

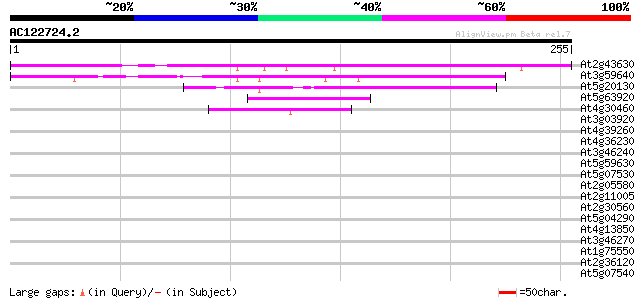
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122724.2 + phase: 0
(255 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g43630 unknown protein 137 4e-33
At3g59640 putative protein 119 2e-27
At5g20130 unknown protein 53 2e-07
At5g63920 DNA topoisomerase III 41 7e-04
At4g30460 glycine-rich protein 41 7e-04
At3g03920 putative GAR1 protein 40 0.001
At4g39260 glycine-rich protein (clone AtGRP8) 40 0.002
At4g36230 putative glycine-rich cell wall protein 40 0.002
At3g46240 putative protein 40 0.002
At5g59630 glycine-rich protein - like 39 0.004
At5g07530 glycine-rich protein atGRP-7 39 0.004
At2g05580 unknown protein 39 0.004
At2g11005 putative protein 38 0.005
At2g30560 putative glycine-rich protein 38 0.006
At5g04290 glycine-rich protein 37 0.010
At4g13850 glycine-rich RNA-binding protein AtGRP2 - like 37 0.010
At3g46270 putative protein 37 0.013
At1g75550 hypothetical protein 37 0.013
At2g36120 unknown protein 36 0.018
At5g07540 glycine-rich protein atGRP-6 36 0.023
>At2g43630 unknown protein
Length = 274
Score = 137 bits (346), Expect = 4e-33
Identities = 95/274 (34%), Positives = 147/274 (52%), Gaps = 31/274 (11%)
Query: 1 MNAIQVAASNPAIYVRKNYQPHHFLSTPKGLSMNANPLSRTFLIPRAKLSSGLYCVRQSA 60
M+ Q P+ Y + +P+ S P S+ + RT RA + S
Sbjct: 1 MSFTQANCFRPSYYPARITRPNCISSVPIRSSVRFDHFPRTSFTLRATAAV-------ST 53
Query: 61 TTAPVLKVYPSNLTQSVPVLKSQKPRHVCLAGGKGMMDNSED-SQRKSLEEAMKS----- 114
+P+L + + +P KS++ VCL GGK D S++ S K++E+AM
Sbjct: 54 QFSPLL-----DHRRRLPTGKSKQSSAVCLFGGKDKPDGSDEISPWKAIEKAMGKKSVED 108
Query: 115 -LQEKIQKGEY--SGGSGSKPPGGRGRGGGGGNSD------GSEEGSTGGMSDETAQIVF 165
L+E+IQK ++ + G+ PP G G GGGGGN + G E+G G++DET Q+V
Sbjct: 109 MLREQIQKKDFYDTDSGGNMPPRGGGSGGGGGNGEERPEGSGGEDGGLAGIADETLQVVL 168
Query: 166 ATIGFMFVYIYVINGVELTKLARDFIKYLLGGTQSVRLKRASYKCSRFYKKITGQNEVDE 225
AT+GF+F+Y Y+I G EL KLARD+I++L+G ++VRL RA + F +K++ Q DE
Sbjct: 169 ATLGFIFLYTYIITGEELVKLARDYIRFLMGRPKTVRLTRAMDSWNGFLEKMSRQRVYDE 228
Query: 226 DGLETG----PLRWNIADFYRDVLRNYMKPNSNE 255
LE P ++ + YR V++ Y+ NS+E
Sbjct: 229 YWLEKAIINTPTWYDSPEKYRRVIKAYVDSNSDE 262
>At3g59640 putative protein
Length = 246
Score = 119 bits (297), Expect = 2e-27
Identities = 84/238 (35%), Positives = 127/238 (53%), Gaps = 29/238 (12%)
Query: 1 MNAIQVAASNPAIYVRKNYQPHHFLSTP--KGLSMNANPLSRTFLIPRAKLSSGLYCVRQ 58
M++ Q P+++ + Q H S P L + PL + L RA SS +
Sbjct: 1 MSSTQANLCRPSLFCARTTQTRHVSSAPFMSSLRFDYRPLPK--LAIRASASSSM----- 53
Query: 59 SATTAPVLKVYPSNLTQSVPVLKSQKPRHVCLAGGKGMMDNSED--SQRKSLEEAM--KS 114
S+ +P+ N Q PV VCL GGK + S + S +++E+AM KS
Sbjct: 54 SSQFSPLQNHRCRNQRQG-PV--------VCLLGGKDKSNGSNELSSTWEAIEKAMGKKS 104
Query: 115 LQEKIQKGEYSGGSGSKPPGGRGRGGGG----GNSDGSEEGSTGGMS---DETAQIVFAT 167
+++ +++ +G PP GRG GGGG N G G GG++ DET Q+V AT
Sbjct: 105 VEDMLREQIQKKDTGGIPPRGRGGGGGGRNGGNNGSGGSSGEDGGLASFGDETLQVVLAT 164
Query: 168 IGFMFVYIYVINGVELTKLARDFIKYLLGGTQSVRLKRASYKCSRFYKKITGQNEVDE 225
+GF+F+Y Y+ING EL +LARD+I+YL+G +SVRL R SRF++K++ + +E
Sbjct: 165 LGFIFLYFYIINGEELFRLARDYIRYLIGRPKSVRLTRVMEGWSRFFEKMSRKKVYNE 222
>At5g20130 unknown protein
Length = 202
Score = 52.8 bits (125), Expect = 2e-07
Identities = 45/149 (30%), Positives = 64/149 (42%), Gaps = 15/149 (10%)
Query: 80 LKSQKPRHVCLAGGKGMMDNSEDSQRKSLEEAM-------KSLQEKIQKGEYSGGSGSKP 132
+ K R CL G + E+ RKSLE A+ + ++I+K E SGG
Sbjct: 39 ITKSKGRFSCLFSGG---NQREEQARKSLESALGGKKNEFEKWDKEIKKREESGGGN--- 92
Query: 133 PGGRGRGGGGGNSDGSEEGSTGGMSDETAQIVFATIGFMFVYIYVINGVELTKLARDFIK 192
GG+G GGGGG G S E QI F + + VY+ V G + + +
Sbjct: 93 -GGKG-GGGGGWFGGGGWFSGDHFWKEAQQIAFTLLAILAVYMVVAKGEVMAAFVLNPLL 150
Query: 193 YLLGGTQSVRLKRASYKCSRFYKKITGQN 221
Y L GT+ +S R K++G N
Sbjct: 151 YALRGTREGLSSLSSKLMGREASKVSGDN 179
>At5g63920 DNA topoisomerase III
Length = 926
Score = 40.8 bits (94), Expect = 7e-04
Identities = 21/56 (37%), Positives = 27/56 (47%)
Query: 109 EEAMKSLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMSDETAQIV 164
E+++ + G SGGSG + GRGRGG GG S G GS T + V
Sbjct: 842 EDSINNSSGNATTGSNSGGSGRRGSRGRGRGGRGGQSSGGRRGSGTSFVSATGEPV 897
>At4g30460 glycine-rich protein
Length = 162
Score = 40.8 bits (94), Expect = 7e-04
Identities = 24/70 (34%), Positives = 29/70 (41%), Gaps = 5/70 (7%)
Query: 91 AGGKGMMDNSEDSQRKSLEEAMKSLQEKIQKGEYSG-----GSGSKPPGGRGRGGGGGNS 145
+GG G +S S S + G Y+G GSG + GRGRG GGG
Sbjct: 65 SGGGGSSSSSSSSSSSSSSSGGGGGDAGSEAGSYAGSHAGSGSGGRSGSGRGRGSGGGGG 124
Query: 146 DGSEEGSTGG 155
G G GG
Sbjct: 125 HGGGGGGGGG 134
Score = 36.2 bits (82), Expect = 0.018
Identities = 16/30 (53%), Positives = 18/30 (59%)
Query: 126 GGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
GG G GG GRGGGGG+ +G G GG
Sbjct: 123 GGHGGGGGGGGGRGGGGGSGNGEGYGEGGG 152
Score = 33.9 bits (76), Expect = 0.087
Identities = 17/34 (50%), Positives = 19/34 (55%), Gaps = 2/34 (5%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G + GG G GGRG GGG GN +G EG G
Sbjct: 123 GGHGGGGGGG--GGRGGGGGSGNGEGYGEGGGYG 154
Score = 32.3 bits (72), Expect = 0.25
Identities = 15/32 (46%), Positives = 17/32 (52%)
Query: 126 GGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMS 157
GGSGS G G GGGG +S S S+ S
Sbjct: 53 GGSGSGAGAGSGSGGGGSSSSSSSSSSSSSSS 84
Score = 30.8 bits (68), Expect = 0.74
