

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122723.7 + phase: 0 /pseudo
(1982 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
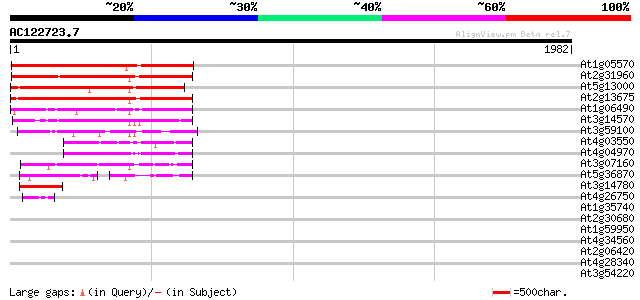
Score E
Sequences producing significant alignments: (bits) Value
At1g05570 putative glucan synthase 977 0.0
At2g31960 glucan synthase like protein 961 0.0
At5g13000 callose synthase catalytic subunit -like protein 956 0.0
At2g13675 callose synthase (1,3-beta-glucan synthase) like protein 633 0.0
At1g06490 glucan synthase, putative 511 e-144
At3g14570 hypothetical protein 504 e-142
At3g59100 putative protein 432 e-121
At4g03550 putative glucan synthase component 285 2e-76
At4g04970 276 9e-74
At3g07160 putative glucan synthase 270 6e-72
At5g36870 putative glucan synthase 205 2e-52
At3g14780 unknown protein 112 3e-24
At4g26750 unknown protein 48 7e-05
At1g35740 hypothetical protein 42 0.005
At2g30680 hypothetical protein 39 0.023
At1g59950 hypothetical protein 35 0.44
At4g34560 unknown protein 35 0.57
At2g06420 unknown protein 32 2.8
At4g28340 hypothetical protein 32 3.7
At3g54220 SCARECROW1 31 6.3
>At1g05570 putative glucan synthase
Length = 1858
Score = 977 bits (2525), Expect = 0.0
Identities = 492/651 (75%), Positives = 551/651 (84%), Gaps = 21/651 (3%)
Query: 7 RGPTPSEPPPRRLVRTQTAGNLGESIFDSEVVPSSLVEIAPILRVANEVEKTHPRVAYLC 66
R P P PP RR++RTQT G+LGE++ DSEVVPSSLVEIAPILRVANEVE ++PRVAYLC
Sbjct: 5 REPDPP-PPQRRILRTQTVGSLGEAMLDSEVVPSSLVEIAPILRVANEVEASNPRVAYLC 63
Query: 67 RFYAFEKAHRLDPTSSGRGVRQFKTALLQRLERENDPTLKGRVKKSDAREMQSFYQHYYK 126
RFYAFEKAHRLDPTSSGRGVRQFKTALLQRLEREN+ TL GR +KSDAREMQSFYQHYYK
Sbjct: 64 RFYAFEKAHRLDPTSSGRGVRQFKTALLQRLERENETTLAGR-QKSDAREMQSFYQHYYK 122
Query: 127 KYIQALQNAADKADRAQLTKAYQTANVLFEVLKAVNMTQSMEVDREILETQDKVAEKTEI 186
KYIQAL NAADKADRAQLTKAYQTA VLFEVLKAVN T+ +EV EILET +KV EKT+I
Sbjct: 123 KYIQALLNAADKADRAQLTKAYQTAAVLFEVLKAVNQTEDVEVADEILETHNKVEEKTQI 182
Query: 187 LVPFNILPLDPDSANQAIMKFPEIQAAVYALRNTRGLPWPNDYKKKKDEDILDWLGSMFG 246
VP+NILPLDPDS NQAIM+ PEIQAAV ALRNTRGLPW +KKK DEDILDWL SMFG
Sbjct: 183 YVPYNILPLDPDSQNQAIMRLPEIQAAVAALRNTRGLPWTAGHKKKLDEDILDWLQSMFG 242
Query: 247 FQKHNVANQREHLILLLANVHIRQFPNPDQQPKLDECALTEVMKKLFKNYKKWCKYLDRK 306
FQK NV NQREHLILLLANVHIRQFP PDQQPKLD+ ALT VMKKLF+NYKKWCKYL RK
Sbjct: 243 FQKDNVLNQREHLILLLANVHIRQFPKPDQQPKLDDRALTIVMKKLFRNYKKWCKYLGRK 302
Query: 307 SSLWLPTIQQEVQQRKLLYMGLYLLIWGEAANLRFMPECLCYIYHHMAFELYGMLAGNVS 366
SSLWLPTIQQEVQQRKLLYMGLYLLIWGEAANLRFMPECLCYIYHHMAFELYGMLAG+VS
Sbjct: 303 SSLWLPTIQQEVQQRKLLYMGLYLLIWGEAANLRFMPECLCYIYHHMAFELYGMLAGSVS 362
Query: 367 PMTGENIKPAYGGEDEAFLRKVVTPIYNVIAELKKAKGEGQSIH---------NGGTMMI 417
PMTGE++KPAYGGEDEAFL+KVVTPIY I++ K G+S H N I
Sbjct: 363 PMTGEHVKPAYGGEDEAFLQKVVTPIYQTISKEAKRSRGGKSKHSVWRNYDDLNEYFWSI 422
Query: 418 *MNISGWPMRADADFFCLPAERVVFDKSNPCCNSNA*DDKPPNRDGWFGKVNFVEIRSFW 477
GWPMRADADFFC AE + ++S NS D W GKVNFVEIRSFW
Sbjct: 423 RCFRLGWPMRADADFFCQTAEELRLERSEIKSNSG---------DRWMGKVNFVEIRSFW 473
Query: 478 HLFRSFDRMWSFFILCLQAMIIVAWNGSGDPTVIFHGDVFKKVLSVFITAAILKFGQAVL 537
H+FRSFDR+WSF+ILCLQAMI++AWNGSG+ + IF GDVF KVLSVFITAAILK QAVL
Sbjct: 474 HIFRSFDRLWSFYILCLQAMIVIAWNGSGELSAIFQGDVFLKVLSVFITAAILKLAQAVL 533
Query: 538 GVILSWKARRSMSLYVKLRYILKVISAAAWVILLSVTYAYTWDNPPGFAETIKSWFGSNS 597
+ LSWKAR SMSLYVKLRY++KV +AA WV++++VTYAY+W N GF++TIK+WFG +S
Sbjct: 534 DIALSWKARHSMSLYVKLRYVMKVGAAAVWVVVMAVTYAYSWKNASGFSQTIKNWFGGHS 593
Query: 598 -SAPSLFIVAVVVYLSPNMLAAIFFMFPFIRRYLERSNYRIVMLMMWWSQI 647
++PSLFIVA+++YLSPNML+A+ F+FPFIRRYLERS+Y+I+MLMMWWSQI
Sbjct: 594 HNSPSLFIVAILIYLSPNMLSALLFLFPFIRRYLERSDYKIMMLMMWWSQI 644
>At2g31960 glucan synthase like protein
Length = 1956
Score = 961 bits (2485), Expect = 0.0
Identities = 482/650 (74%), Positives = 544/650 (83%), Gaps = 24/650 (3%)
Query: 7 RGPTPSEPPPRRLVRTQTAGNLGESIFDSEVVPSSLVEIAPILRVANEVEKTHPRVAYLC 66
+GP P PP RR++RTQTAGNLGE++ DSEVVPSSLVEIAPILRVANEVE ++PRVAYLC
