

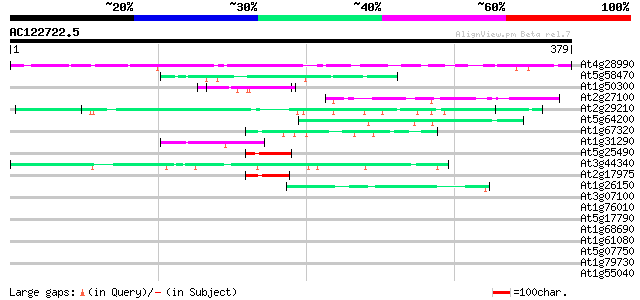
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122722.5 - phase: 0
(379 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g28990 unknown protein 216 2e-56
At5g58470 RNA/ssDNA-binding protein - like 61 1e-09
At1g50300 unknown protein 55 5e-08
At2g27100 unknown protein 50 3e-06
At2g29210 proline-rich protein like 49 3e-06
At5g64200 unknown protein 47 1e-05
At1g67320 unknown protein 47 2e-05
At1g31290 hypothetical protein 46 4e-05
At5g25490 unknown protein 44 1e-04
At3g44340 putative protein 44 1e-04
At2g17975 putative protein 42 4e-04
At1g26150 Pto kinase interactor, putative 42 4e-04
At3g07100 putative Sec24-like COPII protein 41 0.001
At1g76010 unknown protein 41 0.001
At5g17790 putative protein 40 0.003
At1g68690 protein kinase, putative 40 0.003
At1g61080 hypothetical protein 39 0.004
At5g07750 putative protein 38 0.008
At1g79730 unknown protein 38 0.010
At1g55040 putative protein 38 0.010
>At4g28990 unknown protein
Length = 347
Score = 216 bits (550), Expect = 2e-56
Identities = 166/403 (41%), Positives = 197/403 (48%), Gaps = 80/403 (19%)
Query: 1 MSSTSPSQSPPSHHHHHQPLLSSLVVRPSLTENSPPGAAAHSNDYEPGELRRDPPPVNYS 60
M ST Q+ HH P +SSLVVRPS + + G A + DYEPGE+ RD P +++
Sbjct: 1 MGSTEKEQTT----QHHPPHISSLVVRPSGSNDREDGRNA-AGDYEPGEVSRDRP--SFN 53
Query: 61 RSDRYS-DDAGYRFRAGSSSPVHHRRDADHRFPSDYNHF---PRNRGFGGGRDF-GRFRD 115
RSDRY D+ G+R RA SSSP R DH+ SD NH PR R R+ GR RD
Sbjct: 54 RSDRYKGDNGGHRTRASSSSP-GRRGYEDHKHGSDLNHSGVPPRGRELSSRREAPGRHRD 112
Query: 116 PPHPYARGRPGGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPREGDWMCPDALCGNLN 175
P ARG G RP+ R DGP HGR +G+ N V+PREGDW C D LC NLN
Sbjct: 113 YSPPLARGGAGARPYRRGLDGPE--PPHGR-DGMSRNNISKVQPREGDWYCLDPLCRNLN 169
Query: 176 FARRDFCNQCKRPRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLM 235
FARR+ C +CKR R A A SPP PR P PP + SP R NGYRSPP
Sbjct: 170 FARRESCYKCKRHRYAPANSPPL---------PRLLP-PPMNHSPRRDFNGYRSPP---- 215
Query: 236 GRDGLRDYGPAAAALPPLRHEGRFPDPHLHRERMDYMDDAYRGRGKFDRPPPLDWDNRDR 295
R RDY P P R R+R Y D Y R DW + +
Sbjct: 216 -RGWPRDYPPPRHDHPTWRDR--------ERDRPHYSDHEYPP----SRRIASDWAHTE- 261
Query: 296 GRDGFSNERKGFERRPLSPSAPLLPSLPPHRGGDRWSRDVRDRSRS---------PIRGG 346
+ ++RR P L P RGG RW R +R+RSRS P+R G
Sbjct: 262 -----PLPKPHYDRR---------PPLSPPRGG-RWGRVLRERSRSPPLRDVPPPPLRDG 306
Query: 347 PPA----------KDYRRDPVMSRGGRDDRRGGVGRDRIGGMY 379
PP +DYRRD R RDD RG GR R+G Y
Sbjct: 307 PPPSLRGGGPPLHRDYRRDSHFDRERRDDGRG--GRGRMGNSY 347
>At5g58470 RNA/ssDNA-binding protein - like
