

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122722.6 + phase: 0
(461 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
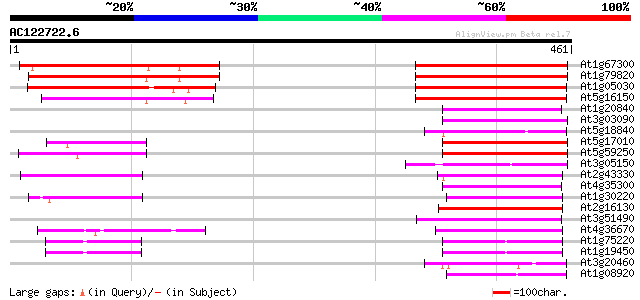
Score E
Sequences producing significant alignments: (bits) Value
At1g67300 transport protein like protein 196 2e-50
At1g79820 hexose transporter like 181 6e-46
At1g05030 sugar transporter like protein 123 2e-28
At5g16150 sugar transporter like protein 122 5e-28
At1g20840 putative sugar transporter protein 75 6e-14
At3g03090 hypothetical protein 72 6e-13
At5g18840 sugar transporter - like protein 72 8e-13
At5g17010 sugar transporter - like protein 71 1e-12
At5g59250 D-xylose-H+ symporter - like protein 70 3e-12
At3g05150 putative sugar transporter 69 5e-12
At2g43330 membrane transporter like protein 69 5e-12
At4g35300 sugar transporter like protein 68 9e-12
At1g30220 unknown protein 65 8e-11
At2g16130 putative sugar transporter 64 2e-10
At3g51490 sugar transporter-like protein 63 3e-10
At4g36670 sugar transporter like protein 62 5e-10
At1g75220 integral membrane protein, putative 61 1e-09
At1g19450 integral membrane protein, putative 61 1e-09
At3g20460 sugar transporter, putative 61 1e-09
At1g08920 putative sugar transport protein 60 3e-09
>At1g67300 transport protein like protein
Length = 493
Score = 196 bits (499), Expect = 2e-50
Identities = 105/176 (59%), Positives = 125/176 (70%), Gaps = 12/176 (6%)
Query: 9 LLDNFIDKE--TTNPSWKLSLPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLA 66
LL+N +D E TTNPSWK SLPHVLVATI+SFLFGYHLGVVNEPLESIS DLGF+G+TLA
Sbjct: 32 LLENDVDNEMETTNPSWKCSLPHVLVATISSFLFGYHLGVVNEPLESISSDLGFSGDTLA 91
Query: 67 EGLVVSICLGGALFGCLLSGWIADAVGRRRAFQLCALPMIIGAAMS--NKQLVWHACRKI 124
EGLVVS+CLGGA G L SG +AD GRRRAFQ+CALPMI+GA +S + L +
Sbjct: 92 EGLVVSVCLGGAFLGSLFSGGVADGFGRRRAFQICALPMILGAFVSGVSNSLAVMLLGRF 151
Query: 125 ICWNWLGLGPSRCC--------SLCDRAYGALIQIATCFGILGSLFIGIPVKEISG 172
+ +GLGP + YG+ IQIATC G++ +LFIGIPV I+G
Sbjct: 152 LVGTGMGLGPPVAALYVTEVSPAFVRGTYGSFIQIATCLGLMAALFIGIPVHNITG 207
Score = 196 bits (497), Expect = 3e-50
Identities = 99/125 (79%), Positives = 110/125 (87%)
Query: 334 AISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAM 393
A +M +Q S+ LP AL LSVGG L+FV TFALGAGPVPGLLL EIFPSRIRAKAM
Sbjct: 366 AAAMALQVGATSSYLPHFSALCLSVGGTLVFVLTFALGAGPVPGLLLPEIFPSRIRAKAM 425
Query: 394 AFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEI 453
AFCMSVHWVINFFVGLLFLRLLEKLG +LLYSMF+TFC+MAV+FVKRNV+ETKGK+LQEI
Sbjct: 426 AFCMSVHWVINFFVGLLFLRLLEKLGPRLLYSMFSTFCLMAVMFVKRNVIETKGKTLQEI 485
Query: 454 EIALL 458
EI+LL
Sbjct: 486 EISLL 490
>At1g79820 hexose transporter like
Length = 495
Score = 181 bits (460), Expect = 6e-46
Identities = 97/167 (58%), Positives = 117/167 (69%), Gaps = 10/167 (5%)
