITSYNVLLEAYAKSGSIKEAMGVFHQMQAA 346
Query: 311 FCKMSDDEYVCMISSLVKLGDFAGVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMA 370
C + + Y +++ + G + V L+ E +S + D N+L+ + + G +
Sbjct: 347 GCTPNANTYSVLLNLFGQSGRYDDVRQLFLEMKSSNTDPDAATYNILIEVFGEGGYFKEV 406
Query: 371 EIFCNQLVEKGVCLSYSSWELLT----RGYLKKKDVKKFLHYF--------GKAISSVKQ 418
+ +VE+ + ++E + +G L +D +K L Y KA + V +
Sbjct: 407 VTLFHDMVEENIEPDMETYEGIIFACGKGGL-HEDARKILQYMTANDIVPSSKAYTGVIE 465
Query: 419 WIPDPRLVQEAFTV------IQEQAHIEGAEQLLVILRNAGHV----------------- 455
L +EA + IE LL G V
Sbjct: 466 AFGQAALYEEALVAFNTMHEVGSNPSIETFHSLLYSFARGGLVKESEAILSRLVDSGIPR 525
Query: 456 NTNIYNLFLKTYAAAGKMPLVVAERMKKDNVQLDKETHRL 495
N + +N ++ Y GK V + + + D + L
Sbjct: 526 NRDTFNAQIEAYKQGGKFEEAVKTYVDMEKSRCDPDERTL 565
Score = 49.7 bits (117), Expect = 4e-06
Identities = 56/252 (22%), Positives = 109/252 (43%), Gaps = 24/252 (9%)
Query: 142 TALLHAYVQNNLTNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKV 201
T ++ A+ Q L +A + M E G S ++ ++ + G +++ + L
Sbjct: 461 TGVIEAFGQAALYEEALVAFNTMHEVGSNPSIETFHSLLYSFARGGLVKESEAILSRLVD 520
Query: 202 NTSP-DVVTFNLLLTACASENDVETAERVLLQLKKAKVDPDWVTYSTLTNLYIRNASVDD 260
+ P + TFN + A E A + + ++K++ DPD T + ++Y VD+
Sbjct: 521 SGIPRNRDTFNAQIEAYKQGGKFEEAVKTYVDMEKSRCDPDERTLEAVLSVYSFARLVDE 580
Query: 261 CLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYV 320
C E+ +EM+ + + Y +L+++ D+VN + +M + ++S+ V
Sbjct: 581 CREQ----FEEMKASDILPSIMCYCMMLAVYGKTERWDDVNELLEEMLS--NRVSNIHQV 634
Query: 321 CMISSLVKLGDFAGVENLYKEWESVSGTND----------VRVSNLLLTSYVDQGQMEMA 370
I ++K GD+ N W+ V D +R N LL + GQ E A
Sbjct: 635 --IGQMIK-GDYDDDSN----WQIVEYVLDKLNSEGCGLGIRFYNALLDALWWLGQKERA 687
Query: 371 EIFCNQLVEKGV 382
N+ ++G+
Sbjct: 688 ARVLNEATKRGL 699
>At1g62910 unknown protein
Length = 1133
Score = 92.4 bits (228), Expect = 5e-19
Identities = 78/363 (21%), Positives = 161/363 (43%), Gaps = 14/363 (3%)
Query: 140 TCTALLHAYVQNNLTNKAEALMSKMSECGFLRSPVPYNRMM-SLYISNGKLEKVPKLFEE 198
T ++LL+ Y + + A AL+ +M E G+ + ++ L++ N E V + +
Sbjct: 155 TLSSLLNGYCHSKRISDAVALVDQMVEMGYKPDTFTFTTLIHGLFLHNKASEAVALVDQM 214
Query: 199 LKVNTSPDVVTFNLLLTACASENDVETAERVLLQLKKAKVDPDWVTYSTLTNLYIRNASV 258
++ PD+VT+ ++ D++ A +L +++K K++ D V Y+T+ + + +
Sbjct: 215 VQRGCQPDLVTYGTVVNGLCKRGDIDLALSLLKKMEKGKIEADVVIYNTIIDGLCKYKHM 274
Query: 259 DDCLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDE 318
DD L + EM+ + R YSSL+S N G + +R+ M +
