

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122723.6 - phase: 0
(437 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
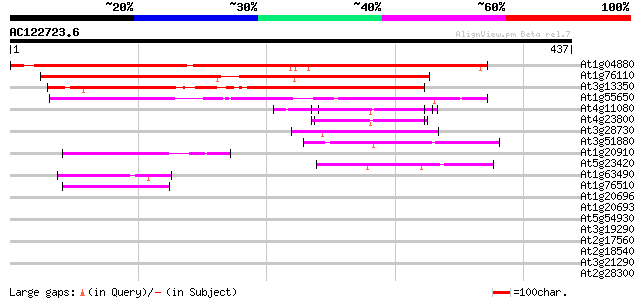
Score E
Sequences producing significant alignments: (bits) Value
At1g04880 unknown protein 379 e-105
At1g76110 hypothetical protein 260 1e-69
At3g13350 unknown protein 238 5e-63
At1g55650 unknown protein 206 3e-53
At4g11080 98b like protein 60 2e-09
At4g23800 98b like protein 56 3e-08
At3g28730 recombination signal sequence recognition protein, put... 56 3e-08
At3g51880 high mobility group protein 2-like 53 3e-07
At1g20910 unknown protein 53 4e-07
At5g23420 unknown protein 49 4e-06
At1g63490 RB-binding protein -like 45 8e-05
At1g76510 putative DNA-binding protein 43 3e-04
At1g20696 unknown protein 41 0.001
At1g20693 unknown protein 39 0.007
At5g54930 unknown protein 36 0.037
At3g19290 abscisic acid responsive elements-binding factor (ABRE... 35 0.082
At2g17560 putative HMG protein 35 0.082
At2g18540 putative vicilin storage protein (globulin-like) 34 0.18
At3g21290 unknown protein 33 0.24
At2g28300 unknown protein 33 0.31
>At1g04880 unknown protein
Length = 448
Score = 379 bits (974), Expect = e-105
Identities = 213/394 (54%), Positives = 268/394 (67%), Gaps = 33/394 (8%)
Query: 1 MASTSCFFNNNEFPMKVVAPMSHSYPEPLAKYDDVVANPKLFMLTLEKLHASMGTKFMVP 60
MAS+SC + PM++ P A Y+ VVA+P+LFM +LE+LH+ +GTKFMVP
Sbjct: 1 MASSSCLKQGS-------VPMNNVCVTPEATYEAVVADPRLFMTSLERLHSLLGTKFMVP 53
Query: 61 IIGGKELDLCRLFIEVTSRGGIEKLMKERRWKEVTAAFSFPSTATNASFVLRKYYSSLLY 120
IIGG++LDL +LF+EVTSRGGI K++ ERRWKEVTA F FP TATNAS+VLRKYY SLL
Sbjct: 54 IIGGRDLDLHKLFVEVTSRGGINKILNERRWKEVTATFVFPPTATNASYVLRKYYFSLLN 113
Query: 121 HYEQIYYFRSKRWTPASSDALQNQSTMSVPASITQFLQPSPGTHPVDFQ-KSGVNASELP 179
+YEQIY+FRS P S QS + P I ++PS + F + +N +E
Sbjct: 114 NYEQIYFFRSNGQIPPDS----MQSPSARPCFIQGAIRPSQELQALTFTPQPKINTAEFL 169
Query: 180 QVSSSGSSLAGVIDGKFESGYLVSVSVGSETLKGVLYE----------SPQS---IKINN 226
S +GS++ GVIDGKFESGYLV+V++GSE LKGVLY+ +PQ + N
Sbjct: 170 GGSLAGSNVVGVIDGKFESGYLVTVTIGSEQLKGVLYQLLPQNTVSYQTPQQSHGVLPNT 229
Query: 227 NNIAS----AALGVQRRRRRRKKSEIKRRDPAHPKPNRSGYNFFFAEQHARLKLLNQTMD 282
NI++ A GV +RRRRRKKSEIKRRDP HPKPNRSGYNFFFAEQHARLK L+ D