Identities = 19/53 (35%), Positives = 22/53 (40%)
Query: 105 RKSLEEAMKSLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMS 157
RK L + + I G GG GS G G G GGG S S S+ S
Sbjct: 30 RKDLGLDLGGIGAGIGIGIGIGGGGSGSGAGAGSGSGGGGSSSSSSSSSSSSS 82
>At3g03920 putative GAR1 protein
Length = 202
Score = 40.0 bits (92), Expect = 0.001
Identities = 23/73 (31%), Positives = 34/73 (46%), Gaps = 3/73 (4%)
Query: 121 KGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMSDETAQIVFATIGFMFVYIYVING 180
+G GG G + GG GR GGGG G G GG DE + +++ G
Sbjct: 14 RGGRDGGGGGRFGGGGGRFGGGGGRFGGGGGRFGGFRDEGPPSEVVEVA---TFVHACEG 70
Query: 181 VELTKLARDFIKY 193
+TKL+++ I +
Sbjct: 71 DAVTKLSQEKIPH 83
>At4g39260 glycine-rich protein (clone AtGRP8)
Length = 169
Score = 39.7 bits (91), Expect = 0.002
Identities = 24/63 (38%), Positives = 31/63 (49%), Gaps = 1/63 (1%)
Query: 94 KGMMDNSEDSQRKSLEEAMKSLQEKIQKGEYSGGSGSKPPGGRGR-GGGGGNSDGSEEGS 152
K M D E+ K L+ + ++ E +G GG G GG R GGGGG S G G
Sbjct: 58 KAMRDAIEEMNGKELDGRVITVNEAQSRGSGGGGGGRGGSGGGYRSGGGGGYSGGGGGGY 117
Query: 153 TGG 155
+GG
Sbjct: 118 SGG 120
Score = 31.6 bits (70), Expect = 0.43
Identities = 16/32 (50%), Positives = 17/32 (53%), Gaps = 2/32 (6%)
Query: 126 GGSGSKPPGGRGR--GGGGGNSDGSEEGSTGG 155
GG G + GG GR GGG G DG G GG
Sbjct: 136 GGGGGRGYGGGGRREGGGYGGGDGGSYGGGGG 167
Score = 31.2 bits (69), Expect = 0.56
Identities = 18/40 (45%), Positives = 20/40 (50%), Gaps = 7/40 (17%)
Query: 120 QKGEYSGGSGSKPPGGRGRGGGG----GNSDGSEEGSTGG 155
+ G Y G G GGRG GGGG G G + GS GG
Sbjct: 128 RSGGYGSGGGG---GGRGYGGGGRREGGGYGGGDGGSYGG 164
Score = 28.9 bits (63), Expect = 2.8
Identities = 18/37 (48%), Positives = 18/37 (48%), Gaps = 7/37 (18%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGG---NSDGSEEGSTGG 155
G YSGG G GG GGGGG S G G GG
Sbjct: 107 GGYSGGGG----GGYSGGGGGGYERRSGGYGSGGGGG 139
>At4g36230 putative glycine-rich cell wall protein
Length = 221
Score = 39.7 bits (91), Expect = 0.002
Identities = 24/64 (37%), Positives = 29/64 (44%), Gaps = 5/64 (7%)
Query: 92 GGKGMMDNSEDSQRKSLEEAMKSLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEG 151
GG+G N +D+ K + + G GG GS GRGRGGGGG G G
Sbjct: 111 GGEGNGGNGKDNSHKRNKSSGGG-----GGGGGGGGGGSGNGSGRGRGGGGGGGGGGGGG 165
Query: 152 STGG 155
GG
Sbjct: 166 GGGG 169
Score = 35.4 bits (80), Expect = 0.030
Identities = 16/34 (47%), Positives = 17/34 (49%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G GG G GG G GGGGG G +G GG
Sbjct: 155 GGGGGGGGGGGGGGGGGGGGGGGGGGGSDGKGGG 188
Score = 33.9 bits (76), Expect = 0.087
Identities = 18/38 (47%), Positives = 19/38 (49%), Gaps = 4/38 (10%)
Query: 121 KGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMSD 158
+G GG G GG G GGGGG G G GG SD
Sbjct: 150 RGRGGGGGG----GGGGGGGGGGGGGGGGGGGGGGGSD 183
Score = 32.7 bits (73), Expect = 0.19
Identities = 15/36 (41%), Positives = 19/36 (52%)
Query: 125 SGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMSDET 160
+GG G GG G GGG G+ G EG+ G D +
Sbjct: 88 AGGGGGGGGGGGGGGGGQGSGGGGGEGNGGNGKDNS 123
Score = 32.3 bits (72), Expect = 0.25
Identities = 18/50 (36%), Positives = 25/50 (50%), Gaps = 3/50 (6%)
Query: 98 DNSEDSQRKSLEEAMKSLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDG 147
+N+ D Q + E +S + G GG G GG+G GGGGG +G
Sbjct: 70 ENALDDQNVNKTEKCES---RAGGGGGGGGGGGGGGGGQGSGGGGGEGNG 116
Score = 31.6 bits (70), Expect = 0.43
Identities = 18/52 (34%), Positives = 23/52 (43%), Gaps = 18/52 (34%)
Query: 122 GEYSGGSGSKPPGGRGR------------------GGGGGNSDGSEEGSTGG 155
G+ SGG G + GG G+ GGGGG+ +GS G GG
Sbjct: 104 GQGSGGGGGEGNGGNGKDNSHKRNKSSGGGGGGGGGGGGGSGNGSGRGRGGG 155
Score = 31.6 bits (70), Expect = 0.43
Identities = 15/34 (44%), Positives = 15/34 (44%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G GG G GG G GGGGG G GG
Sbjct: 154 GGGGGGGGGGGGGGGGGGGGGGGGGGGGSDGKGG 187
Score = 30.0 bits (66), Expect = 1.3
Identities = 14/34 (41%), Positives = 17/34 (49%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G GG GS GG G GG G ++ S+GG
Sbjct: 99 GGGGGGQGSGGGGGEGNGGNGKDNSHKRNKSSGG 132
Score = 29.3 bits (64), Expect = 2.1
Identities = 15/48 (31%), Positives = 20/48 (41%)
Query: 107 SLEEAMKSLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTG 154
+L++ + EK + GG G GG G G G G G G G
Sbjct: 72 ALDDQNVNKTEKCESRAGGGGGGGGGGGGGGGGQGSGGGGGEGNGGNG 119
>At3g46240 putative protein
Length = 441
Score = 39.7 bits (91), Expect = 0.002
Identities = 32/117 (27%), Positives = 49/117 (41%), Gaps = 9/117 (7%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMSD-ETAQIVFATIGFMFVYIYVING 180
G SGGSG GG G G G S G+ +G TGG ++ + + + V++
Sbjct: 316 GSGSGGSGG---GGSGSSGSGSGSGGTSKGGTGGSNEVSPGGSTNGKSSKLPIILGVVSP 372
Query: 181 VELTKLARDFIKYLLGGTQSVRLKRASYKCSRFYKKITGQN-----EVDEDGLETGP 232
V +A FI +L + VRL+ + S TG + ++D D P
Sbjct: 373 VAFVLVAYVFIAIILANRRKVRLQALAMPTSTVATMGTGPSTLFTQQMDNDSTSNQP 429
>At5g59630 glycine-rich protein - like
Length = 253
Score = 38.5 bits (88), Expect = 0.004
Identities = 28/99 (28%), Positives = 38/99 (38%), Gaps = 13/99 (13%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMSDETAQIVFATIGFMFVYIYVINGV 181
G GG G GG G GGGGG G G GG +G
Sbjct: 79 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGKLQLLG------------ 126