Sbjct: 275 DDAL----NLFTEMDNKGIRPDVFTYSSLISCLCNYGRWSDASRLLSDMIERKINPNVVT 330
Query: 319 YVCMISSLVKLGDFAGVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLV 378
+ +I + VK G E LY E S D+ + L+ + +++ A+ ++
Sbjct: 331 FSALIDAFVKEGKLVEAEKLYDEMIKRSIDPDIFTYSSLINGFCMHDRLDEAKHMFELMI 390
Query: 379 EKGVCLSYSSWELLTRGYLKKKDVKKFLHYFGKAISS--VKQWIPDPRLVQEAFTVIQEQ 436
K + ++ L +G+ K K V++ + F + V + L+ F +
Sbjct: 391 SKDCFPNVVTYSTLIKGFCKAKRVEEGMELFREMSQRGLVGNTVTYTTLIHGFF----QA 446
Query: 437 AHIEGAEQLLVILRNAG-HVNTNIYNLFLKTYAAAGKM--PLVVAERMKKDNVQLDKETH 493
+ A+ + + + G H N YN+ L GK+ +VV E +++ ++ D T+
Sbjct: 447 RDCDNAQMVFKQMVSVGVHPNILTYNILLDGLCKNGKLAKAMVVFEYLQRSTMEPDIYTY 506
Query: 494 RLL 496
++
Sbjct: 507 NIM 509
Score = 90.9 bits (224), Expect = 2e-18
Identities = 80/363 (22%), Positives = 157/363 (43%), Gaps = 14/363 (3%)
Query: 140 TCTALLHAYVQNNLTNKAEALMSKMSECGFLRSPVPYNRMM-SLYISNGKLEKVPKLFEE 198
T +LL+ + N + A AL+ +M E G+ V + ++ L++ N E V +
Sbjct: 755 TLNSLLNGFCHGNRISDAVALVDQMVEMGYKPDTVTFTTLIHGLFLHNKASEAVALIDRM 814
Query: 199 LKVNTSPDVVTFNLLLTACASENDVETAERVLLQLKKAKVDPDWVTYSTLTNLYIRNASV 258
++ PD+VT+ ++ D + A +L +++ AK++ + V YST+ + +
Sbjct: 815 VQRGCQPDLVTYGAVVNGLCKRGDTDLALNLLNKMEAAKIEANVVIYSTVIDSLCKYRHE 874
Query: 259 DDCLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDE 318
DD L + EME + R + YSSL+S N G + +R+ M +
Sbjct: 875 DDAL----NLFTEMENKGVRPNVITYSSLISCLCNYGRWSDASRLLSDMIERKINPNLVT 930
Query: 319 YVCMISSLVKLGDFAGVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLV 378
+ +I + VK G E LY+E S ++ + L+ + ++ A+ ++
Sbjct: 931 FSALIDAFVKKGKLVKAEKLYEEMIKRSIDPNIFTYSSLINGFCMLDRLGEAKQMLELMI 990
Query: 379 EKGVCLSYSSWELLTRGYLKKKDVKKFLHYFGKAISS--VKQWIPDPRLVQEAFTVIQEQ 436
K + ++ L G+ K K V K + F + V + L+ F +
Sbjct: 991 RKDCLPNVVTYNTLINGFCKAKRVDKGMELFREMSQRGLVGNTVTYTTLIHGFF----QA 1046
Query: 437 AHIEGAEQLLVILRNAG-HVNTNIYNLFLKTYAAAGKM--PLVVAERMKKDNVQLDKETH 493
+ A+ + + + G H N YN+ L GK+ +VV E +++ ++ D T+
Sbjct: 1047 RDCDNAQMVFKQMVSVGVHPNILTYNILLDGLCKNGKLAKAMVVFEYLQRSTMEPDIYTY 1106
Query: 494 RLL 496
++
Sbjct: 1107 NIM 1109
Score = 65.1 bits (157), Expect = 9e-11
Identities = 66/310 (21%), Positives = 129/310 (41%), Gaps = 12/310 (3%)
Query: 160 LMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEEL---KVNTSPDVVTFNLLLTA 216
L ++M G Y+ ++S + G+ +L ++ K+N P+VVTF+ L+ A
Sbjct: 280 LFTEMDNKGIRPDVFTYSSLISCLCNYGRWSDASRLLSDMIERKIN--PNVVTFSALIDA 337
Query: 217 CASENDVETAERVLLQLKKAKVDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEMEKRT 276