Query: 16 KETTNPSWKLSLPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICL 75
K+ NPSWK SLPHVLVA++TS LFGYHLGVVNE LESIS+DLGF+GNT+AEGLVVS CL
Sbjct: 44 KDCGNPSWKRSLPHVLVASLTSLLFGYHLGVVNETLESISIDLGFSGNTIAEGLVVSTCL 103
Query: 76 GGALFGCLLSGWIADAVGRRRAFQLCALPMIIGAAM--SNKQLVWHACRKIICWNWLGLG 133
GGA G L SG +AD VGRRRAFQL ALPMI+GA++ S + L+ + + +G+G
Sbjct: 104 GGAFIGSLFSGLVADGVGRRRAFQLSALPMIVGASVSASTESLMGMLLGRFLVGIGMGIG 163
Query: 134 PSRCC--------SLCDRAYGALIQIATCFGILGSLFIGIPVKEISG 172
PS + YG+ QIATC G+LGSLF GIP K+ G
Sbjct: 164 PSVTALYVTEVSPAYVRGTYGSSTQIATCIGLLGSLFAGIPAKDNLG 210
Score = 167 bits (422), Expect = 1e-41
Identities = 83/125 (66%), Positives = 105/125 (83%)
Query: 334 AISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAM 393
A+S+ +QA ++L G L+LSVGGMLLFV +FA GAGPVP LLL+EI P R+RA A+
Sbjct: 367 AVSLGLQAIAYTSLPSPFGTLFLSVGGMLLFVLSFATGAGPVPSLLLSEICPGRLRATAL 426
Query: 394 AFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEI 453
A C++VHWVINFFVGLLFLR+LE+LG+ LL ++F FC++AVIFV++NVVETKGKSLQEI
Sbjct: 427 AVCLAVHWVINFFVGLLFLRMLEQLGSVLLNAIFGFFCVVAVIFVQKNVVETKGKSLQEI 486
Query: 454 EIALL 458
EI+LL
Sbjct: 487 EISLL 491
>At1g05030 sugar transporter like protein
Length = 524
Score = 123 bits (308), Expect = 2e-28
Identities = 68/168 (40%), Positives = 102/168 (60%), Gaps = 16/168 (9%)
Query: 15 DKETTNPSWKLSLPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSIC 74
+K + + W + PHV VA++ +FLFGYH+GV+N P+ SI+ +LGF GN++ EGLVVSI
Sbjct: 68 EKFSADLGWLSAFPHVSVASMANFLFGYHIGVMNGPIVSIARELGFEGNSILEGLVVSIF 127
Query: 75 LGGALFGCLLSGWIADAVGRRRAFQLCALPMIIGAAMSNKQLVWHACRKIICWNWL-GLG 133
+ GA G +++G + D G RR FQ+ +P+I+GA +S + H+ +I+C +L GLG
Sbjct: 128 IAGAFIGSIVAGPLVDKFGYRRTFQIFTIPLILGALVSAQA---HSLDEILCGRFLVGLG 184
Query: 134 --------PSRCCSLCDRAY----GALIQIATCFGILGSLFIGIPVKE 169
P + Y G L QI TC GI+ SL +GIP ++
Sbjct: 185 IGVNTVLVPIYISEVAPTKYRGSLGTLCQIGTCLGIIFSLLLGIPAED 232
Score = 107 bits (267), Expect = 1e-23
Identities = 55/124 (44%), Positives = 78/124 (62%)
Query: 334 AISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAM 393
A+SM + L + LS+ G L+++F+FA+GAGPV GL++ E+ +R R K M
Sbjct: 394 AVSMFLIVYAVGFPLDEDLSQSLSILGTLMYIFSFAIGAGPVTGLIIPELSSNRTRGKIM 453
Query: 394 AFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEI 453
F SVHWV NF VGL FL L+EK G +Y+ F + ++A F VETKG+SL+EI
Sbjct: 454 GFSFSVHWVSNFLVGLFFLDLVEKYGVGTVYASFGSVSLLAAAFSHLFTVETKGRSLEEI 513
Query: 454 EIAL 457
E++L
Sbjct: 514 ELSL 517
>At5g16150 sugar transporter like protein
Length = 546
Score = 122 bits (305), Expect = 5e-28
Identities = 61/124 (49%), Positives = 88/124 (70%)
Query: 334 AISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAM 393
A+SM++ + + A + L+V G +L+V +F+LGAGPVP LLL EIF SRIRAKA+