Query: 182 ELTKLARDFIKYLLGGTQSVRLKRASYKCSRFYKKITGQ 220
L ++ K L+ GT+ V +K S+ S+ YK+ +
Sbjct: 127 -LKPYSKKMKKKLVNGTEQVAIKILSHSSSQGYKQFKAE 164
Score = 35.0 bits (79), Expect = 0.039
Identities = 17/34 (50%), Positives = 17/34 (50%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G SGG G GG G GGGGG G G GG
Sbjct: 68 GGSSGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 101
Score = 33.5 bits (75), Expect = 0.11
Identities = 16/31 (51%), Positives = 16/31 (51%)
Query: 125 SGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
SGGS GG G GGGGG G G GG
Sbjct: 67 SGGSSGGGGGGGGGGGGGGGGGGGGGGGGGG 97
Score = 33.1 bits (74), Expect = 0.15
Identities = 16/34 (47%), Positives = 16/34 (47%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G GG G GG G GGGGG G G GG
Sbjct: 74 GGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGG 107
Score = 33.1 bits (74), Expect = 0.15
Identities = 17/44 (38%), Positives = 19/44 (42%)
Query: 112 MKSLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
+K + G S SG GG G GGGGG G G GG
Sbjct: 51 LKHVSRLDDNGLDSSNSGGSSGGGGGGGGGGGGGGGGGGGGGGG 94
Score = 32.0 bits (71), Expect = 0.33
Identities = 15/31 (48%), Positives = 15/31 (48%)
Query: 125 SGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
S GS GG G GGGGG G G GG
Sbjct: 65 SNSGGSSGGGGGGGGGGGGGGGGGGGGGGGG 95
Score = 28.5 bits (62), Expect = 3.7
Identities = 14/41 (34%), Positives = 19/41 (46%)
Query: 115 LQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
L + + + + +G S G G GGGGG G G GG
Sbjct: 50 LLKHVSRLDDNGLDSSNSGGSSGGGGGGGGGGGGGGGGGGG 90
>At5g07530 glycine-rich protein atGRP-7
Length = 543
Score = 38.5 bits (88), Expect = 0.004
Identities = 30/81 (37%), Positives = 38/81 (46%), Gaps = 14/81 (17%)
Query: 80 LKSQKPRHVCLAGGKGMMDNSEDSQRKSLEEAMKSLQEKIQKGEYSGGSGSKPPGGRGRG 139
LK+ + C++GG M SE +S + I G SGGSGSK G G+
Sbjct: 378 LKANLGKKKCMSGG---MSGSEGGMSRS--------EGGISGGGMSGGSGSKHKIGGGKH 426
Query: 140 GGGGNSDGSE---EGSTGGMS 157
GG G G + GS GGMS
Sbjct: 427 GGLGGKFGKKRGMSGSGGGMS 447
Score = 34.7 bits (78), Expect = 0.051
Identities = 25/76 (32%), Positives = 35/76 (45%), Gaps = 10/76 (13%)
Query: 91 AGGKGMMDNSEDSQRKSLEEAMKSLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDGSE- 149
AG G + S+ KS + +K+ +K+ K ++ G G G GG GG S+G E
Sbjct: 162 AGAGGAQSLIKKSKAKS-KGGLKAWCKKMLKSKFGGKKGKSGGGKSKFGGKGGKSEGEEG 220
Query: 150 --------EGSTGGMS 157
GS GGMS
Sbjct: 221 MSSGDEGMSGSEGGMS 236
Score = 32.7 bits (73), Expect = 0.19
Identities = 31/95 (32%), Positives = 42/95 (43%), Gaps = 28/95 (29%)
Query: 90 LAGGKGMMDNSEDSQRKSLEEAMKSLQEKIQ---------------KGEYSGGSGSKPPG 134
++G +G M E + KS + +K+ EK + +G SGG GSK
Sbjct: 228 MSGSEGGMSGGEGGKSKSGKGKLKAKLEKKKGMSGGSESEEGMSGSEGGMSGGGGSKSKS 287
Query: 135 ---------GRGRGGGGGNSDGSEEG---STGGMS 157
G+ +G GG S GSEEG S GGMS
Sbjct: 288 KKSKLKAKLGKKKGMSGGMS-GSEEGMSGSEGGMS 321
Score = 32.7 bits (73), Expect = 0.19
Identities = 26/79 (32%), Positives = 33/79 (40%), Gaps = 18/79 (22%)
Query: 90 LAGGKGMMDNSEDSQRKSLEEAMKSLQEKIQKGEYSGGSGSKPPGGRGRGG--------- 140
++G G M SE S + + G SGGSGSK G G+ G
Sbjct: 439 MSGSGGGMSGSEGGVSGS--------EGSMSGGGMSGGSGSKHKIGGGKHGGLRGKFGKK 490
Query: 141 -GGGNSDGSEEGSTGGMSD 158
G S+G GS GGMS+
Sbjct: 491 RGMSGSEGGMSGSEGGMSE 509
Score = 30.4 bits (67), Expect = 0.96
Identities = 27/90 (30%), Positives = 35/90 (38%), Gaps = 24/90 (26%)
Query: 90 LAGGKGMMDNSEDSQRKSLEEAMKSLQEKIQKGEYSGGSGSKPPGGRGR-----GGGGGN 144
++G +G M + S+ KS + +K+ K K SGG G G GGGGG
Sbjct: 313 MSGSEGGMSSGGGSKSKSKKSKLKAKLGK--KKSMSGGMSGSEEGMSGSEGGMSGGGGGK 370
Query: 145 S-----------------DGSEEGSTGGMS 157
S G GS GGMS
Sbjct: 371 SKSRKSKLKANLGKKKCMSGGMSGSEGGMS 400
Score = 28.1 bits (61), Expect = 4.8
Identities = 21/63 (33%), Positives = 32/63 (50%), Gaps = 2/63 (3%)
Query: 90 LAGGKGMMDNSEDSQRKSLEEAMKSLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDGSE 149
++G +G M S+ KS + +K+ K +KG G SGS+ G G GG + GS+
Sbjct: 270 MSGSEGGMSGGGGSKSKSKKSKLKAKLGK-KKGMSGGMSGSEE-GMSGSEGGMSSGGGSK 327
Query: 150 EGS 152
S
Sbjct: 328 SKS 330
Score = 27.3 bits (59), Expect = 8.1
Identities = 21/70 (30%), Positives = 32/70 (45%), Gaps = 5/70 (7%)
Query: 90 LAGGKGMMDNSEDSQRKSLEEAMKSL--QEKIQKGEYSGGSG--SKPPGGRGRGGGGGNS 145
++G +G M + KS + +K+ ++K G SG G S+ GG GGG
Sbjct: 356 MSGSEGGMSGGGGGKSKSRKSKLKANLGKKKCMSGGMSGSEGGMSRSEGGIS-GGGMSGG 414
Query: 146 DGSEEGSTGG 155
GS+ GG
Sbjct: 415 SGSKHKIGGG 424
>At2g05580 unknown protein
Length = 302
Score = 38.5 bits (88), Expect = 0.004
Identities = 17/35 (48%), Positives = 19/35 (53%)
Query: 121 KGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
+G + GG G GG GRGGGGG G G GG
Sbjct: 255 QGGHKGGGGGGHVGGGGRGGGGGGRGGGGSGGGGG 289
Score = 37.7 bits (86), Expect = 0.006
Identities = 18/38 (47%), Positives = 22/38 (57%), Gaps = 2/38 (5%)
Query: 120 QKGEYSGGSGSKPPGGRGRGG--GGGNSDGSEEGSTGG 155
QKG GG G + GG G+GG GGG G ++G GG
Sbjct: 98 QKGGGGGGQGGQKGGGGGQGGQKGGGGGQGGQKGGGGG 135
Score = 35.8 bits (81), Expect = 0.023
Identities = 23/67 (34%), Positives = 34/67 (50%), Gaps = 4/67 (5%)
Query: 96 MMDNSEDSQRKSLEEAMKSLQEKIQKGEYSGG-SGSKPPGGRGRGG--GGGNSDGSEEGS 152
++D E+ + K+ + + Q QKG GG G K GG G+GG GGG G ++G
Sbjct: 64 VLDGEEEEESKNNQGGIGRGQGG-QKGGGGGGQGGQKGGGGGGQGGQKGGGGGQGGQKGG 122
Query: 153 TGGMSDE 159