Sbjct: 5 KGPDPP-PPQRRILRTQTAGNLGEAMLDSEVVPSSLVEIAPILRVANEVEASNPRVAYLC 63
Query: 67 RFYAFEKAHRLDPTSSGRGVRQFKTALLQRLERENDPTLKGRVKKSDAREMQSFYQHYYK 126
RFYAFEKAHRLDPTSSGRGVRQFKTALLQRLEREN+ TL GR +KSDAREMQSFYQHYYK
Sbjct: 64 RFYAFEKAHRLDPTSSGRGVRQFKTALLQRLERENETTLAGR-QKSDAREMQSFYQHYYK 122
Query: 127 KYIQALQNAADKADRAQLTKAYQTANVLFEVLKAVNMTQSMEVDREILETQDKVAEKTEI 186
KYIQALQNAADKADRAQLTKAYQTA VLFEVLKAVN T+ +EV E KV EK++I
Sbjct: 123 KYIQALQNAADKADRAQLTKAYQTAAVLFEVLKAVNQTEDVEVADE---AHTKVEEKSQI 179
Query: 187 LVPFNILPLDPDSANQAIMKFPEIQAAVYALRNTRGLPWPNDYKKKKDEDILDWLGSMFG 246
VP+NILPLDPDS NQAIM+FPEIQA V ALRNTRGLPWP +KKK DED+LDWL +MFG
Sbjct: 180 YVPYNILPLDPDSQNQAIMRFPEIQATVSALRNTRGLPWPAGHKKKLDEDMLDWLQTMFG 239
Query: 247 FQKHNVANQREHLILLLANVHIRQFPNPDQQPKLDECALTEVMKKLFKNYKKWCKYLDRK 306
FQK NV+NQREHLILLLANVHIRQFP P+QQP+LD+ ALT VMKKLFKNYKKWCKYL RK
Sbjct: 240 FQKDNVSNQREHLILLLANVHIRQFPRPEQQPRLDDRALTIVMKKLFKNYKKWCKYLGRK 299
Query: 307 SSLWLPTIQQEVQQRKLLYMGLYLLIWGEAANLRFMPECLCYIYHHMAFELYGMLAGNVS 366
SSLWLPTIQQEVQQRKLLYMGLYLLIWGEAANLRF+PECLCYIYHHMAFELYGMLAG+VS
Sbjct: 300 SSLWLPTIQQEVQQRKLLYMGLYLLIWGEAANLRFLPECLCYIYHHMAFELYGMLAGSVS 359
Query: 367 PMTGENIKPAYGGEDEAFLRKVVTPIYNVIAELKKAKGEGQSIHNGGTMMI*MN------ 420
PMTGE++KPAYGGEDEAFL+KVVTPIY IA+ K G+S H+ +N
Sbjct: 360 PMTGEHVKPAYGGEDEAFLQKVVTPIYKTIAKEAKRSRGGKSKHSEWRNYDDLNEYFWSI 419
Query: 421 ---ISGWPMRADADFFCLPAERVVFDKSNPCCNSNA*DDKPPNRDGWFGKVNFVEIRSFW 477
GWPMRADADFFC AE + D+S ++KP D W GKVNFVEIRSFW
Sbjct: 420 RCFRLGWPMRADADFFCQTAEELRLDRS---------ENKPKTGDRWMGKVNFVEIRSFW 470
Query: 478 HLFRSFDRMWSFFILCLQAMIIVAWNGSGDPTVIFHGDVFKKVLSVFITAAILKFGQAVL 537
H+FRSFDRMWSF+IL LQAMII+AWNGSG + IF GDVF KVLS+FITAAILK QAVL
Sbjct: 471 HIFRSFDRMWSFYILSLQAMIIIAWNGSGKLSGIFQGDVFLKVLSIFITAAILKLAQAVL 530
Query: 538 GVILSWKARRSMSLYVKLRYILKVISAAAWVILLSVTYAYTWDNPPGFAETIKSWFGSN- 596
+ LSWK+R SMS +VKLR+I K ++AA WV+L+ +TYAY+W P GFAETIK+WFG +
Sbjct: 531 DIALSWKSRHSMSFHVKLRFIFKAVAAAIWVVLMPLTYAYSWKTPSGFAETIKNWFGGHQ 590
Query: 597 SSAPSLFIVAVVVYLSPNMLAAIFFMFPFIRRYLERSNYRIVMLMMWWSQ 646
+S+PS FI+ +++YLSPNML+ + F FPFIRRYLERS+Y+IVMLMMWWSQ
Sbjct: 591 NSSPSFFIIVILIYLSPNMLSTLLFAFPFIRRYLERSDYKIVMLMMWWSQ 640
>At5g13000 callose synthase catalytic subunit -like protein
Length = 1963
Score = 956 bits (2472), Expect = 0.0
Identities = 492/639 (76%), Positives = 539/639 (83%), Gaps = 33/639 (5%)
Query: 3 SSSSRGPT--PSEPPPRRLVRTQTAGNLGESIFDSEVVPSSLVEIAPILRVANEVEKTHP 60
S++ GP PS+P RR++RTQTAGNLGES FDSEVVPSSLVEIAPILRVANEVE ++P
Sbjct: 2 SATRGGPDQGPSQPQQRRIIRTQTAGNLGES-FDSEVVPSSLVEIAPILRVANEVESSNP 60
Query: 61 RVAYLCRFYAFEKAHRLDPTSSGRGVRQFKTALLQRLERENDPTLKGRVKKSDAREMQSF 120
RVAYLCRFYAFEKAHRLDPTSSGRGVRQFKTALLQRLERE+DPTL GRVKKSDAREMQSF
Sbjct: 61 RVAYLCRFYAFEKAHRLDPTSSGRGVRQFKTALLQRLEREHDPTLMGRVKKSDAREMQSF 120
Query: 121 YQHYYKKYIQALQNAADKADRAQLTKAYQTANVLFEVLKAVNMTQSMEVDREILETQDKV 180
YQHYYKKYIQAL NAADKADRAQLTKAYQTANVLFEVLKAVN+TQS+EVDREILE QDKV
Sbjct: 121 YQHYYKKYIQALHNAADKADRAQLTKAYQTANVLFEVLKAVNLTQSIEVDREILEAQDKV 180
Query: 181 AEKTEILVPFNILPLDPDSANQAIMKFPEIQAAVYALRNTRGLPWPNDYKKKKDEDILDW 240
AEKT++ VP+NILPLDPDSANQAIM++PEIQAAV ALRNTRGLPWP +KKKKDED+LDW
Sbjct: 181 AEKTQLYVPYNILPLDPDSANQAIMRYPEIQAAVLALRNTRGLPWPEGHKKKKDEDMLDW 240
Query: 241 LGSMFGFQKHNVANQREHLILLLANVHIRQFPNPDQQPK------------LDECALTEV 288
L MFGFQK NVANQREHLILLLANVHIRQFP PDQQPK LD+ ALTEV
Sbjct: 241 LQEMFGFQKDNVANQREHLILLLANVHIRQFPKPDQQPKFILSFVLIVPSQLDDQALTEV 300
Query: 289 MKKLFKNYKKWCKYLDRKSSLWLPTIQQEVQQRKLLYMGLYLLIWGEAANLRFMPECLCY 348
MKKLFKNYKKWCKYL RKSSLWLPTIQQE+QQRKLLYM LYLLIWGEAANLRFMPECLCY
Sbjct: 301 MKKLFKNYKKWCKYLGRKSSLWLPTIQQEMQQRKLLYMALYLLIWGEAANLRFMPECLCY 360
Query: 349 IYHHMAFELYGMLAGNVSPMTGENIKPAYGGEDEAFLRKVVTPIYNVIAELKKAKGEGQS 408
IYHHMAFELYGMLAGNVSPMTGEN+KPAYGGE++AFLRKVVTPIY VI + +G+S
Sbjct: 361 IYHHMAFELYGMLAGNVSPMTGENVKPAYGGEEDAFLRKVVTPIYEVIQMEAQRSKKGKS 420
Query: 409 IHNGGTMMI*MN---------ISGWPMRADADFFCLPAERVVFDKSNPCCNSNA*DDKP- 458