Sbjct: 230 LNISANPQGVAGGVTKRRRRRKKSEIKRRDPDHPKPNRSGYNFFFAEQHARLKPLHPGKD 289
Query: 283 KDISRMIGELWNNLKESEKTVYQEKAIKDKERYQAEMEDYRDKMKTSIVTDNAGPLQQRF 342
+DISRMIGELWN L E EK +YQ KA++DKERY+ EMEDYR+K K + NA PLQQR
Sbjct: 290 RDISRMIGELWNKLNEDEKLIYQGKAMEDKERYRTEMEDYREKKKNGQLISNAVPLQQRL 349
Query: 343 PEGDSALVDVDIKMHDSCQTPEE----SSSGESD 372
PE + + + D+ + + + EE SSGES+
Sbjct: 350 PEQNVDMAEADLPIDEVEEDDEEGDSSGSSGESE 383
>At1g76110 hypothetical protein
Length = 338
Score = 260 bits (664), Expect = 1e-69
Identities = 134/313 (42%), Positives = 193/313 (60%), Gaps = 23/313 (7%)
Query: 25 YPEPLAKYDDVVANPKLFMLTLEKLHASMGTKFMVPIIGGKELDLCRLFIEVTSRGGIEK 84
YPEPLA ++ VV + +F TL + H+ M TKFM+P+IGGKELDL L++EVT RGG EK
Sbjct: 27 YPEPLALHEVVVKDSSVFWDTLRRFHSIMSTKFMIPVIGGKELDLHVLYVEVTRRGGYEK 86
Query: 85 LMKERRWKEVTAAFSFPSTATNASFVLRKYYSSLLYHYEQIYYFRSKRWTPASSDALQNQ 144
++ E++W+EV F F +T T+ASFVLRK+Y +LL+HYEQ++ F ++
Sbjct: 87 VVVEKKWREVGGVFRFSATTTSASFVLRKHYLNLLFHYEQVHLFTARGPLLHPIATFHAN 146
Query: 145 STMSVPASITQFLQPS---PGTHPVDFQKSGVNASELPQVSSSGSSLAGVIDGKFESGYL 201
+ S ++ ++ PS THP P SS + G I+GKF+ GYL
Sbjct: 147 PSTSKEMALVEYTPPSIRYHNTHP-------------PSQGSSSFTAIGTIEGKFDCGYL 193
Query: 202 VSVSVGSETLKGVLYESPQ-------SIKINNNNIASAALGVQRRRRRRKKSEIKRRDPA 254
V V +GSE L GVLY S Q + +NN + G +RRR +++ +R DP
Sbjct: 194 VKVKLGSEILNGVLYHSAQPGPSSSPTAVLNNAVVPYVETGRRRRRLGKRRRSRRREDPN 253
Query: 255 HPKPNRSGYNFFFAEQHARLKLLNQTMDKDISRMIGELWNNLKESEKTVYQEKAIKDKER 314
+PKPNRSGYNFFFAE+H +LK L +++ +++IGE W+NL E+ VYQ+ +KDKER
Sbjct: 254 YPKPNRSGYNFFFAEKHCKLKSLYPNKEREFTKLIGESWSNLSTEERMVYQDIGLKDKER 313
Query: 315 YQAEMEDYRDKMK 327
YQ E+ +YR+ ++
Sbjct: 314 YQRELNEYRETLR 326
>At3g13350 unknown protein
Length = 319
Score = 238 bits (607), Expect = 5e-63
Identities = 140/298 (46%), Positives = 184/298 (60%), Gaps = 30/298 (10%)
Query: 30 AKYDDVVANPKLFMLTLEKLHASMGTK---FMVPIIGGKELDLCRLFIEVTSRGGIEKLM 86
AKYDD+V N LF EKL A +G VP +GG LDL RLFIEVTSRGGIE+++
Sbjct: 34 AKYDDLVRNSALFW---EKLRAFLGLTSKTLKVPTVGGNTLDLHRLFIEVTSRGGIERVV 90
Query: 87 KERRWKEVTAAFSFPSTATNASFVLRKYYSSLLYHYEQIYYFRSKRWTPASSDALQNQST 146
K+R+WKEV AFSFP+T T+ASFVLRKYY L+ E +YY P S S
Sbjct: 91 KDRKWKEVIGAFSFPTTITSASFVLRKYYLKFLFQLEHVYYLEK----PVS-------SL 139
Query: 147 MSVPASITQFLQPSPGTHPVDFQKSGVNASELPQVSSSGSSLAGVIDGKFESGYLVSVSV 206