GG +
Sbjct: 123 GGGQGGQ 129
Score = 35.4 bits (80), Expect = 0.030
Identities = 18/39 (46%), Positives = 21/39 (53%), Gaps = 3/39 (7%)
Query: 120 QKGEYSGGSGSKPPGGRGRG---GGGGNSDGSEEGSTGG 155
QKG G G K GG G+G GGGG G ++G GG
Sbjct: 119 QKGGGGGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGG 157
Score = 35.4 bits (80), Expect = 0.030
Identities = 15/35 (42%), Positives = 18/35 (50%)
Query: 121 KGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
+G + GG G GG GGGGG G +G GG
Sbjct: 220 QGGHKGGGGGGGQGGHKGGGGGGQGGGGHKGGGGG 254
Score = 35.0 bits (79), Expect = 0.039
Identities = 33/132 (25%), Positives = 55/132 (41%), Gaps = 16/132 (12%)
Query: 31 LSMNANPLSRTFLIPRAKLSSGLYCVRQSATTAPVLKVYPSNLTQSVPVLKSQKPRHVCL 90
+ MN+ L FL L S ++ ++ A+ +V + + +S K C
Sbjct: 1 MKMNSRALIVWFL-----LLSAVFAIKVVASIDDTKQVGENQVEESKSTCKYGN----CA 51
Query: 91 AGGKGMMDNSEDSQRKSLEEAMKSLQEKIQKGEYSGGSGSKPPGGRGRG---GGGGNSDG 147
G+ +++ EE K+ Q I +G+ G K GG G+G GGGG G
Sbjct: 52 QYGENHFVETDEVLDGEEEEESKNNQGGIGRGQ----GGQKGGGGGGQGGQKGGGGGGQG 107
Query: 148 SEEGSTGGMSDE 159
++G GG +
Sbjct: 108 GQKGGGGGQGGQ 119
Score = 34.7 bits (78), Expect = 0.051
Identities = 19/41 (46%), Positives = 22/41 (53%), Gaps = 5/41 (12%)
Query: 120 QKGEYSGGS--GSKPPGGRGRG---GGGGNSDGSEEGSTGG 155
Q G+ GG G K GGRG+G GGGG G +G GG
Sbjct: 177 QGGQKGGGGQGGQKGGGGRGQGGMKGGGGGGQGGHKGGGGG 217
Score = 34.3 bits (77), Expect = 0.067
Identities = 15/35 (42%), Positives = 18/35 (50%)
Query: 121 KGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
+G + GG G GG GGGGG G + G GG
Sbjct: 208 QGGHKGGGGGGGQGGHKGGGGGGGQGGHKGGGGGG 242
Score = 33.9 bits (76), Expect = 0.087
Identities = 18/39 (46%), Positives = 22/39 (56%), Gaps = 4/39 (10%)
Query: 120 QKGEYSGGSGSKPPGGRGRG---GGGGNSDGSEEGSTGG 155
QKG GG G + GG G+G GGGG G ++G GG
Sbjct: 109 QKGG-GGGQGGQKGGGGGQGGQKGGGGGGQGGQKGGGGG 146
Score = 33.9 bits (76), Expect = 0.087
Identities = 17/35 (48%), Positives = 18/35 (50%), Gaps = 2/35 (5%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGM 156
G GG G + GG G GGGGG G G GGM
Sbjct: 270 GGRGGGGGGR--GGGGSGGGGGGGSGRGGGGGGGM 302
Score = 32.0 bits (71), Expect = 0.33
Identities = 17/35 (48%), Positives = 17/35 (48%), Gaps = 1/35 (2%)
Query: 122 GEYSGGSGSKPPGGRGRGG-GGGNSDGSEEGSTGG 155
G GG GGRG GG GGG GS G GG
Sbjct: 265 GHVGGGGRGGGGGGRGGGGSGGGGGGGSGRGGGGG 299
Score = 32.0 bits (71), Expect = 0.33
Identities = 17/37 (45%), Positives = 19/37 (50%), Gaps = 1/37 (2%)
Query: 120 QKGEYSGG-SGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
QKG GG G K GG G+GG G G + G GG
Sbjct: 129 QKGGGGGGQGGQKGGGGGGQGGQKGGGGGGQGGQKGG 165
Score = 31.6 bits (70), Expect = 0.43
Identities = 16/36 (44%), Positives = 18/36 (49%), Gaps = 2/36 (5%)
Query: 120 QKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
Q G GG G + GG G GGGG G + G GG
Sbjct: 232 QGGHKGGGGGGQ--GGGGHKGGGGGQGGHKGGGGGG 265
Score = 31.6 bits (70), Expect = 0.43
Identities = 16/36 (44%), Positives = 18/36 (49%), Gaps = 2/36 (5%)
Query: 120 QKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
Q G GG G G +G GGGGG G +G GG
Sbjct: 208 QGGHKGGGGGGGQGGHKGGGGGGG--QGGHKGGGGG 241
Score = 31.2 bits (69), Expect = 0.56
Identities = 16/39 (41%), Positives = 19/39 (48%), Gaps = 3/39 (7%)
Query: 120 QKGEYSGGSGSKPPGGRGRGG---GGGNSDGSEEGSTGG 155
Q G+ GG GG G+GG GGG G +G GG
Sbjct: 168 QGGQKGGGGQGGQKGGGGQGGQKGGGGRGQGGMKGGGGG 206
Score = 30.8 bits (68), Expect = 0.74
Identities = 16/36 (44%), Positives = 18/36 (49%), Gaps = 2/36 (5%)
Query: 120 QKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
Q G GG G + GG GGGGG G + G GG
Sbjct: 197 QGGMKGGGGGGQ--GGHKGGGGGGGQGGHKGGGGGG 230
Score = 30.0 bits (66), Expect = 1.3
Identities = 14/35 (40%), Positives = 17/35 (48%)
Query: 120 QKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTG 154
QKG GG G + GG G GG G ++G G
Sbjct: 151 QKGGGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGG 185
Score = 30.0 bits (66), Expect = 1.3
Identities = 16/38 (42%), Positives = 18/38 (47%), Gaps = 4/38 (10%)
Query: 122 GEYSGGSGSKPPGGRGRGG----GGGNSDGSEEGSTGG 155
G G G K GG G+GG GGG G +G GG
Sbjct: 227 GGGGGQGGHKGGGGGGQGGGGHKGGGGGQGGHKGGGGG 264
Score = 29.6 bits (65), Expect = 1.6
Identities = 16/35 (45%), Positives = 17/35 (47%), Gaps = 1/35 (2%)
Query: 121 KGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
KG GG G GG G GG GG+ G G GG
Sbjct: 201 KGGGGGGQGGHKGGGGG-GGQGGHKGGGGGGGQGG 234
Score = 29.6 bits (65), Expect = 1.6
Identities = 15/35 (42%), Positives = 16/35 (44%)
Query: 121 KGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
KG GG GG G GG GG+ G G GG
Sbjct: 212 KGGGGGGGQGGHKGGGGGGGQGGHKGGGGGGQGGG 246
Score = 29.6 bits (65), Expect = 1.6
Identities = 14/35 (40%), Positives = 16/35 (45%)
Query: 120 QKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTG 154
Q G GG G G +G GGGG G + G G
Sbjct: 220 QGGHKGGGGGGGQGGHKGGGGGGQGGGGHKGGGGG 254
Score = 29.3 bits (64), Expect = 2.1
Identities = 14/42 (33%), Positives = 18/42 (42%)
Query: 121 KGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMSDETAQ 162
+G GG G G +G GG GG G +G G + Q
Sbjct: 148 QGGQKGGGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGQGGQ 189
Score = 28.9 bits (63), Expect = 2.8