Length = 422
Score = 60.8 bits (146), Expect = 1e-09
Identities = 59/192 (30%), Positives = 72/192 (36%), Gaps = 45/192 (23%)
Query: 103 GFGGGRDFGRFRDPPHPYARGRPGGRPFG-------------RAFDGPGY--GHRHGRGE 147
G+GGG +G D + RG GG +G R G GY G R GRG
Sbjct: 22 GYGGGGGYGG-GDAGYG-GRGASGGGSYGGRGGYGGGGGRGNRGGGGGGYQGGDRGGRGS 79
Query: 148 GLHGRNNPNVRPREGDWMCPDALCGNLNFARRDFCNQCKRPRPAAAGSPPR--------- 198
G GR +GDW CP+ CGN+NFARR CN+C P+ S
Sbjct: 80 GGGGR--------DGDWRCPNPSCGNVNFARRVECNKCGALAPSGTSSGANDRGGGGYSR 131
Query: 199 --------RGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDGLRDYGPAAAAL 250
G + R Y +D R Y S R G G P AAA+
Sbjct: 132 GGGDSDRGGGRGGRNDSGRSYESSRYD-GGSRSGGSYGSGSQRENGSYG--QAPPPAAAI 188
Query: 251 PPLRHEGRFPDP 262
P G +P P
Sbjct: 189 PSYDGSGSYPPP 200
Score = 31.2 bits (69), Expect = 0.98
Identities = 25/84 (29%), Positives = 31/84 (36%), Gaps = 6/84 (7%)
Query: 116 PPHPYARGRP--GGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPREGDWMCPDALCGN 173
PP Y+ G P GG G D P G R GR G G + PR + DA
Sbjct: 210 PPTSYSGGPPSYGGPRGGYGSDAPSTGGRGGRSGGYDGGS----APRRQEASYEDAATEK 265
Query: 174 LNFARRDFCNQCKRPRPAAAGSPP 197
+ D + C R + PP
Sbjct: 266 VKQCDADCDDNCDNARIYISNLPP 289
>At1g50300 unknown protein
Length = 372
Score = 55.5 bits (132), Expect = 5e-08
Identities = 27/62 (43%), Positives = 36/62 (57%), Gaps = 3/62 (4%)
Query: 134 FDGPGYGHRHGRGEGLHGRNNPNVRP--REGDWMCPDALCGNLNFARRDFCNQCKRPRPA 191
FDG G G G G+ + + +P ++GDWMCP+ C N+NFA R CN+C RPA
Sbjct: 108 FDGGAEETNGGAGRG-RGQADSSAKPWQQDGDWMCPNTSCTNVNFAFRGVCNRCGTARPA 166
Query: 192 AA 193
A
Sbjct: 167 GA 168
Score = 43.9 bits (102), Expect = 1e-04
Identities = 26/70 (37%), Positives = 33/70 (47%), Gaps = 9/70 (12%)
Query: 128 RPFGRAFDGPGYGHRHGRGEGLHGR---NNPNVRPR----EGDWMCPDALCGNLNFARRD 180
RP G + G G GRG G G P+ P DW CP +CGN+N+A+R
Sbjct: 164 RPAGASGGSMGAGRGRGRGGGADGGAPGKQPSGAPTGLFGPNDWACP--MCGNVNWAKRL 221
Query: 181 FCNQCKRPRP 190
CN C +P
Sbjct: 222 KCNICNTNKP 231
>At2g27100 unknown protein
Length = 720
Score = 49.7 bits (117), Expect = 3e-06
Identities = 56/165 (33%), Positives = 69/165 (40%), Gaps = 44/165 (26%)
Query: 214 PPFD----RSPERPMNGYRSPPPRLMGRDGLRDYGPAAAALPPLRHEGRFPDPHLHRERM 269
PP D R PE+ + SPPP P +++LP E L RER
Sbjct: 7 PPSDSVDNRLPEKSTSS--SPPPP-----------PPSSSLPQQEQEQDQQQLPLRRER- 52
Query: 270 DYMDDAYRGRGKFD--RPPPLDWDNRDRGRDGFSNERKGFERRP-LSPSAPLLPSLPPHR 326
D+ R + D RPPP + R+R R R+ ++RRP LSP PP+R
Sbjct: 53 ----DSRERRDERDIERPPP---NRRERDRSPLPPPRRDYKRRPSLSPP-------PPYR 98