E + AE++ ++ K +DPD TYS+L N + + D L++A + M +
Sbjct: 338 FVKEGKLVEAEKLYDEMIKRSIDPDIFTYSSLINGF----CMHDRLDEAKHMFELMISKD 393
Query: 277 SRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCMISSLVKLGDFAGVE 336
V YS+L+ V+E ++ +M + Y +I + D +
Sbjct: 394 CFPNVVTYSTLIKGFCKAKRVEEGMELFREMSQRGLVGNTVTYTTLIHGFFQARDCDNAQ 453
Query: 337 NLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKGVCLSYSSWELLTRGY 396
++K+ SV ++ N+LL G++ A + L + ++ ++ G
Sbjct: 454 MVFKQMVSVGVHPNILTYNILLDGLCKNGKLAKAMVVFEYLQRSTMEPDIYTYNIMIEGM 513
Query: 397 LKKKDVKKFLHYFGKAISSVKQWIPDPRLVQEAFTVIQEQAHIEGAEQLLVILRNAGHV- 455
K V+ F S+K P+ + + E A+ LL ++ G +
Sbjct: 514 CKAGKVEDGWELFCNL--SLKGVSPNVIAYNTMISGFCRKGSKEEADSLLKKMKEDGPLP 571
Query: 456 NTNIYNLFLK 465
N+ YN ++
Sbjct: 572 NSGTYNTLIR 581
Score = 63.9 bits (154), Expect = 2e-10
Identities = 58/263 (22%), Positives = 116/263 (44%), Gaps = 7/263 (2%)
Query: 144 LLHAYVQNNLTNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEEL-KVN 202
LL A + N + + KM G + YN +++ + +L L ++ K+
Sbjct: 689 LLSAIAKMNKFDLVISFGEKMEILGISHNLYTYNILINCFCRCSRLSLALALLGKMMKLG 748
Query: 203 TSPDVVTFNLLLTACASENDVETAERVLLQLKKAKVDPDWVTYSTLTN-LYIRNASVDDC 261
PD+VT N LL N + A ++ Q+ + PD VT++TL + L++ N +
Sbjct: 749 YEPDIVTLNSLLNGFCHGNRISDAVALVDQMVEMGYKPDTVTFTTLIHGLFLHNKA---- 804
Query: 262 LEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVC 321
+A + + M +R + V Y ++++ G+ D + KM+A + + Y
Sbjct: 805 -SEAVALIDRMVQRGCQPDLVTYGAVVNGLCKRGDTDLALNLLNKMEAAKIEANVVIYST 863
Query: 322 MISSLVKLGDFAGVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKG 381
+I SL K NL+ E E+ +V + L++ + G+ A + ++E+
Sbjct: 864 VIDSLCKYRHEDDALNLFTEMENKGVRPNVITYSSLISCLCNYGRWSDASRLLSDMIERK 923
Query: 382 VCLSYSSWELLTRGYLKKKDVKK 404
+ + ++ L ++KK + K
Sbjct: 924 INPNLVTFSALIDAFVKKGKLVK 946
Score = 56.6 bits (135), Expect = 3e-08
Identities = 50/240 (20%), Positives = 102/240 (41%), Gaps = 9/240 (3%)
Query: 121 LNSAEKFFEDLPDK--MRGQPTCTALLHAYVQNNLTNKAEALMSKMSECGFLRSPVPYNR 178
L+ A+ FE + K T + L+ + + + L +MS+ G + + V Y
Sbjct: 379 LDEAKHMFELMISKDCFPNVVTYSTLIKGFCKAKRVEEGMELFREMSQRGLVGNTVTYTT 438
Query: 179 MMSLYISNGKLEKVPKLFEEL-KVNTSPDVVTFNLLLTACASENDVETAERVLLQLKKAK 237
++ + + +F+++ V P+++T+N+LL + A V L+++
Sbjct: 439 LIHGFFQARDCDNAQMVFKQMVSVGVHPNILTYNILLDGLCKNGKLAKAMVVFEYLQRST 498
Query: 238 VDPDWVTYSTLTNLYIRNASVDDCLEKAAS-TLKEMEKRTSRETRVAYSSLLSLHANMGN 296
++PD TY+ + + V+D E + +LK + +AY++++S G+