Sbjct: 419 ALSMLLLSLSFTWKALAAYSGTLAVVGTVLYVLSFSLGAGPVPALLLPEIFASRIRAKAV 478
Query: 394 AFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEI 453
A + +HW+ NF +GL FL ++ K G +Y FA C++AV+++ NVVETKG+SL+EI
Sbjct: 479 ALSLGMHWISNFVIGLYFLSVVTKFGISSVYLGFAGVCVLAVLYIAGNVVETKGRSLEEI 538
Query: 454 EIAL 457
E+AL
Sbjct: 539 ELAL 542
Score = 85.9 bits (211), Expect = 4e-17
Identities = 54/151 (35%), Positives = 80/151 (52%), Gaps = 10/151 (6%)
Query: 27 LPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLGGALFGCLLSG 86
LP V VA + + LFGYHLGVVN LE ++ DLG NT+ +G +VS L GA G G
Sbjct: 106 LPFVGVACLGAILFGYHLGVVNGALEYLAKDLGIAENTVLQGWIVSSLLAGATVGSFTGG 165
Query: 87 WIADAVGRRRAFQLCALPMIIGAAM--SNKQLVWHACRKIICWNWLGLGPSRCCSLCDR- 143
+AD GR R FQL A+P+ IGA + + + + +++ +G+ +
Sbjct: 166 ALADKFGRTRTFQLDAIPLAIGAFLCATAQSVQTMIVGRLLAGIGIGISSAIVPLYISEI 225
Query: 144 -------AYGALIQIATCFGILGSLFIGIPV 167
A G++ Q+ C GIL +L G+P+
Sbjct: 226 SPTEIRGALGSVNQLFICIGILAALIAGLPL 256
>At1g20840 putative sugar transporter protein
Length = 734
Score = 75.5 bits (184), Expect = 6e-14
Identities = 37/98 (37%), Positives = 58/98 (58%)
Query: 356 LSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLFLRLL 415
LS G ++L+ F +G GP+P +L +EIFP+R+R +A C V W+ + V LL
Sbjct: 618 LSTGCVVLYFCFFVMGYGPIPNILCSEIFPTRVRGLCIAICAMVFWIGDIIVTYSLPVLL 677
Query: 416 EKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEI 453
+G ++S++A C+++ IFV V ETKG L+ I
Sbjct: 678 SSIGLVGVFSIYAAVCVISWIFVYMKVPETKGMPLEVI 715
Score = 37.0 bits (84), Expect = 0.023
Identities = 32/98 (32%), Positives = 50/98 (50%), Gaps = 15/98 (15%)
Query: 31 LVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLGGALFGCLLSGWIAD 90
L ATI +FL G+ + + I+ DL N T +GLVV++ L GA SG I+D
Sbjct: 9 LAATIGNFLQGWDNATIAGAMVYINKDL--NLPTSVQGLVVAMSLIGATVITTCSGPISD 66
Query: 91 AVGRRRAFQLCALPMIIGAAMSNKQLVWHACRKIICWN 128
+GRR PM+I ++ +++ C I+ W+
Sbjct: 67 WLGRR--------PMLILSS-----VMYFVCGLIMLWS 91
>At3g03090 hypothetical protein
Length = 342
Score = 72.0 bits (175), Expect = 6e-13
Identities = 36/103 (34%), Positives = 62/103 (59%)
Query: 356 LSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLFLRLL 415
++V +LL+V + L GP+ L+++EIFP ++R + ++ + V++ N V F L
Sbjct: 240 VAVAALLLYVGCYQLSFGPIGWLMISEIFPLKLRGRGISLAVLVNFGANALVTFAFSPLK 299
Query: 416 EKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIEIALL 458
E LGA +L+ F C++++ F+ V ETKG +L+EIE L
Sbjct: 300 ELLGAGILFCAFGVICVVSLFFIYYIVPETKGLTLEEIEAKCL 342
>At5g18840 sugar transporter - like protein
Length = 482
Score = 71.6 bits (174), Expect = 8e-13
Identities = 42/121 (34%), Positives = 68/121 (55%), Gaps = 6/121 (4%)
Query: 342 TGASTLLPTAGALY-----LSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFC 396
TG S LL L L+VGG+L++V F++G GPVP ++++EIFP ++ A +