S ++ SP + G++ PQV G + G IDGKF+SGYLV++ +
Sbjct: 140 QSTDEALKSLANESPN------PEEGIDE---PQV---GYEVQGFIDGKFDSGYLVTMKL 187
Query: 207 GSETLKGVLYESPQSIKINNNNIASAALGVQ-RRRRRRKKSEIKRRDPAHPKPNRSGYNF 265
GS+ LKGVLY PQ+ + + + + VQ +RR RKKS++ D PK +RSGYNF
Sbjct: 188 GSQELKGVLYHIPQTPSQSQQTMETPSAIVQSSQRRHRKKSKLAVVDTQKPKCHRSGYNF 247
Query: 266 FFAEQHARLKLLNQTMDKDISRMIGELWNNLKESEKTVYQEKAIKDKERYQAEMEDYR 323
FFAEQ+ARLK ++ I++ IG +W+NL ESEK VYQ+K +KD ERY+ EM +Y+
Sbjct: 248 FFAEQYARLKPEYHGQERSITKKIGHMWSNLTESEKQVYQDKGVKDVERYRIEMLEYK 305
>At1g55650 unknown protein
Length = 337
Score = 206 bits (523), Expect = 3e-53
Identities = 127/346 (36%), Positives = 185/346 (52%), Gaps = 46/346 (13%)
Query: 32 YDDVVANPKLFMLTLEKLHASMGTKFMVPIIGGKELDLCRLFIEVTSRGGIEKLMKERRW 91
Y D+V NP+LF L H S KF +PI+GGK LDL RLF EVTSRGG+EK++K+RR
Sbjct: 30 YQDIVRNPELFWEMLRDFHESSDKKFKIPIVGGKSLDLHRLFNEVTSRGGLEKVIKDRRC 89
Query: 92 KEVTAAFSFPSTATNASFVLRKYYSSLLYHYEQIYYFRSKRWTPASSDALQNQSTMSVPA 151
KEV AF+F +T TN++FVLRK Y +L+ +E +YYF+ A
Sbjct: 90 KEVIDAFNFKTTITNSAFVLRKSYLKMLFEFEHLYYFQ---------------------A 128
Query: 152 SITQFLQPSPGTHPVDFQKSGVNASELPQVSSSGSSLAGVIDGKFESGYLVSVSVGSETL 211
++ F + + +KS N + Q G+ + G+IDGKFESGYL+S VGSE L
Sbjct: 129 PLSTFWEKEKALKLL-IEKS-ANRDKDSQELKPGTVITGIIDGKFESGYLISTKVGSEKL 186
Query: 212 KGVLYE-SPQSIKINNNNIASAALGVQRRRRRRKKSEIKRRDPAHPKPNRSGYNFFFAEQ 270
KG+LY SP++ +R +++ K S+ P PK R+GYNFF AEQ
Sbjct: 187 KGMLYHISPET---------------KRGKKKAKSSQGDSHKP--PKRQRTGYNFFVAEQ 229
Query: 271 HARLKLLNQTMDKDISRMIGELWNNLKESEKTVYQEKAIKDKERYQAEMEDYRDKMKTS- 329
R+K N + G +W NL ES++ VY EK+ +D +RY+ E+ YR M++
Sbjct: 230 SVRIKAENAGQKVSSPKNFGNMWTNLSESDRKVYYEKSREDGKRYKMEILQYRSLMESRV 289
Query: 330 ---IVTDNAGPLQQRFPEGDSALVDVDIKMHDSCQTPEESSSGESD 372
+ +AG D A + ++ D+C + ++ E +
Sbjct: 290 AEIVAATDAGTSASAAETADEASQE-NLAKTDACTSASSAAETEDE 334
>At4g11080 98b like protein
Length = 446
Score = 60.1 bits (144), Expect = 2e-09
Identities = 34/88 (38%), Positives = 48/88 (53%), Gaps = 1/88 (1%)
Query: 236 VQRRRRRRKKSEIKRRDPAHPKPNRSGYNFFFAEQHARLKLLNQTMDKDISRMIGELWNN 295
VQ KK K +DP PK S Y + E+ A LK N+++ ++++M GE W N
Sbjct: 226 VQEAEHDNKKKAKKIKDPLKPKQPISAYLIYANERRAALKGENKSVI-EVAKMAGEEWKN 284
Query: 296 LKESEKTVYQEKAIKDKERYQAEMEDYR 323