Identities = 14/34 (41%), Positives = 15/34 (43%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G + GG G GG GGG G G G GG
Sbjct: 264 GGHVGGGGRGGGGGGRGGGGSGGGGGGGSGRGGG 297
Score = 28.5 bits (62), Expect = 3.7
Identities = 16/35 (45%), Positives = 16/35 (45%), Gaps = 2/35 (5%)
Query: 121 KGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
KG G G K GG G GGGG G G GG
Sbjct: 249 KGGGGGQGGHKGGGGGGHVGGGGRGGGG--GGRGG 281
Score = 28.1 bits (61), Expect = 4.8
Identities = 16/43 (37%), Positives = 19/43 (43%), Gaps = 2/43 (4%)
Query: 120 QKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMSDETAQ 162
QKG GG G + GG G G GG G +G G + Q
Sbjct: 140 QKGGGGGGQGGQKGGGGG--GQGGQKGGGGQGGQKGGGGQGGQ 180
Score = 28.1 bits (61), Expect = 4.8
Identities = 14/35 (40%), Positives = 15/35 (42%)
Query: 120 QKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTG 154
Q G+ GG G G G GG GG G G G
Sbjct: 116 QGGQKGGGGGQGGQKGGGGGGQGGQKGGGGGGQGG 150
Score = 28.1 bits (61), Expect = 4.8
Identities = 14/36 (38%), Positives = 18/36 (49%), Gaps = 1/36 (2%)
Query: 120 QKGEYSGGSGSKPPGGRGRGGG-GGNSDGSEEGSTG 154
Q G+ GG GG G+GG GG G ++G G
Sbjct: 159 QGGQKGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGG 194
Score = 28.1 bits (61), Expect = 4.8
Identities = 15/37 (40%), Positives = 18/37 (48%), Gaps = 2/37 (5%)
Query: 121 KGEYSGGSGSKPPGGRGRGGGG--GNSDGSEEGSTGG 155
+G GG G G +G GGGG G+ G G GG
Sbjct: 186 QGGQKGGGGRGQGGMKGGGGGGQGGHKGGGGGGGQGG 222
Score = 27.3 bits (59), Expect = 8.1
Identities = 14/40 (35%), Positives = 18/40 (45%), Gaps = 6/40 (15%)
Query: 121 KGEYSGGSGSKPPGGRGRGGG------GGNSDGSEEGSTG 154
+G GG G G +G GGG GG G ++G G
Sbjct: 137 QGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGQGGQKGGGG 176
Score = 27.3 bits (59), Expect = 8.1
Identities = 15/34 (44%), Positives = 19/34 (55%), Gaps = 2/34 (5%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G + GG G + G +G GGGGG+ G G GG
Sbjct: 246 GGHKGGGGGQG-GHKG-GGGGGHVGGGGRGGGGG 277
>At2g11005 putative protein
Length = 170
Score = 38.1 bits (87), Expect = 0.005
Identities = 19/37 (51%), Positives = 21/37 (56%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMSD 158
G GSGS GG G GGG +SDGS + S GG D
Sbjct: 22 GGSGDGSGSGDGGGSGDGGGSRDSDGSGDSSGGGSGD 58
Score = 34.3 bits (77), Expect = 0.067
Identities = 15/40 (37%), Positives = 19/40 (47%)
Query: 119 IQKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMSD 158
++K + G G GG G G G G+ DG G GG D
Sbjct: 5 VKKWWFGSGDGGGSGGGGGSGDGSGSGDGGGSGDGGGSRD 44
Score = 28.5 bits (62), Expect = 3.7
Identities = 18/38 (47%), Positives = 19/38 (49%), Gaps = 4/38 (10%)
Query: 125 SGGSGSKPPGGRGRGGG-GGNSDG---SEEGSTGGMSD 158
S GSG GG G GG G NSD S + S GG D
Sbjct: 45 SDGSGDSSGGGSGDSGGFGDNSDNNSVSSDSSGGGSRD 82
>At2g30560 putative glycine-rich protein
Length = 171
Score = 37.7 bits (86), Expect = 0.006
Identities = 16/30 (53%), Positives = 17/30 (56%)
Query: 126 GGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
GG G GG G+GGGGG S G G GG
Sbjct: 2 GGKGGSGSGGGGKGGGGGGSGGGRGGGGGG 31
Score = 34.7 bits (78), Expect = 0.051
Identities = 16/34 (47%), Positives = 17/34 (49%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G SGG G G G GGGGG G G +GG
Sbjct: 103 GGKSGGGGGGGKNGGGCGGGGGGKGGKSGGGSGG 136
Score = 33.5 bits (75), Expect = 0.11
Identities = 17/36 (47%), Positives = 18/36 (49%), Gaps = 1/36 (2%)
Query: 121 KGEYSGGSGSKPPGGRGRGGG-GGNSDGSEEGSTGG 155
KG G G K GG G GGG GG G +G GG
Sbjct: 4 KGGSGSGGGGKGGGGGGSGGGRGGGGGGGAKGGCGG 39
Score = 33.1 bits (74), Expect = 0.15
Identities = 16/34 (47%), Positives = 16/34 (47%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G GG G GGRG GGGGG G G G
Sbjct: 11 GGGKGGGGGGSGGGRGGGGGGGAKGGCGGGGKSG 44
Score = 32.3 bits (72), Expect = 0.25
Identities = 16/29 (55%), Positives = 18/29 (61%), Gaps = 3/29 (10%)
Query: 126 GGSGSKPPGGRGRGGGGGNSDGSEEGSTG 154
GGSG GG+G GGGGG S G G +G
Sbjct: 75 GGSGG---GGKGGGGGGGISGGGAGGKSG 100
Score = 32.3 bits (72), Expect = 0.25
Identities = 15/34 (44%), Positives = 17/34 (49%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G+ GSG GG G G GGG G G+ GG
Sbjct: 3 GKGGSGSGGGGKGGGGGGSGGGRGGGGGGGAKGG 36
Score = 31.6 bits (70), Expect = 0.43
Identities = 16/34 (47%), Positives = 17/34 (49%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G+ GG G GG GGGGG S GS GG
Sbjct: 104 GKSGGGGGGGKNGGGCGGGGGGKGGKSGGGSGGG 137
Score = 30.8 bits (68), Expect = 0.74
Identities = 16/36 (44%), Positives = 19/36 (52%), Gaps = 2/36 (5%)
Query: 122 GEYSGGSGSKPP--GGRGRGGGGGNSDGSEEGSTGG 155
G GG+G K GG+ GGGGG +G G GG
Sbjct: 89 GISGGGAGGKSGCGGGKSGGGGGGGKNGGGCGGGGG 124
Score = 30.0 bits (66), Expect = 1.3
Identities = 16/32 (50%), Positives = 16/32 (50%), Gaps = 1/32 (3%)
Query: 126 GGSGSKPPGGRGRGGGG-GNSDGSEEGSTGGM 156
GG G K GG G GGGG G G G G M
Sbjct: 109 GGGGGKNGGGCGGGGGGKGGKSGGGSGGGGYM 140
Score = 29.3 bits (64), Expect = 2.1
Identities = 17/39 (43%), Positives = 19/39 (48%), Gaps = 3/39 (7%)
Query: 122 GEYSGGSGSKPPGGRG---RGGGGGNSDGSEEGSTGGMS 157
G SG G K GG G GGG G G + G +GG S
Sbjct: 96 GGKSGCGGGKSGGGGGGGKNGGGCGGGGGGKGGKSGGGS 134
Score = 29.3 bits (64), Expect = 2.1
Identities = 15/29 (51%), Positives = 15/29 (51%), Gaps = 1/29 (3%)
Query: 130 SKPPGGRGRGG-GGGNSDGSEEGSTGGMS 157