Query: 327 GGDRWSRDVRDRSRSPIRGGPPAKDYRRDPVMSRGGRDDRRGGVG 371
DR R P R PP K YRRD G R RGG G
Sbjct: 99 --DR-------RHSPPQRRSPPQKRYRRDDNGYDGRRGSPRGGYG 134
Score = 33.9 bits (76), Expect = 0.15
Identities = 43/127 (33%), Positives = 47/127 (36%), Gaps = 25/127 (19%)
Query: 27 RPSLTENSPPGAAAHSNDYEPGELRRDPPPVNYSRSDRYSDDAGYRFRAGSSSPVHHRRD 86
RPSL+ P HS RR PP Y R DD GY R G SP
Sbjct: 88 RPSLSPPPPYRDRRHSPPQ-----RRSPPQKRYRR-----DDNGYDGRRG--SPRGGYGP 135
Query: 87 ADHRFPSDYNHFPRNRGFGGGRDFGRFRDPPHPYARGRPGGRPFGRAFDGPGYGHRHGRG 146
D RF D+ GGG D R Y RP GR GR D G R G G
Sbjct: 136 PDRRFGYDH---------GGGYD--REMGGRPGYGDERPHGRFMGRYQDWE--GGRGGYG 182
Query: 147 EGLHGRN 153
+ + N
Sbjct: 183 DASNSGN 189
>At2g29210 proline-rich protein like
Length = 878
Score = 49.3 bits (116), Expect = 3e-06
Identities = 91/307 (29%), Positives = 104/307 (33%), Gaps = 58/307 (18%)
Query: 49 ELRRD--PP--------PVNYSRSDRYSDDAGYRFRAGSSSPVHHRRDADHRFPSDYNHF 98
E RRD PP P+ SRS S + +GS S R+ SD +
Sbjct: 194 ERRRDLIPPRRGDASRSPLRGSRSRSIS-----KTNSGSKSYSGERKSRSTSQSSDASIS 248
Query: 99 PRNRGFGGGRDFGRFRDPPHPYA-RGRPGGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNV 157
PR R R R R + R R PF P HR EG P+
Sbjct: 249 PRKRRLSNSRRRSRSRSVRRSLSPRRRRIHSPFRSRSRSPIRRHRRPTHEGRRQSPAPSR 308
Query: 158 RPREGDWMCPDALCGNLNFARRDFCNQCKRPRPAAAGSPP--RRGSPPLHAPPRRYPGPP 215
R R P A R + SPP RR SP A R P PP
Sbjct: 309 RRRSPS---PPA-------------------RRRRSPSPPARRRRSPSPPARRHRSPTPP 346
Query: 216 FD--RSPERPMNGYRSPPPRLMGRD----GLRDYGPAAAAL-----PPLRHEGRFPDPHL 264
RSP P +RSPPP R R P+ A PL R P P L
Sbjct: 347 ARQRRSPSPPARRHRSPPPARRRRSPSPPARRRRSPSPPARRRRSPSPLYRRNRSPSP-L 405
Query: 265 HRERMDYMDDAYRGRGKFD--RPPPLDWDNRDRGRDGFSNERKGFERR-PLSPSAPLLPS 321
+R A RGR P P+ R R G E+R P P A LPS
Sbjct: 406 YRRNRSRSPLAKRGRSDSPGRSPSPV---ARLRDPTGARLPSPSIEQRLPSPPVAQRLPS 462
Query: 322 LPPHRGG 328
PP R G
Sbjct: 463 PPPRRAG 469
Score = 44.3 bits (103), Expect = 1e-04
Identities = 94/372 (25%), Positives = 121/372 (32%), Gaps = 42/372 (11%)
Query: 5 SPSQSPPSHHHHHQPLLSSLVVRPSLTENSP-PGAAAHSNDYEPGELRRDP-PPVNYSRS 62
S S+SP H PS SP P A + P RR P PP RS
Sbjct: 283 SRSRSPIRRHRRPTHEGRRQSPAPSRRRRSPSPPARRRRSPSPPARRRRSPSPPARRHRS 342
Query: 63 DRYSDDAGYRFRAGSSSPVHHRRDADHRFPSDYNHFPRNRGFGGGRDFGRFRDPPHPYAR 122
R R S P HR P P R R R P P AR
Sbjct: 343 PT----PPARQRRSPSPPARR-----HRSP------PPARRRRSPSPPARRRRSPSPPAR 387
Query: 123 GRPGGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPREGDWMCPDALCGNLNFARRDFC 182
R P R P +R R + + P G P A + AR
Sbjct: 388 RRRSPSPLYRRNRSPSPLYRRNRSRSPLAKRGRSDSP--GRSPSPVARLRDPTGARLPSP 445