L E +K Y + A K+KE Y EME Y+
Sbjct: 285 LSEEKKAPYDQMAKKNKEIYLQEMEGYK 312
Score = 42.0 bits (97), Expect = 7e-04
Identities = 32/92 (34%), Positives = 46/92 (49%), Gaps = 8/92 (8%)
Query: 241 RRRKKSEIKRRDPAHPKPNRSGYNFFFAEQHARLKLLNQ---TMDKDISRMIGELWNNLK 297
+ +KK+E DP PK S Y F + AR +L + + ++ I W L
Sbjct: 359 KNKKKNE--NVDPNKPKKPTSSYFLFCKD--ARKSVLEEHPGINNSTVTAHISLKWMELG 414
Query: 298 ESEKTVYQEKAIKDKERYQAEMEDYRDKMKTS 329
E EK VY KA + E Y+ E+E+Y +K KTS
Sbjct: 415 EEEKQVYNSKAAELMEAYKKEVEEY-NKTKTS 445
Score = 41.6 bits (96), Expect = 9e-04
Identities = 35/130 (26%), Positives = 57/130 (42%), Gaps = 3/130 (2%)
Query: 206 VGSETLKGVLYESPQSIKINNNNIASAALGVQRRRRRRKKSEIKRRDPAHPKPNRSGYNF 265
V E LK L + Q +K N+ A + KK + K++D A K + Y
Sbjct: 80 VEQEKLKTEL-KKLQKMKEFKPNMTFAFSQSLAQTEEEKKGKKKKKDCAETKRPSTPYIL 138
Query: 266 FFAEQHARLKLLNQTMD-KDISRMIGELWNNLKESEKTVYQEKAIKDKERY-QAEMEDYR 323
+ + +K N D K+ S ++G W + EK Y+EK DKE Y Q ++ R
Sbjct: 139 WCKDNWNEVKKQNPEADFKETSNILGAKWKGISAEEKKPYEEKYQADKEAYLQVITKEKR 198
Query: 324 DKMKTSIVTD 333
++ ++ D
Sbjct: 199 EREAMKLLDD 208
>At4g23800 98b like protein
Length = 456
Score = 56.2 bits (134), Expect = 3e-08
Identities = 29/88 (32%), Positives = 50/88 (55%), Gaps = 1/88 (1%)
Query: 236 VQRRRRRRKKSEIKRRDPAHPKPNRSGYNFFFAEQHARLKLLNQTMDKDISRMIGELWNN 295
VQ + KK K +DP PK S + + E+ A L+ N+++ +++++ GE W N
Sbjct: 235 VQEAEQDNKKKNKKEKDPLKPKHPVSAFLVYANERRAALREENKSV-VEVAKITGEEWKN 293
Query: 296 LKESEKTVYQEKAIKDKERYQAEMEDYR 323
L + +K Y++ A K+KE Y ME+Y+
Sbjct: 294 LSDKKKAPYEKVAKKNKETYLQAMEEYK 321
Score = 47.8 bits (112), Expect = 1e-05
Identities = 30/91 (32%), Positives = 44/91 (47%), Gaps = 5/91 (5%)
Query: 238 RRRRRRKKSEIKRRDPAHPKPNRSGYNFFFAEQHARLKLLNQ---TMDKDISRMIGELWN 294
++ + KK + + DP PK S Y F ++ R KL + T + ++ +I W
Sbjct: 361 KKEKATKKKKNENVDPNKPKKPASSYFLFSKDE--RKKLTEERPGTNNATVTALISLKWK 418
Query: 295 NLKESEKTVYQEKAIKDKERYQAEMEDYRDK 325
L E EK VY KA K E Y+ E+E Y K
Sbjct: 419 ELSEEEKQVYNGKAAKLMEAYKKEVEAYNKK 449
Score = 40.0 bits (92), Expect = 0.003
Identities = 25/95 (26%), Positives = 47/95 (49%), Gaps = 2/95 (2%)
Query: 242 RRKKSEIKRRDPAHPKPNRSGYNFFFAEQHARLKLLNQTMD-KDISRMIGELWNNLKESE 300
+ K ++ K++D K S Y + +Q +K N D K+ S ++G W +L +
Sbjct: 124 QEKANKKKKKDCPETKRPSSSYVLWCKDQWTEVKKENPEADFKETSNILGAKWKSLSAED 183