Sbjct: 359 TGTSFLLKGQSLLLEWVPSLAVGGVLIYVAAFSIGMGPVPWVIMSEIFPINVKGIAGSLV 418
Query: 397 MSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIEIA 456
+ V+W + V F L+ Y +++ F +IFV + V ETKGK+L+EI+
Sbjct: 419 VLVNWSGAWAVSYTFNFLMSWSSPGTFY-LYSAFAAATIIFVAKMVPETKGKTLEEIQAC 477
Query: 457 L 457
+
Sbjct: 478 I 478
Score = 37.4 bits (85), Expect = 0.018
Identities = 34/96 (35%), Positives = 47/96 (48%), Gaps = 5/96 (5%)
Query: 15 DKET-TNPSWKLSLPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAE-GLVVS 72
+KE+ N S+ + L VA SF FG +G SI DL + LAE + S
Sbjct: 32 EKESENNESYLMVLFSTFVAVCGSFEFGSCVGYSAPTQSSIRQDLNLS---LAEFSMFGS 88
Query: 73 ICLGGALFGCLLSGWIADAVGRRRAFQLCALPMIIG 108
I GA+ G ++SG I+D GR+ A + A I G
Sbjct: 89 ILTIGAMLGAVMSGKISDFSGRKGAMRTSACFCITG 124
>At5g17010 sugar transporter - like protein
Length = 503
Score = 71.2 bits (173), Expect = 1e-12
Identities = 36/103 (34%), Positives = 63/103 (60%)
Query: 356 LSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLFLRLL 415
++V +LL+V + L GP+ L+++EIFP ++R + ++ + V++ N V F L
Sbjct: 401 VAVVALLLYVGCYQLSFGPIGWLMISEIFPLKLRGRGLSLAVLVNFGANALVTFAFSPLK 460
Query: 416 EKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIEIALL 458
E LGA +L+ F C+++++F+ V ETKG +L+EIE L
Sbjct: 461 ELLGAGILFCGFGVICVLSLVFIFFIVPETKGLTLEEIEAKCL 503
Score = 46.2 bits (108), Expect = 4e-05
Identities = 29/87 (33%), Positives = 48/87 (54%), Gaps = 5/87 (5%)
Query: 31 LVATITSFLFGYHLGV-----VNEPLESISVDLGFNGNTLAEGLVVSICLGGALFGCLLS 85
L + L+GY +G ++ S+S +N +++ GLV S L GALFG +++
Sbjct: 52 LFPALGGLLYGYEIGATSCATISLQSPSLSGISWYNLSSVDVGLVTSGSLYGALFGSIVA 111
Query: 86 GWIADAVGRRRAFQLCALPMIIGAAMS 112
IAD +GRR+ L AL ++GA ++
Sbjct: 112 FTIADVIGRRKELILAALLYLVGALVT 138
>At5g59250 D-xylose-H+ symporter - like protein
Length = 558
Score = 69.7 bits (169), Expect = 3e-12
Identities = 37/103 (35%), Positives = 63/103 (60%)
Query: 356 LSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLFLRLL 415
++VG +LL+V + + GP+ L+++EIFP R R + ++ + ++ N V F L
Sbjct: 455 VAVGALLLYVGCYQISFGPISWLMVSEIFPLRTRGRGISLAVLTNFGSNAIVTFAFSPLK 514
Query: 416 EKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIEIALL 458
E LGA+ L+ +F +++++FV V ETKG SL+EIE +L
Sbjct: 515 EFLGAENLFLLFGGIALVSLLFVILVVPETKGLSLEEIESKIL 557
Score = 42.7 bits (99), Expect = 4e-04
Identities = 30/110 (27%), Positives = 52/110 (47%), Gaps = 5/110 (4%)
Query: 8 DLLDNFIDKETTNPSWKLSLPHVLVATITSFLFGYHLGVVNEPLESI-----SVDLGFNG 62
++ D+ + SW + + + LFGY +G + S+ S FN
Sbjct: 80 EVADSLASDAPESFSWSSVILPFIFPALGGLLFGYDIGATSGATLSLQSPALSGTTWFNF 139
Query: 63 NTLAEGLVVSICLGGALFGCLLSGWIADAVGRRRAFQLCALPMIIGAAMS 112
+ + GLVVS L GAL G + +AD +GRRR + A+ ++G+ ++
Sbjct: 140 SPVQLGLVVSGSLYGALLGSISVYGVADFLGRRRELIIAAVLYLLGSLIT 189
>At3g05150 putative sugar transporter