Sbjct: 499 MEPDIYTYNIMIEGMCKAGKVEDGWELFCNLSLKGVSPNV-----IAYNTMISGFCRKGS 553
Query: 297 VDEVNRIWGKMKACFCKMSDDEYVCMISSLVKLGDFAGVENLYKEWESVSGTNDVRVSNL 356
+E + + KMK + Y +I + ++ GD L KE S D L
Sbjct: 554 KEEADSLLKKMKEDGPLPNSGTYNTLIRARLRDGDREASAELIKEMRSCGFAGDASTIGL 613
Score = 39.7 bits (91), Expect = 0.004
Identities = 61/323 (18%), Positives = 122/323 (36%), Gaps = 12/323 (3%)
Query: 188 KLEKVPKLFEEL-KVNTSPDVVTFNLLLTACASENDVETAERVLLQLKKAKVDPDWVTYS 246
K++ LF ++ K P +V FN LL+A A N E + Q++ + D TYS
Sbjct: 63 KVDDAVDLFGDMVKSRPFPSIVEFNKLLSAVAKMNKFELVISLGEQMQTLGISHDLYTYS 122
Query: 247 TLTNLYIRNASVDDCLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGK 306
N + R + L A + L +M K V SSLL+ + + + + + +
Sbjct: 123 IFINCFCRRSQ----LSLALAVLAKMMKLGYEPDIVTLSSLLNGYCHSKRISDAVALVDQ 178
Query: 307 MKACFCKMSDDEYVCMISSLVKLGDFAGVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQ 366
M K + +I L + L + D+ ++ +G
Sbjct: 179 MVEMGYKPDTFTFTTLIHGLFLHNKASEAVALVDQMVQRGCQPDLVTYGTVVNGLCKRGD 238
Query: 367 MEMAEIFCNQLVEKGVCLSYSSWELLTRGYLKKKDVKKFLHYFGKAISSVKQWIPDPRLV 426
+++A ++ + + + + G K K + L+ F + + K PD
Sbjct: 239 IDLALSLLKKMEKGKIEADVVIYNTIIDGLCKYKHMDDALNLFTEMDN--KGIRPDVFTY 296
Query: 427 QEAFTVIQEQAHIEGAEQLLV-ILRNAGHVNTNIYNLFLKTYAAAGKMPLVVAERMKKDN 485
+ + A +LL ++ + N ++ + + GK LV AE++ +
Sbjct: 297 SSLISCLCNYGRWSDASRLLSDMIERKINPNVVTFSALIDAFVKEGK--LVEAEKLYDEM 354
Query: 486 VQ--LDKETHRLLDLTSKMCVSD 506
++ +D + L + C+ D
Sbjct: 355 IKRSIDPDIFTYSSLINGFCMHD 377
>At1g28000 hypothetical protein
Length = 349
Score = 92.0 bits (227), Expect = 7e-19
Identities = 70/254 (27%), Positives = 120/254 (46%), Gaps = 11/254 (4%)
Query: 56 INKWKEEGHTLPRKYQLNRMIRELRKNKRYKHALEVCEWMKLQNDIKLVEGDYAVQLDLI 115
+++W + G L L +I L +++R+ HAL+V EW+ + L D+A +L L+
Sbjct: 14 LDEWLKRGKDL-NPAVLRGLINSLCESQRFNHALQVSEWITKRGIFDLSTEDFASRLCLV 72
Query: 116 TKVRGLNSAEKFFEDLPDKMRGQPTCTALLHAY-VQNNLTNKAEALMSKMSECGFLRSPV 174
GL AEKFF+ +P+ MR T LL Y + ++AEA M L P
Sbjct: 73 EISTGLKEAEKFFKSIPENMRDDSVHTTLLSLYTISKKTRHEAEATYQTMRVQNMLLKPY 132
Query: 175 PYNRMMSLYISNGKLEKVPKLFEELKVN--TSPDVVTFNLLLTACASENDVETAERVL-- 230
PY M+ LY G+ + ++ ++K N +T N +L A AS DVE E L
Sbjct: 133 PYYSMIYLYALLGEKNMIDEILRQMKENGVEHDKNLTANNVLKAYASLPDVEAMEMFLMG 192
Query: 231 LQLKKAKVDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEMEKRTSRETR-VAYSSLLS 289
L++++ + W T ++ Y++ S KA L+ E +++ A L+
Sbjct: 193 LEVEEPRFSLAWQTGISIAKAYLKGGS----SRKAVEMLRRTELVVDTKSKDSANKVLMM 248