H+ +N GWPMRADADFFCLP + P + + KP
Sbjct: 421 KHSQWRNYDDLNEYFWSVDCFRLGWPMRADADFFCLPV-------AVPNTEKDGDNSKPI 473
Query: 459 PNRDGWFGKVNFVEIRSFWHLFRSFDRMWSFFILCLQAMIIVAWNGSGDPTVIFHGDVFK 518
RD W GKVNFVEIRSFWH+FRSFDRMWSF+ILCLQAMII+AW+G G P+ +F DVFK
Sbjct: 474 VARDRWVGKVNFVEIRSFWHVFRSFDRMWSFYILCLQAMIIMAWDG-GQPSSVFGADVFK 532
Query: 519 KVLSVFITAAILKFGQAVLGVILSWKARRSMSLYVKLRYILKVISAAAWVILLSVTYAYT 578
KVLSVFITAAI+K GQAVL VIL++KA +SM+L+VKLRYILKV SAAAWVI+L VTYAY+
Sbjct: 533 KVLSVFITAAIMKLGQAVLDVILNFKAHQSMTLHVKLRYILKVFSAAAWVIILPVTYAYS 592
Query: 579 WDNPPGFAETIKSWFGSNSSAPSLFIVAVVVYLSPNMLA 617
W +PP FA TIKSWFGS +PSLFI+AVV YLSPNMLA
Sbjct: 593 WKDPPAFARTIKSWFGSAMHSPSLFIIAVVSYLSPNMLA 631
>At2g13675 callose synthase (1,3-beta-glucan synthase) like protein
Length = 1939
Score = 633 bits (1632), Expect = 0.0
Identities = 340/659 (51%), Positives = 437/659 (65%), Gaps = 32/659 (4%)
Query: 2 SSSSSRGPTPSEPPPRRLVRTQTAGNLGESIFDSEVVPSSLVEIAPILRVANEVEKTHPR 61
S+S GP P R T + +FD EVVP+SL IAPILRVA E+E PR
Sbjct: 5 STSHDSGPQGLMRRPSRSAAT----TVSIEVFDHEVVPASLGTIAPILRVAAEIEHERPR 60
Query: 62 VAYLCRFYAFEKAHRLDPTSSGRGVRQFKTALLQRLERENDPTLKGRVKKSDAREMQSFY 121
VAYLCRFYAFEKAHRLDP+S GRGVRQFKT L QRLER+N +L RVKK+D RE++SFY
Sbjct: 61 VAYLCRFYAFEKAHRLDPSSGGRGVRQFKTLLFQRLERDNASSLASRVKKTDGREVESFY 120
Query: 122 QHYYKKYIQALQNAADKADRAQLTKAYQTANVLFEVLKAVNMTQSME-VDREILETQDKV 180
Q YY+ Y++AL D+ADRAQL KAYQTA VLFEVL AVN ++ +E V EI+ V
Sbjct: 121 QQYYEHYVRALDQG-DQADRAQLGKAYQTAGVLFEVLMAVNKSEKVEAVAPEIIAAARDV 179
Query: 181 AEKTEILVPFNILPLDPDSANQAIMKFPEIQAAVYALRNTRGLPWPNDY----KKKKDED 236
EK EI P+NILPLD A+Q++M+ E++AAV AL NTRGL WP+ + KK + D
Sbjct: 180 QEKNEIYAPYNILPLDSAGASQSVMQLEEVKAAVAALGNTRGLNWPSGFEQHRKKTGNLD 239
Query: 237 ILDWLGSMFGFQKHNVANQREHLILLLANVHIRQFPNPDQQPKLDECALTEVMKKLFKNY 296
+LDWL +MFGFQ +NV NQREHL+ L A+ HIR P P+ KLD+ A+ VM KLFKNY
Sbjct: 240 LLDWLRAMFGFQANNVRNQREHLVCLFADNHIRLTPKPEPLNKLDDRAVDTVMSKLFKNY 299
Query: 297 KKWCKYLDRKSSLWLPTIQQEVQQRKLLYMGLYLLIWGEAANLRFMPECLCYIYHHMAFE 356
K WCK+L RK SL LP Q++QQRK+LYMGLYLLIWGEAAN+RFMPECLCYI+H+MA+E
Sbjct: 300 KNWCKFLGRKHSLRLPQAAQDIQQRKILYMGLYLLIWGEAANIRFMPECLCYIFHNMAYE 359
Query: 357 LYGMLAGNVSPMTGENIKPAYGGEDEAFLRKVVTPIYNVIAELKKAKGEGQSIHNGGTMM 416
L+G+LAGNVS +TGENIKP+YGG+DEAFLRKV+TPIY V+ G++ H+ +
Sbjct: 360 LHGLLAGNVSIVTGENIKPSYGGDDEAFLRKVITPIYRVVQTEANKNANGKAAHSDWSNY 419
Query: 417 I*MN---------ISGWPMRADADFFCLPAERVVFDKSNPCCNSNA*DDKPPNRDGWFGK 467
+N GWPMR D D F + K + + G GK
Sbjct: 420 DDLNEYFWTPDCFSLGWPMRDDGDLFKSTRDTTQGKKGS------------FRKAGRTGK 467
Query: 468 VNFVEIRSFWHLFRSFDRMWSFFILCLQAMIIVAWNGSGDPTVIFHGDVFKKVLSVFITA 527
NF E R+FWH++ SFDR+W+F++L LQAMII+A+ + I DV + S+FITA
Sbjct: 468 SNFTETRTFWHIYHSFDRLWTFYLLALQAMIILAFE-RVELREILRKDVLYALSSIFITA 526
Query: 528 AILKFGQAVLGVILSWKARRSMSLYVKLRYILKVISAAAWVILLSVTYAYTWDNPPGFAE 587
A L+F Q+VL VIL++ LR ILK++ + AW ++L + YA + PG +
Sbjct: 527 AFLRFLQSVLDVILNFPGFHRWKFTDVLRNILKIVVSLAWCVVLPLCYAQSVSFAPGKLK 586
Query: 588 TIKSWFGSNSSAPSLFIVAVVVYLSPNMLAAIFFMFPFIRRYLERSNYRIVMLMMWWSQ 646
S+ P L+I+AV +YL PN+LAAI F+FP +RR++E S++ I L++WWSQ
Sbjct: 587 QWLSFLPQVKGVPPLYIMAVALYLLPNVLAAIMFIFPMLRRWIENSDWHIFRLLLWWSQ 645
>At1g06490 glucan synthase, putative
Length = 1933
Score = 511 bits (1317), Expect = e-144
Identities = 302/680 (44%), Positives = 410/680 (59%), Gaps = 53/680 (7%)
Query: 1 MSSSSSRGPTPSEPPP---------RRLVRTQTA----GNLGESIFDSEVVPSSLVEIAP 47
M+S+SS G PP R++ R T N E DSE+VPSSL IAP
Sbjct: 1 MASTSSGGRGEDGRPPQMQPVRSMSRKMTRAGTMMIEHPNEDERPIDSELVPSSLASIAP 60
Query: 48 ILRVANEVEKTHPRVAYLCRFYAFEKAHRLDPTSSGRGVRQFKTALLQRLERENDPTLKG 107
ILRVAN++++ + RVAYLCRF+AFEKAHR+DPTSSGRGVRQFKT LL +LE E + T +
Sbjct: 61 ILRVANDIDQDNARVAYLCRFHAFEKAHRMDPTSSGRGVRQFKTYLLHKLEEEEEIT-EH 119
Query: 108 RVKKSDAREMQSFYQHYYKKYIQALQNAADKADRAQLTKAYQTANVLFEVLKAVNMTQSM 167
+ KSD RE+Q +YQ +Y+ IQ + K ++ K YQ A VL++VLK V
Sbjct: 120 MLAKSDPREIQLYYQTFYENNIQ---DGEGKKTPEEMAKLYQIATVLYDVLKTV--VPQA 174