Query: 301 KTVYQEKAIKDKERY-QAEMEDYRDKMKTSIVTDN 334
K Y+E+ +KE Y Q ++ R+K ++ D+
Sbjct: 184 KKPYEERYQVEKEAYLQVIAKEKREKEAMKLLEDD 218
>At3g28730 recombination signal sequence recognition protein,
putative
Length = 646
Score = 56.2 bits (134), Expect = 3e-08
Identities = 32/124 (25%), Positives = 64/124 (50%), Gaps = 9/124 (7%)
Query: 220 QSIKINNNNIASAALGVQRRRRR--------RKKSEIKRRDPAHPKPNRSGYNFFFAEQH 271
+SIK AS++ G+ +R+ ++K K++DP PK SG+ FF +
Sbjct: 518 KSIKKEPKKEASSSKGLPPKRKTVAADEGSSKRKKPKKKKDPNAPKRAMSGFMFFSQMER 577
Query: 272 ARLKLLNQTMD-KDISRMIGELWNNLKESEKTVYQEKAIKDKERYQAEMEDYRDKMKTSI 330
+K + + ++ +++G+ W + +K Y+ KA DK+RY+ E+ DY++ ++
Sbjct: 578 DNIKKEHPGIAFGEVGKVLGDKWRQMSADDKEPYEAKAQVDKQRYKDEISDYKNPQPMNV 637
Query: 331 VTDN 334
+ N
Sbjct: 638 DSGN 641
>At3g51880 high mobility group protein 2-like
Length = 178
Score = 53.1 bits (126), Expect = 3e-07
Identities = 37/154 (24%), Positives = 67/154 (43%), Gaps = 7/154 (4%)
Query: 230 ASAALGVQRRRRRRKKSEIKRRDPAHPKPNRSGYNFFFAEQHARLKLLNQTMD--KDISR 287
A A +R R+ KK+ ++DP PK S + F + K N + + +
Sbjct: 30 APAEKPTKRETRKEKKA---KKDPNKPKRAPSAFFVFLEDFRVTFKKENPNVKAVSAVGK 86
Query: 288 MIGELWNNLKESEKTVYQEKAIKDKERYQAEMEDYRDKMKTSIVTDNAGPLQQRFPEGDS 347
G+ W ++ ++EK Y+EKA K K Y+ +M+ Y ++ +D + + + D
Sbjct: 87 AGGQKWKSMSQAEKAPYEEKAAKRKAEYEKQMDAYNKNLEEG--SDESEKSRSEINDEDE 144
Query: 348 ALVDVDIKMHDSCQTPEESSSGESDYVADDINMD 381
A + ++ ++ EE E D DD D
Sbjct: 145 ASGEEELLEKEAAGDDEEEEEEEDDDDDDDEEED 178
>At1g20910 unknown protein
Length = 398
Score = 52.8 bits (125), Expect = 4e-07
Identities = 34/131 (25%), Positives = 57/131 (42%), Gaps = 16/131 (12%)
Query: 42 FMLTLEKLHASMGTKFMVPIIGGKELDLCRLFIEVTSRGGIEKLMKERRWKEVTAAFSFP 101
F+ +E + +F P G+ L++ +L+ V + GG E + + W++V +F+ P
Sbjct: 112 FLREVEAFYKESFLEFKPPKFYGQPLNILKLWRAVVNLGGYEVVTTNKLWRQVGESFNPP 171
Query: 102 STATNASFVLRKYYSSLLYHYEQIYYFRSKRWTPASSDALQNQSTMSVPASITQFLQPSP 161
T T S+ R +Y L YE+ L+N +++P S T L S
Sbjct: 172 KTCTTVSYTFRNFYEKALLEYEK---------------CLRNNGELNLPGS-TLILSSSV 215
Query: 162 GTHPVDFQKSG 172
P Q SG
Sbjct: 216 EKEPSSHQGSG 226
>At5g23420 unknown protein
Length = 241
Score = 49.3 bits (116), Expect = 4e-06
Identities = 37/147 (25%), Positives = 66/147 (44%), Gaps = 12/147 (8%)