Query: 290 LHANMGNVDEVNRI 303
++ + G + +R+
Sbjct: 249 MYWDAGAKQDASRL 262
>At1g09680 hypothetical protein
Length = 607
Score = 91.3 bits (225), Expect = 1e-18
Identities = 68/276 (24%), Positives = 127/276 (45%), Gaps = 8/276 (2%)
Query: 133 DKMRGQP---TCTALLHAYVQNNLTNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKL 189
+K R +P T +AL++A + N + A L +M + G + + V + ++ + NG++
Sbjct: 302 EKSRTRPDVFTYSALINALCKENKMDGAHGLFDEMCKRGLIPNDVIFTTLIHGHSRNGEI 361
Query: 190 EKVPKLFEE-LKVNTSPDVVTFNLLLTACASENDVETAERVLLQLKKAKVDPDWVTYSTL 248
+ + + +++ L PD+V +N L+ D+ A ++ + + + PD +TY+TL
Sbjct: 362 DLMKESYQKMLSKGLQPDIVLYNTLVNGFCKNGDLVAARNIVDGMIRRGLRPDKITYTTL 421
Query: 249 TNLYIRNASVDDCLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMK 308
+ + R V+ LE KEM++ RV +S+L+ G V + R +M
Sbjct: 422 IDGFCRGGDVETALE----IRKEMDQNGIELDRVGFSALVCGMCKEGRVIDAERALREML 477
Query: 309 ACFCKMSDDEYVCMISSLVKLGDFAGVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQME 368
K D Y M+ + K GD L KE +S V N+LL GQM+
Sbjct: 478 RAGIKPDDVTYTMMMDAFCKKGDAQTGFKLLKEMQSDGHVPSVVTYNVLLNGLCKLGQMK 537
Query: 369 MAEIFCNQLVEKGVCLSYSSWELLTRGYLKKKDVKK 404
A++ + ++ GV ++ L G+ + + K
Sbjct: 538 NADMLLDAMLNIGVVPDDITYNTLLEGHHRHANSSK 573
Score = 74.3 bits (181), Expect = 1e-13
Identities = 74/338 (21%), Positives = 139/338 (40%), Gaps = 14/338 (4%)
Query: 141 CTALLHAYVQNNLTNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEEL- 199
C LL ++ N T ++ + GF + +N +M+ + G + K+F+E+
Sbjct: 208 CGNLLDRMMKLNPTGTIWGFYMEILDAGFPLNVYVFNILMNKFCKEGNISDAQKVFDEIT 267
Query: 200 KVNTSPDVVTFNLLLTACASENDVETAERVLLQLKKAKVDPDWVTYSTLTNLYIRNASVD 259
K + P VV+FN L+ +++ R+ Q++K++ PD TYS L N + +D
Sbjct: 268 KRSLQPTVVSFNTLINGYCKVGNLDEGFRLKHQMEKSRTRPDVFTYSALINALCKENKMD 327
Query: 260 DCLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEY 319
A EM KR V +++L+ H+ G +D + + KM + + Y
Sbjct: 328 G----AHGLFDEMCKRGLIPNDVIFTTLIHGHSRNGEIDLMKESYQKMLSKGLQPDIVLY 383
Query: 320 VCMISSLVKLGDFAGVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVE 379
+++ K GD N+ D L+ + G +E A ++ +
Sbjct: 384 NTLVNGFCKNGDLVAARNIVDGMIRRGLRPDKITYTTLIDGFCRGGDVETALEIRKEMDQ 443
Query: 380 KGVCLSYSSWELLTRGYLKKK---DVKKFLHYFGKAISSVKQWIPDPRLVQEAFTVIQEQ 436
G+ L + L G K+ D ++ L +A +K PD ++
Sbjct: 444 NGIELDRVGFSALVCGMCKEGRVIDAERALREMLRA--GIK---PDDVTYTMMMDAFCKK 498
Query: 437 AHIEGAEQLLVILRNAGHVNTNI-YNLFLKTYAAAGKM 473
+ +LL +++ GHV + + YN+ L G+M
Sbjct: 499 GDAQTGFKLLKEMQSDGHVPSVVTYNVLLNGLCKLGQM 536