Query: 168 EVDREILETQDKVAEKTEILVPFNILPLDPDSANQAIMKFPEIQAAVYALRNTRGLPWPN 227
+D + L +V K E +NILPL A A+M+ PEI+AA+ A+ N LP P
Sbjct: 175 RIDDKTLRYAKEVERKKEQYEHYNILPLYALGAKTAVMELPEIKAAILAVCNVDNLPRPR 234
Query: 228 DYKKKKD------------EDILDWLGSMFGFQKHNVANQREHLILLLANVHIRQFPNPD 275
+ + DIL+WL +FGFQ+ NVANQREHLILLLAN+ +R+ + +
Sbjct: 235 FHSASANLDEVDRERGRSFNDILEWLALVFGFQRGNVANQREHLILLLANIDVRK-RDLE 293
Query: 276 QQPKLDECALTEVMKKLFKNYKKWCKYLDRKSSLWLPTIQQEVQQRKLLYMGLYLLIWGE 335
++ + ++M+K FKNY WCKYL S L P + QQ LLY+GLYLLIWGE
Sbjct: 294 NYVEIKPSTVRKLMEKYFKNYNSWCKYLRCDSYLRFPA-GCDKQQLSLLYIGLYLLIWGE 352
Query: 336 AANLRFMPECLCYIYHHMAFELYGMLAGNVSPMTGENIKPAYGGEDEAFLRKVVTPIYNV 395
A+N+RFMPECLCYI+H+MA E++G+L GNV P+TG+ + A ++EAFLR V+TPIY V
Sbjct: 353 ASNVRFMPECLCYIFHNMANEVHGILFGNVYPVTGDTYE-AGAPDEEAFLRNVITPIYQV 411
Query: 396 IAELKKAKGEGQSIHNGGTMMI*MNIS---------GWPMRADADFFCLPAERVVFDKSN 446
+ + + G++ H+ +N WPM ADFF E + N
Sbjct: 412 LRKEVRRNKNGKASHSKWRNYDDLNEYFWDKRCFRLKWPMNFKADFFIHTDE--ISQVPN 469
Query: 447 PCCNSNA*DDKPPNRDGWFGKVNFVEIRSFWHLFRSFDRMWSFFILCLQAMIIVAWNGSG 506
+ + + P K NFVE R+FW+L+RSFDRMW F +L LQ MIIVAW+ SG
Sbjct: 470 QRHDQVSHGKRKP-------KTNFVEARTFWNLYRSFDRMWMFLVLSLQTMIIVAWHPSG 522
Query: 507 DPTVIFHGDVFKKVLSVFITAAILKFGQAVLGVILSWKARRSMSLYVKLRYILKVISAAA 566
IF DVF+ VL++FIT+A L QA L ++LS+ A +S+ +RYI K + AA
Sbjct: 523 SILAIFTEDVFRNVLTIFITSAFLNLLQATLDLVLSFGAWKSLKFSQIMRYITKFLMAAM 582
Query: 567 WVILLSVTYAYTWDNPPGFAETIKSWFGSNSSAPSLFIVAVVVYLSPNMLAAIFFMFPFI 626
W I+L +TY+ + NP G + SW GS SL+ A+ +Y+ PN+LAA+FF+ P +
Sbjct: 583 WAIMLPITYSKSVQNPTGLIKFFSSWVGSWLHR-SLYDYAIALYVLPNILAAVFFLLPPL 641
Query: 627 RRYLERSNYRIVMLMMWWSQ 646
RR +ERSN RIV L+MWW+Q
Sbjct: 642 RRIMERSNMRIVTLIMWWAQ 661
>At3g14570 hypothetical protein
Length = 1973
Score = 504 bits (1299), Expect = e-142
Identities = 285/674 (42%), Positives = 399/674 (58%), Gaps = 61/674 (9%)
Query: 11 PSEPPPRRLVRTQTAGNLGESI----FDSEVVPSSLV-EIAPILRVANEVEKTHPRVAYL 65
P E P R + E + FDSE +P++L EI LR+AN VE PR+AYL
Sbjct: 25 PREDSPERATEFTRSLTFREHVSSEPFDSERLPATLASEIQRFLRIANLVESEEPRIAYL 84
Query: 66 CRFYAFEKAHRLDPTSSGRGVRQFKTALLQRLERENDPTLKGRVKKSDAREMQSFYQHYY 125
CRF+AFE AH +D S+GRG +F T++ R +KSD RE++ Y H Y
Sbjct: 85 CRFHAFEIAHHMDRNSTGRGDEEF--------------TVRRRKEKSDVRELKRVY-HAY 129
Query: 126 KKYIQALQNAA----DKADRAQLTKAYQTANVLFEVLKAVNMTQSMEVDREILETQDKVA 181
K+YI +++ A D + R +L A + A+VL+EVLK V + + ++ +
Sbjct: 130 KEYI--IRHGAAFNLDNSQREKLINARRIASVLYEVLKTVTSGAGPQA----IADRESIR 183
Query: 182 EKTEILVPFNILPLDPDSANQAIMKFPEIQAAVYALRNTRGLPWPNDYKKKKDE-DILDW 240
K+E VP+NILPLD +QAIM PEI+AAV +RNTRGLP P ++++ + D+ ++
Sbjct: 184 AKSEFYVPYNILPLDKGGVHQAIMHLPEIKAAVAIVRNTRGLPPPEEFQRHQPFLDLFEF 243
Query: 241 LGSMFGFQKHNVANQREHLILLLANVHIRQFPNPDQQPKLDECALTEVMKKLFKNYKKWC 300
L FGFQ NVANQREHLILLL+N IRQ PK + A+ +MKK FKNY WC
Sbjct: 244 LQYAFGFQNGNVANQREHLILLLSNTIIRQPQKQSSAPKSGDEAVDALMKKFFKNYTNWC 303
Query: 301 KYLDRKSSLWLPTIQQEVQQRKLLYMGLYLLIWGEAANLRFMPECLCYIYHHMAFELYGM 360
K+L RK+++ LP ++QE Q K LY+GLYLLIWGEA+NLRFMPECLCYI+HHMA+EL+G+
Sbjct: 304 KFLGRKNNIRLPYVKQEALQYKTLYIGLYLLIWGEASNLRFMPECLCYIFHHMAYELHGV 363
Query: 361 LAGNVSPMTGENIKPAYGGEDEAFLRKVVTPIYNVIAELKKAKGEGQSIHNGGTMMI*MN 420
L G VS +TGE + PAYGG E+FL VVTPIY V+ + + G + H+ +N
Sbjct: 364 LTGAVSMITGEKVAPAYGGGHESFLADVVTPIYMVVQKEAEKNKNGTADHSMWRNYDDLN 423
Query: 421 ---------ISGWPMRADADFFCLPAER----------VVFDKSNPCCNSNA*DD----- 456
GWPMR + DFFC+ + + F K + DD
Sbjct: 424 EFFWSLECFEIGWPMRPEHDFFCVESSETSKPGRWRGMLRFRKQTKKTDEEIEDDEELGV 483
Query: 457 ----KPPNRDGWFGKVNFVEIRSFWHLFRSFDRMWSFFILCLQAMIIVAWNGSGDPTVIF 512
+P W GK NFVE RSFW +FRSFDRMWSFF+L LQA+II+A + G P +F
Sbjct: 484 LSEEQPKPTSRWLGKTNFVETRSFWQIFRSFDRMWSFFVLSLQALIIMACHDVGSPLQVF 543
Query: 513 HGDVFKKVLSVFITAAILKFGQAVLGVILSWKARRSMSLYVKLRYILKVISAAAWVILLS 572
+ ++F+ V+S+FIT+AILK + +L +I WKAR +M + K + ++K+ AA W I+L
Sbjct: 544 NANIFEDVMSIFITSAILKLIKGILDIIFKWKARNTMPINEKKKRLVKLGFAAMWTIILP 603