Length = 463
Score = 68.9 bits (167), Expect = 5e-12
Identities = 37/134 (27%), Positives = 77/134 (56%), Gaps = 7/134 (5%)
Query: 326 LYIFCTV*AISMIIQATG-ASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIF 384
+ I C + S +++A G A ++P L+V G+L+++ +F++G G +P ++++EIF
Sbjct: 336 MLIGCLLIGNSFLLKAHGLALDIIPA-----LAVSGVLVYIGSFSIGMGAIPWVIMSEIF 390
Query: 385 PSRIRAKAMAFCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVE 444
P ++ A V+W+ ++ V F L + ++ C++A+IF+ + V E
Sbjct: 391 PINLKGTAGGLVTVVNWLSSWLVSFTF-NFLMIWSPHGTFYVYGGVCVLAIIFIAKLVPE 449
Query: 445 TKGKSLQEIEIALL 458
TKG++L+EI+ ++
Sbjct: 450 TKGRTLEEIQAMMM 463
Score = 31.6 bits (70), Expect = 0.96
Identities = 25/87 (28%), Positives = 43/87 (48%), Gaps = 3/87 (3%)
Query: 22 SWKLSLPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLGGALFG 81
SW + L ++A S+ FG +G I +L + + + + SI GA+ G
Sbjct: 25 SWMVYLS-TIIAVCGSYEFGTCVGYSAPTQFGIMEELNLSYSQFS--VFGSILNMGAVLG 81
Query: 82 CLLSGWIADAVGRRRAFQLCALPMIIG 108
+ SG I+D +GR+ A +L ++ IG
Sbjct: 82 AITSGKISDFIGRKGAMRLSSVISAIG 108
>At2g43330 membrane transporter like protein
Length = 509
Score = 68.9 bits (167), Expect = 5e-12
Identities = 38/105 (36%), Positives = 58/105 (55%), Gaps = 2/105 (1%)
Query: 352 GALY--LSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGL 409
G LY L+V G+ L++ FA G GPVP + +EI+P + R +V+W+ N V
Sbjct: 372 GGLYGWLAVLGLALYIVFFAPGMGPVPWTVNSEIYPQQYRGICGGMSATVNWISNLIVAQ 431
Query: 410 LFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIE 454
FL + E G + + + A ++AVIFV V ET+G + E+E
Sbjct: 432 TFLTIAEAAGTGMTFLILAGIAVLAVIFVIVFVPETQGLTFSEVE 476
Score = 42.4 bits (98), Expect = 5e-04
Identities = 30/101 (29%), Positives = 45/101 (43%), Gaps = 1/101 (0%)
Query: 10 LDNFIDKETTNPSWKLSLPHVLVATITSFLFGYHLGVVNEPLESISVDLGF-NGNTLAEG 68
LD F ++ + L + A I LFGY GV++ L I D ++ +
Sbjct: 15 LDMFPERRMSYFGNSYILGLTVTAGIGGLLFGYDTGVISGALLYIKDDFEVVKQSSFLQE 74
Query: 69 LVVSICLGGALFGCLLSGWIADAVGRRRAFQLCALPMIIGA 109
+VS+ L GA+ G GWI D GR++A + GA
Sbjct: 75 TIVSMALVGAMIGAAAGGWINDYYGRKKATLFADVVFAAGA 115
>At4g35300 sugar transporter like protein
Length = 739
Score = 68.2 bits (165), Expect = 9e-12
Identities = 34/98 (34%), Positives = 54/98 (54%)
Query: 356 LSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLFLRLL 415
LS ++L+ F +G GP P +L +EIFP+R+R +A C W+ + V LL
Sbjct: 625 LSTVSVVLYFCFFVMGFGPAPNILCSEIFPTRVRGICIAICALTFWICDIIVTYSLPVLL 684
Query: 416 EKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEI 453
+ +G ++ M+A C ++ +FV V ETKG L+ I
Sbjct: 685 KSIGLAGVFGMYAIVCCISWVFVFIKVPETKGMPLEVI 722
Score = 35.4 bits (80), Expect = 0.067
Identities = 21/79 (26%), Positives = 37/79 (46%)
Query: 31 LVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLGGALFGCLLSGWIAD 90
+ A + + L G+ + + I + N EGL+V++ L GA SG +AD
Sbjct: 9 IAAAVGNLLQGWDNATIAGAVLYIKKEFNLESNPSVEGLIVAMSLIGATLITTCSGGVAD 68
Query: 91 AVGRRRAFQLCALPMIIGA 109
+GRR L ++ +G+
Sbjct: 69 WLGRRPMLILSSILYFVGS 87
>At1g30220 unknown protein
Length = 580
Score = 65.1 bits (157), Expect = 8e-11
Identities = 32/95 (33%), Positives = 52/95 (54%)
Query: 360 GMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLFLRLLEKLG 419
G+ L++ F+ G G VP ++ +EI+P R R + +W+ N V FL L E +G
Sbjct: 459 GLGLYIIFFSPGMGTVPWIVNSEIYPLRFRGICGGIAATANWISNLIVAQSFLSLTEAIG 518
Query: 420 AQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIE 454
+ +F ++A++FV V ETKG ++EIE
Sbjct: 519 TSWTFLIFGVISVIALLFVMVCVPETKGMPMEEIE 553
Score = 51.6 bits (122), Expect = 9e-07
Identities = 34/99 (34%), Positives = 51/99 (51%), Gaps = 7/99 (7%)
Query: 16 KETTNPSWKLSLPHVL----VATITSFLFGYHLGVVNEPLESISVDL-GFNGNTLAEGLV 70
KE + +WK P+VL A I LFGY GV++ L I D + NT + ++
Sbjct: 16 KECFSLTWKN--PYVLRLAFSAGIGGLLFGYDTGVISGALLYIRDDFKSVDRNTWLQEMI 73
Query: 71 VSICLGGALFGCLLSGWIADAVGRRRAFQLCALPMIIGA 109
VS+ + GA+ G + GW D +GRR A + ++GA
Sbjct: 74 VSMAVAGAIVGAAIGGWANDKLGRRSAILMADFLFLLGA 112
>At2g16130 putative sugar transporter
Length = 511
Score = 63.5 bits (153), Expect = 2e-10
Identities = 36/102 (35%), Positives = 62/102 (60%)
Query: 353 ALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLFL 412
A+ L+V ++ FV TF+LGAGPV + +EIFP R+RA+ + + ++ +++ +G+ FL
Sbjct: 385 AIGLAVTTVMTFVATFSLGAGPVTWVYASEIFPVRLRAQGASLGVMLNRLMSGIIGMTFL 444
Query: 413 RLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIE 454
L + L + +FA + A +F + ET+G L+EIE
Sbjct: 445 SLSKGLTIGGAFLLFAGVAVAAWVFFFTFLPETRGVPLEEIE 486
Score = 30.0 bits (66), Expect = 2.8
Identities = 22/81 (27%), Positives = 39/81 (47%), Gaps = 2/81 (2%)
Query: 31 LVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLGGALFGCLLSGWIAD 90
++A++TS + GY +GV++ I DL + L +++ I +L G +G +D
Sbjct: 31 ILASMTSIILGYDIGVMSGAAIFIKDDLKLSDVQLE--ILMGILNIYSLIGSGAAGRTSD 88
Query: 91 AVGRRRAFQLCALPMIIGAAM 111
+GRR L GA +
Sbjct: 89 WIGRRYTIVLAGFFFFCGALL 109
>At3g51490 sugar transporter-like protein
Length = 729
Score = 63.2 bits (152), Expect = 3e-10
Identities = 31/119 (26%), Positives = 60/119 (50%)
Query: 335 ISMIIQATGASTLLPTAGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMA 394
+S++ G+ L + +S + +++ F +G G +P +L +EIFP+ +R +
Sbjct: 590 LSLVTLVIGSLVNLGGSINALISTASVTVYLSCFVMGFGAIPNILCSEIFPTSVRGLCIT 649
Query: 395 FCMSVHWVINFFVGLLFLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEI 453
C W+ + V +L+ +G ++ ++A C +A +FV V ETKG L+ I
Sbjct: 650 ICALTFWICDIIVTYTLPVMLKSIGIAGVFGIYAIVCAVAWVFVYLKVPETKGMPLEVI 708
Score = 30.4 bits (67), Expect = 2.1
Identities = 20/65 (30%), Positives = 30/65 (45%)