>At1g09900 hypothetical protein
Length = 598
Score = 90.9 bits (224), Expect = 2e-18
Identities = 80/359 (22%), Positives = 155/359 (42%), Gaps = 11/359 (3%)
Query: 141 CTALLHAYVQNNLTNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELK 200
CT L+ + + T KA ++ + G + + YN M+S Y G++ + + +
Sbjct: 140 CTTLIRGFCRLGKTRKAAKILEILEGSGAVPDVITYNVMISGYCKAGEINNALSVLDRMS 199
Query: 201 VNTSPDVVTFNLLLTACASENDVETAERVLLQLKKAKVDPDWVTYSTLTNLYIRNASVDD 260
V SPDVVT+N +L + ++ A VL ++ + PD +TY+ L R++ V
Sbjct: 200 V--SPDVVTYNTILRSLCDSGKLKQAMEVLDRMLQRDCYPDVITYTILIEATCRDSGVGH 257
Query: 261 CLEKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYV 320
++ L EM R V Y+ L++ G +DE + M + C+ + +
Sbjct: 258 AMK----LLDEMRDRGCTPDVVTYNVLVNGICKEGRLDEAIKFLNDMPSSGCQPNVITHN 313
Query: 321 CMISSLVKLGDFAGVENLYKEWESVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEK 380
++ S+ G + E L + + V N+L+ +G + A ++ +
Sbjct: 314 IILRSMCSTGRWMDAEKLLADMLRKGFSPSVVTFNILINFLCRKGLLGRAIDILEKMPQH 373
Query: 381 GVCLSYSSWELLTRGYLKKKDVKKFLHYFGKAISSVKQWIPDPRLVQEAFTVIQEQAHIE 440
G + S+ L G+ K+K + + + Y + +S + PD T + + +E
Sbjct: 374 GCQPNSLSYNPLLHGFCKEKKMDRAIEYLERMVS--RGCYPDIVTYNTMLTALCKDGKVE 431
Query: 441 GAEQLLVILRNAGHVNTNI-YNLFLKTYAAAGK--MPLVVAERMKKDNVQLDKETHRLL 496
A ++L L + G I YN + A AGK + + + M+ +++ D T+ L
Sbjct: 432 DAVEILNQLSSKGCSPVLITYNTVIDGLAKAGKTGKAIKLLDEMRAKDLKPDTITYSSL 490
Score = 67.0 bits (162), Expect = 2e-11
Identities = 62/292 (21%), Positives = 123/292 (41%), Gaps = 9/292 (3%)
Query: 108 YAVQLDLITKVRGLNSAEKFFEDLPDKMRGQP---TCTALLHAYVQNNLTNKAEALMSKM 164
Y V ++ I K L+ A KF D+P QP T +L + AE L++ M
Sbjct: 277 YNVLVNGICKEGRLDEAIKFLNDMPSS-GCQPNVITHNIILRSMCSTGRWMDAEKLLADM 335
Query: 165 SECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKVN-TSPDVVTFNLLLTACASENDV 223
GF S V +N +++ G L + + E++ + P+ +++N LL E +
Sbjct: 336 LRKGFSPSVVTFNILINFLCRKGLLGRAIDILEKMPQHGCQPNSLSYNPLLHGFCKEKKM 395
Query: 224 ETAERVLLQLKKAKVDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEMEKRTSRETRVA 283
+ A L ++ PD VTY+T+ ++ V+D +E L ++ + +
Sbjct: 396 DRAIEYLERMVSRGCYPDIVTYNTMLTALCKDGKVEDAVE----ILNQLSSKGCSPVLIT 451
Query: 284 YSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCMISSLVKLGDFAGVENLYKEWE 343
Y++++ A G + ++ +M+A K Y ++ L + G + E+E
Sbjct: 452 YNTVIDGLAKAGKTGKAIKLLDEMRAKDLKPDTITYSSLVGGLSREGKVDEAIKFFHEFE 511
Query: 344 SVSGTNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKGVCLSYSSWELLTRG 395
+ + N ++ Q + A F ++ +G + +S+ +L G
Sbjct: 512 RMGIRPNAVTFNSIMLGLCKSRQTDRAIDFLVFMINRGCKPNETSYTILIEG 563
Score = 59.7 bits (143), Expect = 4e-09
Identities = 44/188 (23%), Positives = 79/188 (41%), Gaps = 5/188 (2%)
Query: 144 LLHAYVQNNLTNKAEALMSKMSECGFLRSPVPYNRMMSLYISNGKLEKVPKLFEELKVN- 202
LLH + + ++A + +M G V YN M++ +GK+E ++ +L
Sbjct: 385 LLHGFCKEKKMDRAIEYLERMVSRGCYPDIVTYNTMLTALCKDGKVEDAVEILNQLSSKG 444
Query: 203 TSPDVVTFNLLLTACASENDVETAERVLLQLKKAKVDPDWVTYSTLTNLYIRNASVDDCL 262
SP ++T+N ++ A A ++L +++ + PD +TYS+L R VD+
Sbjct: 445 CSPVLITYNTVIDGLAKAGKTGKAIKLLDEMRAKDLKPDTITYSSLVGGLSREGKVDE-- 502
Query: 263 EKAASTLKEMEKRTSRETRVAYSSLLSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCM 322
A E E+ R V ++S++ D M CK ++ Y +
Sbjct: 503 --AIKFFHEFERMGIRPNAVTFNSIMLGLCKSRQTDRAIDFLVFMINRGCKPNETSYTIL 560
Query: 323 ISSLVKLG 330
I L G
Sbjct: 561 IEGLAYEG 568
Score = 46.6 bits (109), Expect = 3e-05
Identities = 40/214 (18%), Positives = 84/214 (38%), Gaps = 8/214 (3%)
Query: 169 FLRSPVPYNRMMSLYISNGKLEKVPKLFEELKVNTS-PDVVTFNLLLTACASENDVETAE 227
F V N + + G+LE+ K E + + + PD++ L+ A
Sbjct: 98 FALEDVESNNHLRQMVRTGELEEGFKFLENMVYHGNVPDIIPCTTLIRGFCRLGKTRKAA 157
Query: 228 RVLLQLKKAKVDPDWVTYSTLTNLYIRNASVDDCLEKAASTLKEMEKRTSRETRVAYSSL 287
++L L+ + PD +TY+ + + Y + ++ + L +++ + V Y+++
Sbjct: 158 KILEILEGSGAVPDVITYNVMISGYCKAGEIN-------NALSVLDRMSVSPDVVTYNTI 210
Query: 288 LSLHANMGNVDEVNRIWGKMKACFCKMSDDEYVCMISSLVKLGDFAGVENLYKEWESVSG 347
L + G + + + +M C Y +I + + L E
Sbjct: 211 LRSLCDSGKLKQAMEVLDRMLQRDCYPDVITYTILIEATCRDSGVGHAMKLLDEMRDRGC 270
Query: 348 TNDVRVSNLLLTSYVDQGQMEMAEIFCNQLVEKG 381
T DV N+L+ +G+++ A F N + G
Sbjct: 271 TPDVVTYNVLVNGICKEGRLDEAIKFLNDMPSSG 304
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.132 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,970,659
Number of Sequences: 26719
Number of extensions: 458529
Number of successful extensions: 5399
Number of sequences better than 10.0: 457
Number of HSP's better than 10.0 without gapping: 293
Number of HSP's successfully gapped in prelim test: 164
Number of HSP's that attempted gapping in prelim test: 1491
Number of HSP's gapped (non-prelim): 1737
length of query: 512
length of database: 11,318,596
effective HSP length: 104
effective length of query: 408
effective length of database: 8,539,820
effective search space: 3484246560
effective search space used: 3484246560
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC122725.14