Query: 573 VTYAYTWDNPPGFAETIKSWFGSNSSAPSLFIVAVVVYLSPNMLAAIFFMFPFIRRYLER 632
V Y+++ + K+W G +P ++VAV +YL+ + + + F P I +Y+E
Sbjct: 604 VLYSHSRRKYICYFTNYKTWLGEWCFSP--YMVAVTIYLTGSAIELVLFFVPAISKYIET 661
Query: 633 SNYRIVMLMMWWSQ 646
SN+ I + WW Q
Sbjct: 662 SNHGIFKTLSWWGQ 675
>At3g59100 putative protein
Length = 1808
Score = 432 bits (1111), Expect = e-121
Identities = 272/675 (40%), Positives = 372/675 (54%), Gaps = 109/675 (16%)
Query: 27 NLGESIFDSEVVPSSLVEIAPILRVANEVEKTHPRVAYLCRFYAFEKAHRLDPTSSGRGV 86
N S DSE+VPSSL IAPILRVANE+EK +PRVAYLCRF+AFEKAHR+D TSSGRGV
Sbjct: 32 NEDASAMDSELVPSSLASIAPILRVANEIEKDNPRVAYLCRFHAFEKAHRMDATSSGRGV 91
Query: 87 RQFKTALLQRLERENDPTLKGRVKKSDAREMQSFYQHYYKKYIQALQNAADKADRAQLTK 146
RQFKT LL RLE+E + T K ++ K+D RE+Q++YQ++Y+KYI+ + + + A+L
Sbjct: 92 RQFKTYLLHRLEKEEEET-KPQLAKNDPREIQAYYQNFYEKYIKEGETSRKPEEMARL-- 148
Query: 147 AYQTANVLFEVLKAVNMTQSMEVDREILETQDKVAEKTEILVPFNILPLDPDSANQAIMK 206
YQ A+VL++VLK V S +VD E ++V K + +NILPL AI++
Sbjct: 149 -YQIASVLYDVLKTV--VPSPKVDYETRRYAEEVERKRDRYEHYNILPLYAVGTKPAIVE 205
Query: 207 FPEIQAAVYALRNTRGLP---------WPNDYKKKKDE--DILDWLGSMFGFQKHNVANQ 255
PE++AA A+RN R LP PN+ +K + + DIL+WL S FGFQ+ NVANQ
Sbjct: 206 LPEVKAAFSAVRNVRNLPRRRIHLPSNTPNEMRKARTKLNDILEWLASEFGFQRGNVANQ 265
Query: 256 REHLILLLANVHIRQFPNPDQQPKLDECALTEVMKKLFKNYKKWCKYLDRKSSLWLPT-- 313
REH+ILLLAN IR+ N ++ +L +TE+M K FK+Y WCKYL S+L
Sbjct: 266 REHIILLLANADIRK-RNDEEYDELKPSTVTELMDKTFKSYYSWCKYLHSTSNLKSDVGC 324
Query: 314 ---------IQQEVQQRKLLYMGLYLLIWGEAANLRFMPECLCYIYHHMAFELYGMLAGN 364
+ QQ +L+Y+ LYLLIWGEA+N MA ++YG+L N
Sbjct: 325 FNFILKRFPDDCDKQQLQLIYISLYLLIWGEASN--------------MANDVYGILFSN 370
Query: 365 VSPMTGENIKPAYGGEDEAFLRKVVTPIYNVIAELKKAKGEGQSIHNGGTMMI*MN---- 420
V ++GE + ++E+FLR V+TPIY VI K G + H+ +N
Sbjct: 371 VEAVSGETYETEEVIDEESFLRTVITPIYQVIRNEAKRNKGGTASHSQWRNYDDLNEYFW 430
Query: 421 -----ISGWPMRADADFFCLPAE---------RVVFDKSNPCCNSNA*DDKPPNRDGWFG 466
GWP+ ADFF E +V + KS P
Sbjct: 431 SKKCFKIGWPLDLKADFFLNSDEITPQDERLNQVTYGKSKP------------------- 471
Query: 467 KVNFVEIRSFWHLFRSFDRMWSFFILCLQAMIIVAWNGSGDPTVIFHGDVFKKVLSVFIT 526
K NFVE+R+FW+LFR FDRMW F ++ QAM+IV W+GSG IF DVFK VL++FIT
Sbjct: 472 KTNFVEVRTFWNLFRDFDRMWIFLVMAFQAMVIVGWHGSGSLGDIFDKDVFKTVLTIFIT 531
Query: 527 AAILKFGQAVLGVILSWKARRSMSLYVKLRYILKVISAAAWVILLSVTYAYTWDNPPGFA 586
+A L Q A W +LL + Y+ + P G
Sbjct: 532 SAYLTLLQV----------------------------AFMWAVLLPIAYSKSVQRPTGVV 563
Query: 587 ETIKSWFGSNSSAPSLFIVAVVVYLSPNMLAAIFFMFPFIRRYLERSNYRIVMLMMWWSQ 646
+ +W G + S + AV Y+ PN+LAA+ F+ P RR +E S+ R + ++MWW+Q
Sbjct: 564 KFFSTWTG-DWKDQSFYTYAVSFYVLPNILAALLFLVPPFRRAMECSDMRPIKVIMWWAQ 622
Query: 647 IFRF*LGVYGAL*LP 661
+GV A+ P
Sbjct: 623 ATTNNIGVVIAIWAP 637
>At4g03550 putative glucan synthase component
Length = 1780
Score = 285 bits (729), Expect = 2e-76
Identities = 172/467 (36%), Positives = 250/467 (52%), Gaps = 31/467 (6%)
Query: 189 PFNILPLDPDSANQAIMKFPEIQAAVYALRNTRGLPWPNDYKKKKDEDILDWLGSMFGFQ 248
P+NI+P++ A+ ++FPE++AA AL+ L P + + D+LDWL FGFQ
Sbjct: 27 PYNIIPVNNLLADHPSLRFPEVRAAAAALKTVGDLRRPPYVQWRSHYDLLDWLALFFGFQ 86
Query: 249 KHNVANQREHLILLLANVHIRQFPNPDQQPKLDECALTEVMKKLFKNYKKWCKYLDRKSS 308
K NV NQREH++L LAN +R P PD LD + +KL NY WC YL +KS+
Sbjct: 87 KDNVRNQREHMVLHLANAQMRLSPPPDNIDSLDSAVVRRFRRKLLANYSSWCSYLGKKSN 146
Query: 309 LWLPTIQQEVQQRKLLYMGLYLLIWGEAANLRFMPECLCYIYHHMAFELYGMLAGNVSPM 368
+W+ + +R+LLY+GLYLLIWGEAANLRFMPEC+CYI+H+MA EL +L +
Sbjct: 147 IWISDRNPD-SRRELLYVGLYLLIWGEAANLRFMPECICYIFHNMASELNKILEDCLDEN 205
Query: 369 TGENIKPAYGGEDEAFLRKVVTPIYNVI-AELKKAKGEGQSIHNGGTMMI*MNISGWPMR 427
TG+ P+ GE+ AFL VV PIY+ I AE+ ++K G H +N W R
Sbjct: 206 TGQPYLPSLSGEN-AFLTGVVKPIYDTIQAEIDESK-NGTVAHCKWRNYDDINEYFWTDR 263
Query: 428 ADADFFCLPAERVVFDKSNPCCNSNA*DDKPPNRDGWFGKVNFVEIRSFWHLFRSFDRMW 487
C + D + S R GK FVE R+F++L+RSFDR+W
Sbjct: 264 ------CFSKLKWPLDLGSNFFKS---------RGKSVGKTGFVERRTFFYLYRSFDRLW 308