S P GG G GG GGG G G GG S
Sbjct: 71 SDPKGGSGGGGKGGGGGGGISGGGAGGKS 99
Score = 28.9 bits (63), Expect = 2.8
Identities = 15/32 (46%), Positives = 16/32 (49%), Gaps = 1/32 (3%)
Query: 125 SGGSGSKPPGGRG-RGGGGGNSDGSEEGSTGG 155
SGG G GG G GGG G G G +GG
Sbjct: 77 SGGGGKGGGGGGGISGGGAGGKSGCGGGKSGG 108
Score = 28.5 bits (62), Expect = 3.7
Identities = 16/36 (44%), Positives = 18/36 (49%), Gaps = 2/36 (5%)
Query: 122 GEYSGGSGSKPPGGRGRGGG--GGNSDGSEEGSTGG 155
G+ GG GS G G GGG GG G + G GG
Sbjct: 13 GKGGGGGGSGGGRGGGGGGGAKGGCGGGGKSGGGGG 48
Score = 27.3 bits (59), Expect = 8.1
Identities = 15/36 (41%), Positives = 16/36 (43%), Gaps = 2/36 (5%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNS--DGSEEGSTGG 155
G GG G GG GG GG S G + G GG
Sbjct: 76 GSGGGGKGGGGGGGISGGGAGGKSGCGGGKSGGGGG 111
>At5g04290 glycine-rich protein
Length = 1493
Score = 37.0 bits (84), Expect = 0.010
Identities = 16/33 (48%), Positives = 18/33 (54%)
Query: 126 GGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMSD 158
GG G GG+G G GGG +GS G G SD
Sbjct: 398 GGKGEGSGGGKGEGSGGGKGEGSRGGKGEGSSD 430
Score = 28.1 bits (61), Expect = 4.8
Identities = 18/40 (45%), Positives = 19/40 (47%), Gaps = 8/40 (20%)
Query: 126 GGSGSKP-----PGGRGRG---GGGGNSDGSEEGSTGGMS 157
G GSKP GGRG G GGGG G + GG S
Sbjct: 1318 GDGGSKPWNEHSGGGRGFGERRGGGGFRGGRNQSGRGGRS 1357
Score = 27.7 bits (60), Expect = 6.2
Identities = 12/36 (33%), Positives = 19/36 (52%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMS 157
G+ GG G G + GGG G++ G + ++ G S
Sbjct: 1242 GKQDGGGGGSSWGKQDGGGGSGSAWGKQNETSNGSS 1277
Score = 27.3 bits (59), Expect = 8.1
Identities = 13/30 (43%), Positives = 16/30 (53%)
Query: 128 SGSKPPGGRGRGGGGGNSDGSEEGSTGGMS 157
SG G+ GGGGG+S G + GG S
Sbjct: 1284 SGGGSSWGKQDGGGGGSSWGKQNDGGGGSS 1313
Score = 27.3 bits (59), Expect = 8.1
Identities = 13/41 (31%), Positives = 19/41 (45%)
Query: 120 QKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMSDET 160
Q G G S K GG G G G + + GS+ G +++
Sbjct: 1244 QDGGGGGSSWGKQDGGGGSGSAWGKQNETSNGSSWGKQNDS 1284
>At4g13850 glycine-rich RNA-binding protein AtGRP2 - like
Length = 158
Score = 37.0 bits (84), Expect = 0.010
Identities = 17/33 (51%), Positives = 18/33 (54%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTG 154
G YSGG G GG G GGGGG G +G G
Sbjct: 125 GGYSGGGGGYGGGGGGYGGGGGGYGGGGDGGGG 157
Score = 31.6 bits (70), Expect = 0.43
Identities = 15/30 (50%), Positives = 15/30 (50%)
Query: 126 GGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
GG G GG G GGGGG G G GG
Sbjct: 122 GGGGGYSGGGGGYGGGGGGYGGGGGGYGGG 151
Score = 28.9 bits (63), Expect = 2.8
Identities = 16/35 (45%), Positives = 16/35 (45%), Gaps = 2/35 (5%)
Query: 124 YSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMSD 158
Y GG G GG GGGGG G G GG D
Sbjct: 121 YGGGGGYSGGGGGYGGGGGGYGGGG--GGYGGGGD 153
>At3g46270 putative protein
Length = 457
Score = 36.6 bits (83), Expect = 0.013
Identities = 20/53 (37%), Positives = 25/53 (46%), Gaps = 1/53 (1%)
Query: 126 GGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMSDETAQIVFATIGFMFVYIYVI 178
GGSG K GG GGGGG S G G T + + +G F ++VI
Sbjct: 360 GGSGGKSGGG-DNGGGGGQSGGGNNGGTNLKGGKKKNSLPLILGVTFASVFVI 411
>At1g75550 hypothetical protein
Length = 167
Score = 36.6 bits (83), Expect = 0.013
Identities = 18/36 (50%), Positives = 20/36 (55%)
Query: 120 QKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
+K Y+GGSGS G G GGGGG G G GG
Sbjct: 54 KKRSYNGGSGSYRWGWGGGGGGGGGGGGGGGGGGGG 89
Score = 32.0 bits (71), Expect = 0.33
Identities = 15/32 (46%), Positives = 16/32 (49%)
Query: 124 YSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
+ GG G GG G GGGGG G G GG
Sbjct: 69 WGGGGGGGGGGGGGGGGGGGGGGGWGWGGGGG 100
Score = 30.4 bits (67), Expect = 0.96
Identities = 15/30 (50%), Positives = 15/30 (50%)
Query: 126 GGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
GG G GG G GGGGG G G GG
Sbjct: 73 GGGGGGGGGGGGGGGGGGGGWGWGGGGGGG 102
Score = 30.0 bits (66), Expect = 1.3
Identities = 18/54 (33%), Positives = 21/54 (38%)
Query: 98 DNSEDSQRKSLEEAMKSLQEKIQKGEYSGGSGSKPPGGRGRGGGGGNSDGSEEG 151
+N E+S + K G Y G G GG G GGGGG G G
Sbjct: 39 NNDENSTTTGVVVRDKKRSYNGGSGSYRWGWGGGGGGGGGGGGGGGGGGGGGGG 92
Score = 29.3 bits (64), Expect = 2.1
Identities = 14/30 (46%), Positives = 14/30 (46%)
Query: 126 GGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
GG G GG G GGGGG G GG
Sbjct: 72 GGGGGGGGGGGGGGGGGGGGGWGWGGGGGG 101
Score = 28.9 bits (63), Expect = 2.8
Identities = 15/34 (44%), Positives = 15/34 (44%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G GG G GG G GGGGG G GG
Sbjct: 70 GGGGGGGGGGGGGGGGGGGGGGGWGWGGGGGGGG 103
Score = 28.9 bits (63), Expect = 2.8
Identities = 15/34 (44%), Positives = 16/34 (46%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G GG G GG G GGGGG + G GG
Sbjct: 79 GGGGGGGGGGGGGGWGWGGGGGGGGWYKWGCGGG 112
>At2g36120 unknown protein
Length = 255
Score = 36.2 bits (82), Expect = 0.018
Identities = 16/34 (47%), Positives = 19/34 (55%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G Y GG G+ GG G GG GG+ G G+ GG
Sbjct: 145 GGYGGGQGAGAGGGYGGGGAGGHGGGGGGGNGGG 178
Score = 34.3 bits (77), Expect = 0.067
Identities = 16/32 (50%), Positives = 18/32 (56%), Gaps = 3/32 (9%)