Query: 183 NQCKR-PRPAAA----GSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGR 237
+ +R P P A PPRR P P +R P PP P P R+ G
Sbjct: 446 SIEQRLPSPPVAQRLPSPPPRRAGLPSPPPAQRLPSPP-------PRRAGLPSPMRIGGS 498
Query: 238 DGLRDY-GPAAAALPPLRHEGRFPDPHLHRERMDYMDD---AYRGRGKFDRPPPLDWDN- 292
P+ ++L P + P P + R R D+ + G+ P + D
Sbjct: 499 HAANHLESPSPSSLSPPGRKKVLPSPPVRRRRSLTPDEERVSLSQGGRHTSPSHIKQDGS 558
Query: 293 ----RDRGRDGFSNERKGFERRPLSPSAPLLPSLPPHRGGDRWSRDVRDRSRSPIRGGPP 348
R RG+ S+ R R P+ +P + R R R RSP P
Sbjct: 559 MSPVRGRGKSSPSS-RHQKARSPVRRRSPTPVNRRSRRSSSASRSPDRRRRRSPSSSRSP 617
Query: 349 AKDYRRDPVMSR 360
++ R PV+ R
Sbjct: 618 SRS-RSPPVLHR 628
>At5g64200 unknown protein
Length = 303
Score = 47.4 bits (111), Expect = 1e-05
Identities = 51/179 (28%), Positives = 62/179 (34%), Gaps = 30/179 (16%)
Query: 196 PPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDGLR--DYGPAAAALPPL 253
PP + PRR P RSP R + RS PR RD R DY + +
Sbjct: 109 PPPKSRRSRSRSPRRSRSPRRSRSPPRRRSPRRSRSPRRRSRDDYREKDYRKRSRSRSYD 168
Query: 254 RHEGRFPDPHLHRERMDYM----DDAYRGRGKFDR---------------------PPPL 288
R E HR R D+ R RG++D P
Sbjct: 169 RRERHEEKDRDHRRRTRSRSASPDEKRRVRGRYDNESRSHSRSLSASPARRSPRSSSPQK 228
Query: 289 DWDNRDRGRDGFSNERKGFERRPLSPSAPLLPSLPPHRGGDRWSRDVRDRSRSPIRGGP 347
R+ D SNER RR LSP +P L P + S + R RSP G P
Sbjct: 229 TSPAREVSPDKRSNERSPSPRRSLSPRSPALQKASPSK---EMSPERRSNERSPSPGSP 284
>At1g67320 unknown protein
Length = 856
Score = 47.0 bits (110), Expect = 2e-05
Identities = 48/157 (30%), Positives = 60/157 (37%), Gaps = 46/157 (29%)
Query: 160 REGDWMCPDALCGNLNFARRDFCN--QCKRPRPA-----AAGSPPRR------------- 199
RE DW+CP CGN+NF+ R CN C +PRPA +A P +
Sbjct: 22 REDDWICPS--CGNVNFSFRTTCNMRNCTQPRPADHNGKSAPKPMQHQQGFSSPGAYLGS 79
Query: 200 -GSPPLHAPPRRYPGPPFDRSPERPMNGYRSPP---PRLMGRDGLRDYG---PAAAALPP 252
G PP++ Y P F NG PP P G +Y PA A P
Sbjct: 80 GGPPPVYMGGSPYGSPLF--------NGSSMPPYDVPFSGGSPYHFNYNSRMPAGAHYRP 131
Query: 253 LRHEGRFPDPHLHRERMDYMDDAYRGRGKFDRPPPLD 289
L G P P+ M G + PPP+D
Sbjct: 132 LHMSG--PPPYHGGSMMG-------SGGMYGMPPPID 159
Score = 41.2 bits (95), Expect = 0.001
Identities = 47/156 (30%), Positives = 57/156 (36%), Gaps = 28/156 (17%)
Query: 54 PPPVNYSRSDRYSDDAGYRFRAGSSSPVHHRRDADHRFPSDYNHFPRNRGFGGGRDFGRF 113
PPPV Y Y G GSS P + D S Y HF N G +
Sbjct: 82 PPPV-YMGGSPY----GSPLFNGSSMPPY---DVPFSGGSPY-HFNYNSRMPAGAHYRPL 132
Query: 114 R-DPPHPYARGRP--GGRPFGRA--FDGPGYGHRHGRGEGLHGRNNPNVRP--------- 159
P PY G G +G D G G G G P P