Query: 488 SFFILCLQAMIIVAWNGSGDPTVIFH--------GDVFKKVLSVFITAAILKFGQAVLGV 539
L LQA IIVAW D + + DV ++L+VF+T + ++ QAVL
Sbjct: 309 VMLALFLQAAIIVAWEEKPDTSSVTRQLWNALKARDVQVRLLTVFLTWSGMRLLQAVLDA 368
Query: 540 ILSWKARRSMSLYVKLRYILKVISAAAWVILLSVTYAYTWDNPPGFAETIKSWFGSNSSA 599
+ + R ++KVI+AA W++ +V Y W + W + ++
Sbjct: 369 ASQYPLVSRETKRHFFRMLMKVIAAAVWIVAFTVLYTNIWKQ----KRQDRQWSNAATTK 424
Query: 600 PSLFIVAVVVYLSPNMLAAIFFMFPFIRRYLERSNYRIVMLMMWWSQ 646
F+ AV +L P +LA F+ P++R +LE +N++I + WW Q
Sbjct: 425 IYQFLYAVGAFLVPEILALALFIIPWMRNFLEETNWKIFFALTWWFQ 471
>At4g04970
Length = 1768
Score = 276 bits (706), Expect = 9e-74
Identities = 177/464 (38%), Positives = 242/464 (52%), Gaps = 32/464 (6%)
Query: 190 FNILPLDPDSANQAIMKFPEIQAAVYALRNTRGLPWPNDYKKKKDEDILDWLGSMFGFQK 249
+NI+P+ +++PE++AA ALR LP P D++DWLG +FGFQ
Sbjct: 20 YNIIPIHDFLTEHPSLRYPEVRAAAAALRIVGDLPKPPFADFTPRMDLMDWLGLLFGFQI 79
Query: 250 HNVANQREHLILLLANVHIRQFPNPDQQPKLDECALTEVMKKLFKNYKKWCKYLDRKSSL 309
NV NQRE+L+L LAN +R P P LD L KKL +NY WC +L + +
Sbjct: 80 DNVRNQRENLVLHLANSQMRLQPPPRHPDGLDPTVLRRFRKKLLRNYTNWCSFLGVRCHV 139
Query: 310 WLPTIQQEVQ-------QRKLLYMGLYLLIWGEAANLRFMPECLCYIYHHMAFELYGMLA 362
P IQ Q +R+LLY+ LYLLIWGE+ANLRFMPECLCYI+HHMA EL +LA
Sbjct: 140 TSP-IQSRHQTNAVLNLRRELLYVALYLLIWGESANLRFMPECLCYIFHHMAMELNKVLA 198
Query: 363 GNVSPMTGENIKPAYGGEDEAFLRKVVTPIYNVIAELKKAKGEGQSIHNGGTMMI*MNIS 422
G MTG P++ G D AFL+ VV PIY + ++ G H+ +N
Sbjct: 199 GEFDDMTGMPYWPSFSG-DCAFLKSVVMPIYKTVKTEVESSNNGTKPHSAWRNYDDINEY 257
Query: 423 GWPMRADADFFCLPAERVVFDKSNPCCNSNA*DDKPPNRDGWFGKVNFVEIRSFWHLFRS 482
W RA L + + D + SN D P + GK FVE RSFW+++RS
Sbjct: 258 FWSKRA------LKSLKWPLDYT-----SNFFDTTP--KSSRVGKTGFVEQRSFWNVYRS 304
Query: 483 FDRMWSFFILCLQAMIIVAWNGSGDPTVIFHGDVFKKVLSVFITAAILKFGQAVLGVILS 542
FDR+W +L LQA IIVA + P DV +L+VFI+ A L+ Q+VL
Sbjct: 305 FDRLWILLLLYLQAAIIVATSDVKFPWQ--DRDVEVALLTVFISWAGLRLLQSVLDASTQ 362
Query: 543 WKARRSMSLYVKLRYILKVISAAAWVILLSVTYAYTWD--NPPGFAETIKSWFGSNSSAP 600
+ + ++ +R LK + A AW +L SV YA W N G W + +
Sbjct: 363 YSLVSRETYWLFIRLTLKFVVAVAWTVLFSVFYARIWSQKNKDGV------WSRAANERV 416
Query: 601 SLFIVAVVVYLSPNMLAAIFFMFPFIRRYLERSNYRIVMLMMWW 644
F+ V VY+ P +LA + F+ P IR ++E N +V + WW
Sbjct: 417 VTFLKVVFVYVIPELLALVLFIVPCIRNWVEELNLGVVYFLTWW 460
>At3g07160 putative glucan synthase
Length = 1931
Score = 270 bits (690), Expect = 6e-72
Identities = 199/643 (30%), Positives = 309/643 (47%), Gaps = 72/643 (11%)
Query: 38 VPSSLV---EIAPILRVANEVEKTHPRVAYLCRFYAFEKAHRLDPTSSGRGVRQFKTALL 94
VPSSL +I ILR A+E++ P +A + + + A LDP S GRGV QFKT L+
Sbjct: 36 VPSSLSNNRDIDAILRAADEIQDEDPNIARILCEHGYSLAQNLDPNSEGRGVLQFKTGLM 95
Query: 95 QRLERENDPTLKGRVKKS-DAREMQSFYQHYYKKY-IQALQNA----------ADKADRA 142
++++ G + +S D +Q FY+ Y +K + L+ D+ +R
Sbjct: 96 SVIKQKLAKREVGTIDRSQDILRLQEFYRLYREKNNVDTLKEEEKQLRESGAFTDELERK 155
Query: 143 QLTK--AYQTANVLFEVLKAVNMTQSMEVDREILETQDKVAEKTEILVPFNILPLDPDSA 200
+ + + T VL VL+ + + E+ E+ D A +E + +NI+PLD
Sbjct: 156 TVKRKRVFATLKVLGSVLEQL----AKEIPEELKHVIDSDAAMSEDTIAYNIIPLDAPVT 211
Query: 201 NQAIMKFPEIQAAVYALRNTRGLP-WPNDYK--KKKDEDILDWLGSMFGFQKHNVANQRE 257
A FPE+QAAV AL+ GLP P D+ + D+LD+L +FGFQK +V+NQRE
Sbjct: 212 TNATTTFPEVQAAVAALKYFPGLPKLPPDFPIPATRTADMLDFLHYIFGFQKDSVSNQRE 271
Query: 258 HLILLLANVHIRQFPNPDQQPKLDECALTEVMKKLFKNYKKWCKYLDRKSSLWLPTIQQE 317
H++LLLAN R + +PKLD+ A+ +V K +NY KWC YL + + W ++
Sbjct: 272 HIVLLLANEQSRLNIPEETEPKLDDAAVRKVFLKSLENYIKWCDYLCIQPA-W-SNLEAI 329
Query: 318 VQQRKLLYMGLYLLIWGEAANLRFMPECLCYIYHHMAFELYGMLAGNVSPMTGENIKPAY 377
+KLL++ LY LIWGEAAN+RF+PECLCYI+HHM E+ +L V+ +
Sbjct: 330 NGDKKLLFLSLYFLIWGEAANIRFLPECLCYIFHHMVREMDEILRQQVARPAESCMPVDS 389
Query: 378 GGEDE--AFLRKVVTPIYNVIAELKKAKGEGQSIHNGGTMMI*MN---------ISGWPM 426
G D+ +FL V+ P+Y V++ G++ H+ N GWP
Sbjct: 390 RGSDDGVSFLDHVIAPLYGVVSAEAFNNDNGRAPHSAWRNYDDFNEYFWSLHSFELGWPW 449
Query: 427 RADADFFCLPAERVVFDKSNPCCNSNA*DDKPPNRDGWFGKVNFVEIRSFWHLFRSFDRM 486
R + FF P R R GK +FVE R+F HL+ SF R+