Query: 31 LVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLGGALFGCLLSGWIAD 90
L A I + L G+ + + I + EGL+V++ L GA SG ++D
Sbjct: 9 LAAAIGNMLQGWDNATIAGAVIYIKKEFHLEKEPKIEGLIVAMSLIGATLITTFSGPVSD 68
Query: 91 AVGRR 95
VGRR
Sbjct: 69 KVGRR 73
>At4g36670 sugar transporter like protein
Length = 493
Score = 62.4 bits (150), Expect = 5e-10
Identities = 38/104 (36%), Positives = 59/104 (56%)
Query: 351 AGALYLSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLL 410
A AL LS+ FV F++G GP+ + +E+FP ++RA+ + ++V+ V+N V +
Sbjct: 373 AWALVLSIVAAYSFVAFFSIGLGPITWVYSSEVFPLKLRAQGASLGVAVNRVMNATVSMS 432
Query: 411 FLRLLEKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIE 454
FL L + + MFA +A F + ETKGKSL+EIE
Sbjct: 433 FLSLTSAITTGGAFFMFAGVAAVAWNFFFFLLPETKGKSLEEIE 476
Score = 46.2 bits (108), Expect = 4e-05
Identities = 41/140 (29%), Positives = 68/140 (48%), Gaps = 9/140 (6%)
Query: 24 KLSLPHVLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGL--VVSICLGGALFG 81
+ +L +VA+I S +FGY GV++ + I DL N + E L ++++C AL G
Sbjct: 15 RFALQCAIVASIVSIIFGYDTGVMSGAMVFIEEDLKTN-DVQIEVLTGILNLC---ALVG 70
Query: 82 CLLSGWIADAVGRRRAFQLCALPMIIGAAMSNKQLVWHACRKIICWNWLGLGPSRCCSLC 141
LL+G +D +GRR L ++ ++G+ + + C LG+G +
Sbjct: 71 SLLAGRTSDIIGRRYTIVLASILFMLGSILMGWGPNYPVLLSGRCTAGLGVG---FALMV 127
Query: 142 DRAYGALIQIATCFGILGSL 161
Y A I A+ G+L SL
Sbjct: 128 APVYSAEIATASHRGLLASL 147
>At1g75220 integral membrane protein, putative
Length = 487
Score = 61.2 bits (147), Expect = 1e-09
Identities = 32/99 (32%), Positives = 56/99 (56%), Gaps = 1/99 (1%)
Query: 356 LSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLFLRLL 415
LSV G++ V F+LG GP+P L+++EI P I+ A + +W ++ + + LL
Sbjct: 386 LSVVGVVAMVVFFSLGMGPIPWLIMSEILPVNIKGLAGSIATLANWFFSWLI-TMTANLL 444
Query: 416 EKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIE 454
+ ++++ C V+FV V ETKGK+L+E++
Sbjct: 445 LAWSSGGTFTLYGLVCAFTVVFVTLWVPETKGKTLEELQ 483
Score = 42.4 bits (98), Expect = 5e-04
Identities = 27/80 (33%), Positives = 44/80 (54%), Gaps = 4/80 (5%)
Query: 30 VLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVV-SICLGGALFGCLLSGWI 88
VL+ + FG+ G + +I+ DLG T++E V S+ GA+ G + SG I
Sbjct: 52 VLIVALGPIQFGFTCGYSSPTQAAITKDLGL---TVSEYSVFGSLSNVGAMVGAIASGQI 108
Query: 89 ADAVGRRRAFQLCALPMIIG 108
A+ +GR+ + + A+P IIG
Sbjct: 109 AEYIGRKGSLMIAAIPNIIG 128
>At1g19450 integral membrane protein, putative
Length = 488
Score = 61.2 bits (147), Expect = 1e-09
Identities = 32/99 (32%), Positives = 59/99 (59%), Gaps = 1/99 (1%)
Query: 356 LSVGGMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLFLRLL 415
+SV G++ V + +LG GP+P L+++EI P I+ A + ++W +++ V + +L
Sbjct: 387 VSVVGVVAMVISCSLGMGPIPWLIMSEILPVNIKGLAGSIATLLNWFVSWLV-TMTANML 445
Query: 416 EKLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIE 454