Query: 240 RRRRKKSEIKRRDPAHPKPNRSGYNFFFAEQHARLKLL---NQTMDKDISRMIGELWNNL 296
+R +K ++ K+ KP R FF R N ++ KD +++ GE W +L
Sbjct: 97 KRLKKTNDEKKSSSTSNKPKRPLTAFFIFMSDFRKTFKSEHNGSLAKDAAKIGGEKWKSL 156
Query: 297 KESEKTVYQEKAIKDKERYQAEM------EDYRDKMKTSIVTDNAGPLQQRFPEGDSALV 350
E EK VY +KA + K Y + E+ D+ K S D+A +++ + D +
Sbjct: 157 TEEEKKVYLDKAAELKAEYNKSLESNDADEEEEDEEKQSDDVDDA---EEKQVDDDDEVE 213
Query: 351 DVDIKMHDSCQTPEESSSGESDYVADD 377
+ +++ D + E E + + DD
Sbjct: 214 EKEVENTDDDKKEAEGKEEEEEEILDD 240
>At1g63490 RB-binding protein -like
Length = 1458
Score = 45.1 bits (105), Expect = 8e-05
Identities = 32/95 (33%), Positives = 48/95 (49%), Gaps = 9/95 (9%)
Query: 38 NPKLFMLTLEK-LHASMGTKFMVPII-GGKELDLCRLFIEVTSRGGIEKLMKERRWKEVT 95
N K F L + L +G K ++ G+ELDLC+LF V GG EK++K ++W EV
Sbjct: 96 NSKTFKLEYNRFLEEHLGKKLKKRVVFEGEELDLCKLFNAVKRFGGYEKVVKGKKWGEV- 154
Query: 96 AAFSFPSTATN----ASFVLRKYYSSLLYHYEQIY 126
+ F S+ A VL + Y L+ +E +
Sbjct: 155 --YQFMSSGEKISKCAKHVLCQLYKEHLHDFENYH 187
>At1g76510 putative DNA-binding protein
Length = 434
Score = 43.1 bits (100), Expect = 3e-04
Identities = 22/83 (26%), Positives = 39/83 (46%)
Query: 42 FMLTLEKLHASMGTKFMVPIIGGKELDLCRLFIEVTSRGGIEKLMKERRWKEVTAAFSFP 101
F+ +E + +F P G+ L+ +L+ V GG + + + W++V +F P
Sbjct: 148 FIKEVEAFNKENFLEFKAPKFYGQPLNCLKLWRAVIKLGGYDVVTTSKLWRQVGESFHPP 207
Query: 102 STATNASFVLRKYYSSLLYHYEQ 124
T T S+ R +Y L YE+
Sbjct: 208 KTCTTVSWTFRIFYEKALLEYEK 230
>At1g20696 unknown protein
Length = 141
Score = 40.8 bits (94), Expect = 0.001
Identities = 30/104 (28%), Positives = 46/104 (43%), Gaps = 14/104 (13%)
Query: 231 SAALGVQRRRRRRKKSEIKRRDPAHPKPNRSGYNFF-------FAEQHARLKLLNQTMDK 283
S L V ++ + K K DP PK S + F + E+H + K +
Sbjct: 12 STKLSVTKKPAKGAKGAAK--DPNKPKRPSSAFFVFMEDFRVTYKEEHPKNKSVAA---- 65
Query: 284 DISRMIGELWNNLKESEKTVYQEKAIKDKERYQAEMEDYRDKMK 327
+ + GE W +L +SEK Y KA K K Y+ M+ Y K++
Sbjct: 66 -VGKAGGEKWKSLSDSEKAPYVAKADKRKVEYEKNMKAYNKKLE 108
>At1g20693 unknown protein
Length = 144
Score = 38.5 bits (88), Expect = 0.007
Identities = 22/79 (27%), Positives = 35/79 (43%), Gaps = 2/79 (2%)
Query: 251 RDPAHPKPNRSGYNFFFAEQHARLKLLNQTMDK--DISRMIGELWNNLKESEKTVYQEKA 308
+DP PK S + F + K N + + G+ W +L +SEK Y KA