Sbjct: 450 RTSSSFFQKPIPR---------------KKLKTGRAKHRGKTSFVEHRTFLHLYHSFHRL 494
Query: 487 WSFFILCLQAMIIVAWNGSGDPTVIFHGDVFKKVLSVFITAAILKFGQAVLGVILSWKAR 546
W F + QA+ I+A+N + ++LS+ T ++KF ++VL VI+ + A
Sbjct: 495 WIFLAMMFQALAIIAFNKDD----LTSRKTLLQILSLGPTFVVMKFSESVLEVIMMYGAY 550
Query: 547 RSMSLYVKLRYILKVISAAAWVILLSVTYAYTWDNPPGFAETIKSWFGSNSSAP--SLFI 604
+ R L+ I W L SV ++ + +KS NS +P L++
Sbjct: 551 STTRRLAVSRIFLRFI----WFGLASVFISFLY---------VKSLKAPNSDSPIVQLYL 597
Query: 605 VAVVVYLSPNMLAAIFFMFPFIRRYLER-SNYRIVMLMMWWSQ 646
+ + +Y +I P + + ++ W Q
Sbjct: 598 IVIAIYGGVQFFFSILMRIPTCHNIANKCDRWPVIRFFKWMRQ 640
>At5g36870 putative glucan synthase
Length = 1662
Score = 205 bits (522), Expect = 2e-52
Identities = 131/306 (42%), Positives = 175/306 (56%), Gaps = 33/306 (10%)
Query: 34 DSEVVPSSLVE-IAPILRVANEVEKTHPRVAYLCRF----------------YAFEKAHR 76
DSE+VPSSL E I PILRVA +VE T+PR +L +A +KA+
Sbjct: 24 DSELVPSSLHEDITPILRVAKDVEDTNPRSLFLQDLDIKSVDDSINILSGHSHALDKANE 83
Query: 77 LDPTSSGRGVRQFKTALLQRLERENDPTLKGRVKKSDAREMQSFYQHYYKKYIQALQNAA 136
LDPTSSGR VRQFK +LQ LE+ N+ TLK R K SDA EMQSFYQ Y + I L NA
Sbjct: 84 LDPTSSGRDVRQFKNTILQWLEKNNESTLKARQKSSDAHEMQSFYQQYGDEGINDLLNAG 143
Query: 137 DKADRAQLTKAYQTANVLFEVLKAVNMTQSMEVDREILETQDKVAEKTEILVPFNILPLD 196
+ +Q TK YQTA VL++VL AV+ +++V +ILE+ +V K +I VP+NILPLD
Sbjct: 144 AGSSSSQRTKIYQTAVVLYDVLDAVHRKANIKVAAKILESHAEVEAKNKIYVPYNILPLD 203
Query: 197 PDSANQAIMKFPEIQAAVYALRNTRGLPWPNDYKKKKDEDILDWLGSMFGFQKHNVANQR 256
PDS N A+M+ P+I A + A+R T L W +K DED+LDWL +MF FQ +A +
Sbjct: 204 PDSKNHAMMRDPKIVAVLKAIRYTSDLTWQIGHKINDDEDVLDWLKTMFRFQ---MAFEL 260
Query: 257 EHLILLLANVHIRQFPNPDQQPKLDECALTEVMKKLFK------------NYKKWCKYLD 304
++ + + NP K DE LT+V+ ++K + +W Y D
Sbjct: 261 FEMLESKGSKKKYKPKNPTYSGK-DEDFLTKVVTPVYKTIAEEAKKSGEGKHSEWRNYDD 319
Query: 305 RKSSLW 310
W
Sbjct: 320 LNEYFW 325
Score = 179 bits (455), Expect = 1e-44
Identities = 115/302 (38%), Positives = 153/302 (50%), Gaps = 81/302 (26%)
Query: 353 MAFELYGMLAGNVSPMTGENIKPAYGGEDEAFLRKVVTPIYNVIAELKKAKGEGQ----- 407
MAFEL+ ML S + P Y G+DE FL KVVTP+Y IAE K GEG+
Sbjct: 256 MAFELFEMLESKGSKKKYKPKNPTYSGKDEDFLTKVVTPVYKTIAEEAKKSGEGKHSEWR 315
Query: 408 ---SIHNGGTMMI*MNISGWPMRADADFFCLPAERVVFDKSNPCCNSNA*DDKPPNRDGW 464
++ ++ GWPM+A+ADFFC ++++ +KS
Sbjct: 316 NYDDLNEYFWSKQYLDKLGWPMKANADFFCKTSQQLGLNKS------------------- 356
Query: 465 FGKVNFVEIRSFWHLFRSFDRMWSFFILCLQAMIIVAWNGSGDPTVIFHGDVFKKVLSVF 524
+AMII+AWN + + G VF KVLSVF
Sbjct: 357 ------------------------------EAMIIIAWNETSESG----GAVFHKVLSVF 382
Query: 525 ITAAILKFGQAVLGVILSWKARRSMSLYVKLRYILKVISAAAWVILLSVTYAYTWDNPPG 584
ITAA L QA L + LSWKAR SMS +V+ RYI K ++AA WV+L+ +TYAY
Sbjct: 383 ITAAKLNLFQAFLDIALSWKARHSMSTHVRQRYIFKAVAAAVWVLLMPLTYAY------- 435
Query: 585 FAETIKSWFGSNSSAPSLFIVAVVVYLSPNMLAAIFFMFPFIRRYLERSNYRIVMLMMWW 644
S S+FIVA+++YLSPNML + + P IRR LE+S++R V L+MWW
Sbjct: 436 -------------SHTSIFIVAILIYLSPNMLPEMLLLIPSIRRTLEKSDFRPVKLIMWW 482
Query: 645 SQ 646
SQ
Sbjct: 483 SQ 484
>At3g14780 unknown protein
Length = 347
Score = 112 bits (279), Expect = 3e-24
Identities = 66/169 (39%), Positives = 104/169 (61%), Gaps = 16/169 (9%)
Query: 33 FDSEVVPSSLVE-IAPILRVANEVEKTHPRVAYLCRFYAFEKAHRLDPTSSGRGVRQFKT 91
+DSE +P +L I LRVAN VE PRVAYLCRFY FE+AHR+D S+GRGVRQFK
Sbjct: 47 YDSEKLPETLASGIQRFLRVANLVESDDPRVAYLCRFYTFEEAHRIDSRSNGRGVRQFKN 106
Query: 92 ALLQRLERENDPTLKGRVKKSDAREMQSFYQHYYKKYI--QALQNAADKADRAQLTKAYQ 149
+LL+RLE++++ T++ R + +D +E++ Y H Y +YI D + + +L A
Sbjct: 107 SLLRRLEKDDEFTIRRRKEINDHKELKRVY-HAYNEYIIRHGASFNLDNSQQEKLINARS 165
Query: 150 TANVLFEVLKAVNMT-------QSMEVDR-----EILETQDKVAEKTEI 186
A+VL+E+L+ + + +S++++R E+L+ + E +I
Sbjct: 166 IASVLYEILRKIESSMGRASTPESIQLNRDEEIGELLDMDTSMEETNKI 214
>At4g26750 unknown protein
Length = 421
Score = 47.8 bits (112), Expect = 7e-05
Identities = 30/116 (25%), Positives = 55/116 (46%), Gaps = 10/116 (8%)
Query: 45 IAPILRVANEVEKTHPRVAYLCRFYAFEKAHRLDPTSSGRGVRQFKTALLQRLERENDPT 104