Query: 124 YSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
Y GG+G+ GG G GGGGG G G GG
Sbjct: 203 YGGGAGA---GGHGGGGGGGGGSGGGGGGGGG 231
Score = 34.3 bits (77), Expect = 0.067
Identities = 16/34 (47%), Positives = 16/34 (47%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G Y GG GG G G GGG GS EG G
Sbjct: 157 GGYGGGGAGGHGGGGGGGNGGGGGGGSGEGGAHG 190
Score = 33.1 bits (74), Expect = 0.15
Identities = 15/34 (44%), Positives = 18/34 (52%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G +GG G GG G GGGGG+ +G G G
Sbjct: 161 GGGAGGHGGGGGGGNGGGGGGGSGEGGAHGGGYG 194
Score = 33.1 bits (74), Expect = 0.15
Identities = 15/34 (44%), Positives = 19/34 (55%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G + GG+G GG G G GGG+ +G G GG
Sbjct: 86 GGHGGGAGGGGGGGPGGGYGGGSGEGGGAGYGGG 119
Score = 33.1 bits (74), Expect = 0.15
Identities = 24/63 (38%), Positives = 29/63 (45%), Gaps = 10/63 (15%)
Query: 103 SQRKSLEEAMKSLQEKIQKGEYSG-------GSGSKPPGGRGRGGG---GGNSDGSEEGS 152
S R++L + +S E G SG G G P GG G GGG GG + G EG
Sbjct: 20 SARRALLSSSESEAEVAAYGVNSGLSAGLGVGIGGGPGGGSGYGGGSGEGGGAGGHGEGH 79
Query: 153 TGG 155
GG
Sbjct: 80 IGG 82
Score = 32.3 bits (72), Expect = 0.25
Identities = 16/35 (45%), Positives = 17/35 (47%)
Query: 121 KGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
+G GG G GG G GGGGG G GS G
Sbjct: 77 EGHIGGGGGGGHGGGAGGGGGGGPGGGYGGGSGEG 111
Score = 32.3 bits (72), Expect = 0.25
Identities = 15/34 (44%), Positives = 17/34 (49%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G GG+G GG G GGGG E G+ GG
Sbjct: 158 GYGGGGAGGHGGGGGGGNGGGGGGGSGEGGAHGG 191
Score = 31.6 bits (70), Expect = 0.43
Identities = 17/37 (45%), Positives = 17/37 (45%), Gaps = 3/37 (8%)
Query: 122 GEYSGGSGSKPPGGRGRG---GGGGNSDGSEEGSTGG 155
G GG G P GG G G GGG G E G GG
Sbjct: 90 GGAGGGGGGGPGGGYGGGSGEGGGAGYGGGEAGGHGG 126
Score = 31.6 bits (70), Expect = 0.43
Identities = 15/34 (44%), Positives = 17/34 (49%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G Y GGSG G G G GG+ G G+ GG
Sbjct: 102 GGYGGGSGEGGGAGYGGGEAGGHGGGGGGGAGGG 135
Score = 31.6 bits (70), Expect = 0.43
Identities = 16/34 (47%), Positives = 17/34 (49%), Gaps = 2/34 (5%)
Query: 124 YSGGS--GSKPPGGRGRGGGGGNSDGSEEGSTGG 155
Y GG G GG G GGGGG G+ G GG
Sbjct: 116 YGGGEAGGHGGGGGGGAGGGGGGGGGAHGGGYGG 149
Score = 31.2 bits (69), Expect = 0.56
Identities = 14/30 (46%), Positives = 17/30 (56%)
Query: 126 GGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
GG+G G G GGGGG+ G+ G GG
Sbjct: 70 GGAGGHGEGHIGGGGGGGHGGGAGGGGGGG 99
Score = 31.2 bits (69), Expect = 0.56
Identities = 16/34 (47%), Positives = 16/34 (47%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G GG G GG G G GGG GS EG G
Sbjct: 82 GGGGGGHGGGAGGGGGGGPGGGYGGGSGEGGGAG 115
Score = 30.8 bits (68), Expect = 0.74
Identities = 15/35 (42%), Positives = 18/35 (50%)
Query: 121 KGEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
+G +GG G GG G GG GG + G G GG
Sbjct: 68 EGGGAGGHGEGHIGGGGGGGHGGGAGGGGGGGPGG 102
Score = 30.8 bits (68), Expect = 0.74
Identities = 15/34 (44%), Positives = 17/34 (49%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G + GG G G G GGGGG + S G GG
Sbjct: 210 GGHGGGGGGGGGSGGGGGGGGGYAAASGYGHGGG 243
Score = 30.8 bits (68), Expect = 0.74
Identities = 14/37 (37%), Positives = 18/37 (47%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMSD 158
G + GG G GG G G G G + G G+ GG +
Sbjct: 165 GGHGGGGGGGNGGGGGGGSGEGGAHGGGYGAGGGAGE 201
Score = 30.4 bits (67), Expect = 0.96
Identities = 15/34 (44%), Positives = 15/34 (44%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G GG G GG G GGG G G G GG
Sbjct: 94 GGGGGGPGGGYGGGSGEGGGAGYGGGEAGGHGGG 127
Score = 29.6 bits (65), Expect = 1.6
Identities = 14/34 (41%), Positives = 16/34 (46%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G + GG G G G G GGG + G G GG
Sbjct: 141 GAHGGGYGGGQGAGAGGGYGGGGAGGHGGGGGGG 174
Score = 29.6 bits (65), Expect = 1.6
Identities = 15/34 (44%), Positives = 16/34 (46%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G +GG G GG G GGGGG G S G
Sbjct: 206 GAGAGGHGGGGGGGGGSGGGGGGGGGYAAASGYG 239
Score = 29.6 bits (65), Expect = 1.6
Identities = 15/31 (48%), Positives = 16/31 (51%), Gaps = 1/31 (3%)
Query: 126 GGSGSKPPGGRGRGG-GGGNSDGSEEGSTGG 155
GG+G GG G GG GGG G G GG
Sbjct: 197 GGAGEGYGGGAGAGGHGGGGGGGGGSGGGGG 227
Score = 29.3 bits (64), Expect = 2.1
Identities = 16/36 (44%), Positives = 17/36 (46%), Gaps = 2/36 (5%)
Query: 122 GEYSGGSGSKPPGGRGRGG--GGGNSDGSEEGSTGG 155
G GG G GG G GG GGG G G+ GG
Sbjct: 123 GHGGGGGGGAGGGGGGGGGAHGGGYGGGQGAGAGGG 158
Score = 28.9 bits (63), Expect = 2.8
Identities = 15/30 (50%), Positives = 17/30 (56%), Gaps = 1/30 (3%)
Query: 127 GSGSKPPGGRGRGGG-GGNSDGSEEGSTGG 155
G GS GG G GGG GG+ +G G GG
Sbjct: 57 GGGSGYGGGSGEGGGAGGHGEGHIGGGGGG 86
Score = 28.5 bits (62), Expect = 3.7
Identities = 17/37 (45%), Positives = 17/37 (45%), Gaps = 5/37 (13%)
Query: 124 YSGGSGSKP-PGGRGRG----GGGGNSDGSEEGSTGG 155
Y GGSG GG G G GGGG G G GG
Sbjct: 62 YGGGSGEGGGAGGHGEGHIGGGGGGGHGGGAGGGGGG 98
Score = 28.1 bits (61), Expect = 4.8
Identities = 13/34 (38%), Positives = 15/34 (43%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G Y G G+ G G G GG G G +GG