+ ++++A C V+FV V ETKGK+L+EI+
Sbjct: 446 LAWSSGGTFTLYALVCGFTVVFVSLWVPETKGKTLEEIQ 484
Score = 42.7 bits (99), Expect = 4e-04
Identities = 28/80 (35%), Positives = 44/80 (55%), Gaps = 4/80 (5%)
Query: 30 VLVATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVV-SICLGGALFGCLLSGWI 88
VL+ + FG+ G + +I+ DLG T++E V S+ GA+ G + SG I
Sbjct: 53 VLIVALGPIQFGFTCGYSSPTQAAITKDLGL---TVSEYSVFGSLSNVGAMVGAIASGQI 109
Query: 89 ADAVGRRRAFQLCALPMIIG 108
A+ VGR+ + + A+P IIG
Sbjct: 110 AEYVGRKGSLMIAAIPNIIG 129
>At3g20460 sugar transporter, putative
Length = 488
Score = 60.8 bits (146), Expect = 1e-09
Identities = 38/123 (30%), Positives = 66/123 (52%), Gaps = 10/123 (8%)
Query: 342 TGASTLLPTAGAL--YLSVG---GMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFC 396
TG S L + G L Y + G+L+F+ + +G G +P ++++E+ P I+ A C
Sbjct: 367 TGLSFLFQSYGLLEHYTPISTFMGVLVFLTSITIGIGGIPWVMISEMTPINIKGSAGTLC 426
Query: 397 MSVHWVINFFVGLLFLRLLE--KLGAQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIE 454
W N+FV F L + G +Y+M + + ++FV + V ET+G+SL+EI+
Sbjct: 427 NLTSWSSNWFVSYTFNFLFQWSSSGVFFIYTMISG---VGILFVMKMVPETRGRSLEEIQ 483
Query: 455 IAL 457
A+
Sbjct: 484 AAI 486
>At1g08920 putative sugar transport protein
Length = 470
Score = 60.1 bits (144), Expect = 3e-09
Identities = 29/98 (29%), Positives = 56/98 (56%), Gaps = 1/98 (1%)
Query: 360 GMLLFVFTFALGAGPVPGLLLTEIFPSRIRAKAMAFCMSVHWVINFFVGLLFLRLLEKLG 419
G++ F+ +FA+G G +P ++++EIFP ++ A +W + V + +LE
Sbjct: 369 GVVGFISSFAVGMGGLPWIIMSEIFPMNVKVSAGTLVTLANWSFGWIVAFAYNFMLE-WN 427
Query: 420 AQLLYSMFATFCIMAVIFVKRNVVETKGKSLQEIEIAL 457
A + +F T C ++F+ V ETKG++L++I+ +L
Sbjct: 428 ASGTFLIFFTICGAGIVFIYAMVPETKGRTLEDIQASL 465
Score = 30.8 bits (68), Expect = 1.6
Identities = 25/89 (28%), Positives = 36/89 (40%), Gaps = 4/89 (4%)
Query: 32 VATITSFLFGYHLGVVNEPLESISVDLGFNGNTLAEGLVVSICLGGALFGCLLSGWIADA 91
V+ SF FG G + I DLG + + SI G + G + SG +AD
Sbjct: 38 VSVCGSFCFGCAAGYSSVAQTGIINDLGLS--VAQYSMFGSIMTFGGMIGAIFSGKVADL 95
Query: 92 VGRRRAFQLCALPMIIG--AAMSNKQLVW 118
+GR+ + I G A K +W
Sbjct: 96 MGRKGTMWFAQIFCIFGWVAVALAKDSMW 124
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.341 0.151 0.500
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,420,656
Number of Sequences: 26719
Number of extensions: 374489
Number of successful extensions: 1382
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 56
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 1230
Number of HSP's gapped (non-prelim): 121
length of query: 461
length of database: 11,318,596
effective HSP length: 103
effective length of query: 358
effective length of database: 8,566,539
effective search space: 3066820962
effective search space used: 3066820962
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC122722.6