+ P L+ A+E++K P VAY CR YA E+ ++ + + +L+ +LE++
Sbjct: 11 LLPYLQRADELQKHEPLVAYYCRLYAMERGLKIPQSERTKTTNSILMSLINQLEKDKKSL 70
Query: 105 LKGRVKKSDAREMQSFYQHYYKKYIQALQNAADKADRAQL--TKAYQTANVLFEVL 158
+ D ++ F + K + D+A RA L K + A++ FE+L
Sbjct: 71 ---TLSPDDNMHVEGFALSVFAK-----ADKQDRAGRADLGTAKTFYAASIFFEIL 118
>At1g35740 hypothetical protein
Length = 63
Score = 41.6 bits (96), Expect = 0.005
Identities = 28/63 (44%), Positives = 37/63 (58%), Gaps = 2/63 (3%)
Query: 353 MAFELYGMLAGNVSPMTGENIKPAYGGEDEAFLRKVVTPIYNVIAELKKAKGEGQSIHNG 412
MA+E + NV+ T E KPAY GE + FL+K+VT Y IA K+ + EG S+ G
Sbjct: 1 MAYERLEIWGNNVNEETRELNKPAYCGERKVFLKKLVTTTYQRIATPKEVQ-EG-SMSRG 58
Query: 413 GTM 415
TM
Sbjct: 59 ETM 61
>At2g30680 hypothetical protein
Length = 178
Score = 39.3 bits (90), Expect = 0.023
Identities = 38/134 (28%), Positives = 57/134 (42%), Gaps = 39/134 (29%)
Query: 99 RENDPTLK-GRVKK-SDAREMQSFYQHYYKKYIQ------------------ALQNAADK 138
R ND T + G+ K +D +QSFY YY+K + + +AA +
Sbjct: 33 RRNDETREQGKFKPHTDLPRLQSFYLDYYQKNVMDVIKIIGNQYTILKEIESSAHSAAIR 92
Query: 139 ADRAQLTK-----------------AYQTANVLFEVLKAVNMTQSMEVDREILETQDKVA 181
A A L A TA VLF VL+ +++ +++ E LE V
Sbjct: 93 AIHAALPPHKINGDSRKQADSYIQHACDTAGVLFRVLEL--LSEDVQLPSEFLEADADVK 150
Query: 182 EKTEILVPFNILPL 195
+ EI P+NI+PL
Sbjct: 151 QLAEIFRPYNIIPL 164
>At1g59950 hypothetical protein
Length = 320
Score = 35.0 bits (79), Expect = 0.44
Identities = 18/49 (36%), Positives = 26/49 (52%), Gaps = 2/49 (4%)
Query: 285 LTEVMKKLFKNYKKWCKYLDRKSSLWLPTIQQEVQQRKLLYMGLYLLIW 333
L + +LF K WC D L +P IQ+ ++ KL Y+ LYL+ W
Sbjct: 72 LIQSRSELFVTSKLWCA--DAHGGLVVPAIQRSLETLKLDYLDLYLIHW 118
>At4g34560 unknown protein
Length = 221
Score = 34.7 bits (78), Expect = 0.57
Identities = 24/86 (27%), Positives = 42/86 (47%), Gaps = 11/86 (12%)
Query: 466 GKVNFVEIRSFWHLFRSFDRMWSFFILCLQAMIIVAWNGSGDPTVIFHGDVFKKVLSVFI 525
G +NF IRSF LFR F+ + ++++ + P+V GD+FK+ +
Sbjct: 16 GILNFQAIRSFTSLFRLFELL----------LLVILISKLSFPSVKISGDIFKEAAVFLV 65
Query: 526 TAAILKF-GQAVLGVILSWKARRSMS 550
+ + F G A++ +L+ R S S
Sbjct: 66 SPRFVFFIGNAIVITLLAKSGRYSSS 91
>At2g06420 unknown protein
Length = 249
Score = 32.3 bits (72), Expect = 2.8
Identities = 22/82 (26%), Positives = 36/82 (43%), Gaps = 7/82 (8%)
Query: 228 DYKKK-------KDEDILDWLGSMFGFQKHNVANQREHLILLLANVHIRQFPNPDQQPKL 280
D+KKK K ED+ + L M + + R ++ L+N+ I D+ P++
Sbjct: 119 DFKKKHFKNIVVKHEDVREKLIGMVPLGERSKERLRMMVLYFLSNIIIAPIKTGDKAPQV 178
Query: 281 DECALTEVMKKLFKNYKKWCKY 302
DE L V F +W +Y
Sbjct: 179 DEFCLKAVSDLTFCRNFQWGRY 200
>At4g28340 hypothetical protein
Length = 159
Score = 32.0 bits (71), Expect = 3.7
Identities = 23/78 (29%), Positives = 37/78 (46%), Gaps = 7/78 (8%)
Query: 556 RYILKVISAA-AWVILLSVTYAYTWDNPPGFAETIKSWFGSN----SSAPSLF--IVAVV 608
+ L+VIS WV L V YA + +I +W+G N S P+++ ++
Sbjct: 14 KLFLRVISKRRTWVCLFLVVYAVLLSSSRNSLNSIVNWYGENHQTSSGLPAIYASVLLGA 73
Query: 609 VYLSPNMLAAIFFMFPFI 626
V+ +M AA+F P I
Sbjct: 74 VFGVLSMAAALFIAVPAI 91
>At3g54220 SCARECROW1
Length = 653
Score = 31.2 bits (69), Expect = 6.3
Identities = 13/30 (43%), Positives = 19/30 (63%)
Query: 2 SSSSSRGPTPSEPPPRRLVRTQTAGNLGES 31
SSS++RGP P PPP +VR + A + +
Sbjct: 26 SSSNNRGPPPPPPPPLVMVRKRLASEMSSN 55
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.348 0.154 0.546
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 40,396,953
Number of Sequences: 26719
Number of extensions: 1630070
Number of successful extensions: 5933
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 5824
Number of HSP's gapped (non-prelim): 40
length of query: 1982
length of database: 11,318,596
effective HSP length: 114
effective length of query: 1868
effective length of database: 8,272,630
effective search space: 15453272840
effective search space used: 15453272840
T: 11
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 68 (30.8 bits)
Medicago: description of AC122723.7