Sbjct: 191 GGYGAGGGAGEGYGGGAGAGGHGGGGGGGGGSGG 224
Score = 28.1 bits (61), Expect = 4.8
Identities = 14/34 (41%), Positives = 15/34 (43%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
GE G G G G GGGGG + G G G
Sbjct: 119 GEAGGHGGGGGGGAGGGGGGGGGAHGGGYGGGQG 152
Score = 27.7 bits (60), Expect = 6.2
Identities = 14/34 (41%), Positives = 16/34 (46%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G +GG G G G G GGG G+ G GG
Sbjct: 129 GGGAGGGGGGGGGAHGGGYGGGQGAGAGGGYGGG 162
Score = 27.7 bits (60), Expect = 6.2
Identities = 14/36 (38%), Positives = 16/36 (43%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMS 157
G + GG G+ G G GGG G G GG S
Sbjct: 187 GAHGGGYGAGGGAGEGYGGGAGAGGHGGGGGGGGGS 222
Score = 27.7 bits (60), Expect = 6.2
Identities = 12/27 (44%), Positives = 13/27 (47%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGS 148
G GG G G G GGG G +GS
Sbjct: 224 GGGGGGGGYAAASGYGHGGGAGGGEGS 250
Score = 27.7 bits (60), Expect = 6.2
Identities = 15/34 (44%), Positives = 16/34 (46%), Gaps = 1/34 (2%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G GG G GG GGG G G+ EG GG
Sbjct: 174 GNGGGGGGGSGEGG-AHGGGYGAGGGAGEGYGGG 206
Score = 27.3 bits (59), Expect = 8.1
Identities = 14/34 (41%), Positives = 16/34 (46%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G GGSG G G G GGG +G G+ G
Sbjct: 177 GGGGGGSGEGGAHGGGYGAGGGAGEGYGGGAGAG 210
Score = 27.3 bits (59), Expect = 8.1
Identities = 17/34 (50%), Positives = 17/34 (50%), Gaps = 4/34 (11%)
Query: 126 GGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMSDE 159
GG G GG G GGGGG G G GG S E
Sbjct: 81 GGGGGGGHGG-GAGGGGGGGPG---GGYGGGSGE 110
Score = 27.3 bits (59), Expect = 8.1
Identities = 14/34 (41%), Positives = 14/34 (41%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G GG G GG G G G G G G GG
Sbjct: 133 GGGGGGGGGAHGGGYGGGQGAGAGGGYGGGGAGG 166
>At5g07540 glycine-rich protein atGRP-6
Length = 244
Score = 35.8 bits (81), Expect = 0.023
Identities = 17/38 (44%), Positives = 21/38 (54%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMSDE 159
G SGG+ PGG GG GG S G G++GG S +
Sbjct: 169 GGASGGASGGGPGGASGGGPGGASGGGPGGASGGASGD 206
Score = 34.3 bits (77), Expect = 0.067
Identities = 15/36 (41%), Positives = 20/36 (54%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMS 157
G+ GG+ PGG G GG S G+ G++GG S
Sbjct: 141 GDKPGGASGGGPGGASGGASGGASGGASGGASGGAS 176
Score = 33.9 bits (76), Expect = 0.087
Identities = 17/38 (44%), Positives = 22/38 (57%), Gaps = 2/38 (5%)
Query: 122 GEYSGGS--GSKPPGGRGRGGGGGNSDGSEEGSTGGMS 157
G+ GG+ G PGG GG GG S G+ G++GG S
Sbjct: 131 GDKPGGASGGGDKPGGASGGGPGGASGGASGGASGGAS 168
Score = 31.6 bits (70), Expect = 0.43
Identities = 15/34 (44%), Positives = 18/34 (52%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G SGG+ GG GG GG S G G++GG
Sbjct: 161 GGASGGASGGASGGASGGGPGGASGGGPGGASGG 194
Score = 30.0 bits (66), Expect = 1.3
Identities = 14/34 (41%), Positives = 17/34 (49%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G SGG+ GG G GG S G G++GG
Sbjct: 153 GGASGGASGGASGGASGGASGGASGGGPGGASGG 186
Score = 29.6 bits (65), Expect = 1.6
Identities = 14/36 (38%), Positives = 16/36 (43%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMS 157
G SGG+ GG G GG G+ G GG S
Sbjct: 157 GGASGGASGGASGGASGGASGGGPGGASGGGPGGAS 192
Score = 29.6 bits (65), Expect = 1.6
Identities = 14/34 (41%), Positives = 17/34 (49%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGG 155
G SGG GG G GG S G+ G++GG
Sbjct: 145 GGASGGGPGGASGGASGGASGGASGGASGGASGG 178
Score = 28.5 bits (62), Expect = 3.7
Identities = 13/32 (40%), Positives = 15/32 (46%)
Query: 126 GGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMS 157
GG+ GG G GG S G+ G GG S
Sbjct: 153 GGASGGASGGASGGASGGASGGASGGGPGGAS 184
Score = 28.1 bits (61), Expect = 4.8
Identities = 16/36 (44%), Positives = 20/36 (55%), Gaps = 3/36 (8%)
Query: 125 SGGSGSKPPG--GRGRGGGGGN-SDGSEEGSTGGMS 157
+ G G KP G G G GG G S G+ G++GG S
Sbjct: 137 ASGGGDKPGGASGGGPGGASGGASGGASGGASGGAS 172
Score = 28.1 bits (61), Expect = 4.8
Identities = 14/36 (38%), Positives = 15/36 (40%)
Query: 122 GEYSGGSGSKPPGGRGRGGGGGNSDGSEEGSTGGMS 157
GE SG G G GGG G+ G GG S
Sbjct: 121 GEMSGAGGPSGDKPGGASGGGDKPGGASGGGPGGAS 156
Score = 27.7 bits (60), Expect = 6.2
Identities = 16/42 (38%), Positives = 16/42 (38%), Gaps = 2/42 (4%)
Query: 122 GEYSGGSGSKPPG--GRGRGGGGGNSDGSEEGSTGGMSDETA 161
G G SG P G G G GG G G G G E A
Sbjct: 170 GASGGASGGGPGGASGGGPGGASGGGPGGASGGASGDKPEGA 211
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.133 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,128,251
Number of Sequences: 26719
Number of extensions: 282810
Number of successful extensions: 5526
Number of sequences better than 10.0: 217
Number of HSP's better than 10.0 without gapping: 163
Number of HSP's successfully gapped in prelim test: 60
Number of HSP's that attempted gapping in prelim test: 1637
Number of HSP's gapped (non-prelim): 2076
length of query: 255
length of database: 11,318,596
effective HSP length: 97
effective length of query: 158
effective length of database: 8,726,853
effective search space: 1378842774
effective search space used: 1378842774
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC122724.2