Sbjct: 33 KDPNKPKRPASAFFVFMEDFRETFKKENPKNKSVATVGKAAGDKWKSLSDSEKAPYVAKA 92
Query: 309 IKDKERYQAEMEDYRDKMK 327
K K Y+ ++ Y K++
Sbjct: 93 EKRKVEYEKNIKAYNKKLE 111
>At5g54930 unknown protein
Length = 286
Score = 36.2 bits (82), Expect = 0.037
Identities = 28/99 (28%), Positives = 49/99 (49%), Gaps = 27/99 (27%)
Query: 185 GSSLAGVIDGKFESGYLVSVSVGS--ETLKGVLYESPQSIKIN-NNNIA----------- 230
G ++GVI+ FE+G+L+SV VG+ L+GV+++ ++ +N++A
Sbjct: 63 GQPISGVIEATFEAGFLLSVKVGNSDSMLRGVVFKPGHCDPVSVDNDVAPDVPMIRRNSD 122
Query: 231 ------SAALGVQRRRRRRKKSEIKRR-------DPAHP 256
SA G + R R ++ S ++ R PAHP
Sbjct: 123 VMHHDGSAKRGRKSRFREKRGSGVRSRALVPVPIQPAHP 161
>At3g19290 abscisic acid responsive elements-binding factor
(ABRE/ABF4) / bZip transcription factor AtbZip38
Length = 431
Score = 35.0 bits (79), Expect = 0.082
Identities = 39/178 (21%), Positives = 65/178 (35%), Gaps = 14/178 (7%)
Query: 142 QNQSTMSVPASITQFLQPSPGTH-PVDFQKSGVNASELPQVSSSGSSLAGVIDGKFESGY 200
Q Q + Q QP P P N + V+ + AG + +
Sbjct: 244 QQQLLQQQQQQMQQLNQPHPQQRLPQTIFPKQANVAFSAPVNITNKGFAGAANNSINNNN 303
Query: 201 LVSVSVGSETLKGVLYESPQSIKINNNNIASAALGVQRRRR----------RRKKSEIKR 250
++ G+ V SP + NN+++ + R RR RR++ IK
Sbjct: 304 GLASYGGTGVT--VAATSPGTSSAENNSLSPVPYVLNRGRRSNTGLEKVIERRQRRMIKN 361
Query: 251 RDPA-HPKPNRSGYNFFFAEQHARLKLLNQTMDKDISRMIGELWNNLKESEKTVYQEK 307
R+ A + + Y + +LK NQ + K + M+ N LKE+ K + K
Sbjct: 362 RESAARSRARKQAYTLELEAEIEKLKKTNQELQKKQAEMVEMQKNELKETSKRPWGSK 419
>At2g17560 putative HMG protein
Length = 138
Score = 35.0 bits (79), Expect = 0.082
Identities = 21/90 (23%), Positives = 35/90 (38%), Gaps = 2/90 (2%)
Query: 239 RRRRRKKSEIKRRDPAHPKPNRSGYNFFFAEQHARLKLLNQTMDK--DISRMIGELWNNL 296
+ R RK + ++DP PK S + F + L N + + G W +
Sbjct: 18 KTRGRKAGKKTKKDPNQPKRPPSAFFVFLEDFRKEFNLANPNNKSVATVGKAAGARWKAM 77
Query: 297 KESEKTVYQEKAIKDKERYQAEMEDYRDKM 326
+ +K Y KA K Y ++ Y K+
Sbjct: 78 TDEDKAPYVAKAESRKTEYIKNVQQYNLKL 107
>At2g18540 putative vicilin storage protein (globulin-like)
Length = 699
Score = 33.9 bits (76), Expect = 0.18
Identities = 38/156 (24%), Positives = 72/156 (45%), Gaps = 12/156 (7%)
Query: 179 PQVSSSGSSLAGVIDGKFESGYLVSVSVGSETLKGVLYESPQSIKINNNNIASAALGVQR 238
PQ SS+ V+D + VS ++ +ET+KG+L +S+ + A L
Sbjct: 365 PQFLVGQSSVLKVLDRDVVA---VSFNLSNETIKGLLKAQKESVIFECASCAEGELSKLM 421
Query: 239 R----RRRRKKSEIKRRDPAHPKPNRSGYNFFFAEQHARLKLLNQT----MDKDISRMIG 290
R R+RR++ EI+RR + + E+ A+ + +T +++ +R
Sbjct: 422 REIEERKRREEEEIERRRKEEEEARKREEAKRREEEEAKRREEEETERKKREEEEARKRE 481
Query: 291 ELWNNLKESEKTVYQEKAIKDKERYQA-EMEDYRDK 325
E +E K +E+ +++E QA + E+ R+K
Sbjct: 482 EERKREEEEAKRREEERKKREEEAEQARKREEEREK 517
>At3g21290 unknown protein
Length = 1848
Score = 33.5 bits (75), Expect = 0.24
Identities = 42/173 (24%), Positives = 75/173 (43%), Gaps = 10/173 (5%)
Query: 160 SPGTHPVDFQKSGVNASELPQVSSSGSSLAGVIDGKFESGYLVSVSVGSETLKGVLYESP 219
S G+ P+D Q++ + ++LP+ +G L + + E G L +G +T + E
Sbjct: 879 SIGSSPLDSQRTYL--AKLPK--GNGPVLQKQVS-ELELGELPE-PLGEDTALKPIEEKT 932
Query: 220 QSIKINNNNIASAALGVQRRRRRRKKSEIKRRDPAHPKPNRSGYNFFFAEQHAR-LKLLN 278
+ N S LG+ +RR KKS+ K+ P H +G N E R K
Sbjct: 933 SFRQSNLKPSTSEKLGIDSDKRRSKKSDSKKAAPPH---TVNGSNEHVVEDSERSQKWAL 989
Query: 279 QTMDKDISRMIGELWNNLKESEKTVYQEKAIKDKERYQAEMEDYRDKMKTSIV 331
Q+ ++++ E+ + K E Y+ + + R +E Y + K + V
Sbjct: 990 QSHGQNLTGTDTEISSQNKNLEDAAYKSRQKDSRARVGNSVEGYGETNKKTPV 1042
>At2g28300 unknown protein
Length = 2218
Score = 33.1 bits (74), Expect = 0.31
Identities = 22/91 (24%), Positives = 42/91 (45%), Gaps = 2/91 (2%)
Query: 139 DALQNQSTMSVPASITQFLQPSPGTHPVDFQKSGVNASELPQVSSSGSSLAGVIDGKFES 198
D +QN +++ + + + QPS T P D ++ ++ E +V S V G ES
Sbjct: 1354 DTMQNNTSIDIGITSGKTCQPSSSTQPEDENRNSLSHCEPSEVVEQRDSRDQVCIGSVES 1413
Query: 199 GYLVSVSVGSETLKGVLYESPQSIKINNNNI 229
+S ++ + + PQSI ++ +I
Sbjct: 1414 QVEISSAILENRSADI--QPPQSILVDQKDI 1442
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.129 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,701,027
Number of Sequences: 26719
Number of extensions: 421284
Number of successful extensions: 1229
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 25
Number of HSP's successfully gapped in prelim test: 34
Number of HSP's that attempted gapping in prelim test: 1180
Number of HSP's gapped (non-prelim): 70
length of query: 437
length of database: 11,318,596
effective HSP length: 102
effective length of query: 335
effective length of database: 8,593,258
effective search space: 2878741430
effective search space used: 2878741430
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC122723.6