Sbjct: 133 HMSGPPPYHGGSMMGSGGMYGMPPPIDRYGLGMAMGPGSAAAMMPRPRFYPDEKSQKTDS 192
Query: 160 -REGDWMCPDALCGNLNFARRDFCN--QCKRPRPAA 192
R+ DW CP+ CGN+NF+ R CN +C P+P +
Sbjct: 193 TRDNDWTCPN--CGNVNFSFRTVCNMRKCNTPKPGS 226
Score = 32.3 bits (72), Expect = 0.44
Identities = 16/42 (38%), Positives = 22/42 (52%), Gaps = 6/42 (14%)
Query: 161 EGDWMCPDALCGNLNFARRDFCNQ----CKRPRPAAAGSPPR 198
EG W C + CGN+N+ R CN+ +P + GSP R
Sbjct: 242 EGSWKCDN--CGNINYPFRSKCNRQNCGADKPGDRSNGSPSR 281
>At1g31290 hypothetical protein
Length = 1194
Score = 45.8 bits (107), Expect = 4e-05
Identities = 30/74 (40%), Positives = 34/74 (45%), Gaps = 5/74 (6%)
Query: 103 GFGGGRDFGRFRDPPHPYARGRPGGRPFGRAFDGPGYGHRHG----RGEGLHGRNNPNVR 158
G GGG D GR RGR GG GR + G G GH G RG G GR V+
Sbjct: 51 GRGGGGDRGRGYSGRGD-GRGRGGGGDRGRGYSGRGDGHGRGGGGDRGRGYSGRGRGFVQ 109
Query: 159 PREGDWMCPDALCG 172
R+G W+ P G
Sbjct: 110 DRDGGWVNPGQSSG 123
Score = 34.3 bits (77), Expect = 0.12
Identities = 27/74 (36%), Positives = 32/74 (42%), Gaps = 10/74 (13%)
Query: 100 RNRGFGG-GRDFGRFRDPPHPYA-----RGRPGGRPFGRAFDGPGYGHRHG----RGEGL 149
R RG+ G G GR D Y+ GR GG GR + G G G G RG G
Sbjct: 22 RGRGYSGRGDGRGRGGDGDRGYSGRGDGHGRGGGGDRGRGYSGRGDGRGRGGGGDRGRGY 81
Query: 150 HGRNNPNVRPREGD 163
GR + + R GD
Sbjct: 82 SGRGDGHGRGGGGD 95
Score = 34.3 bits (77), Expect = 0.12
Identities = 24/63 (38%), Positives = 27/63 (42%), Gaps = 1/63 (1%)
Query: 102 RGFGGGRDFGR-FRDPPHPYARGRPGGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPR 160
RG GGG D GR + RG G R + DG G G RG G GR + R
Sbjct: 14 RGRGGGGDRGRGYSGRGDGRGRGGDGDRGYSGRGDGHGRGGGGDRGRGYSGRGDGRGRGG 73
Query: 161 EGD 163
GD
Sbjct: 74 GGD 76
>At5g25490 unknown protein
Length = 170
Score = 44.3 bits (103), Expect = 1e-04
Identities = 18/31 (58%), Positives = 21/31 (67%), Gaps = 2/31 (6%)
Query: 160 REGDWMCPDALCGNLNFARRDFCNQCKRPRP 190
R GDW C LC +LNF RRD C +C+ PRP
Sbjct: 3 RPGDWNC--RLCSHLNFQRRDSCQRCREPRP 31
Score = 39.3 bits (90), Expect = 0.004
Identities = 15/37 (40%), Positives = 20/37 (53%)
Query: 160 REGDWMCPDALCGNLNFARRDFCNQCKRPRPAAAGSP 196
+ GDW+CP + C NFA R C +C P+ A P
Sbjct: 133 KSGDWICPRSGCNEHNFASRSECFRCNAPKELATEPP 169
Score = 36.2 bits (82), Expect = 0.030
Identities = 27/87 (31%), Positives = 36/87 (41%), Gaps = 19/87 (21%)
Query: 126 GGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPREGDWMCPDALCGNLNFARRDFCNQC 185
GGRP +F G G P+VRP GDW C CG NFA R C +C
Sbjct: 43 GGRPVSSSF-----GFNTG----------PDVRP--GDWYCNLGDCGTHNFANRSSCFKC 85
Query: 186 KRPRP--AAAGSPPRRGSPPLHAPPRR 210
+ + + + G ++ PRR
Sbjct: 86 GAAKDEFSCSSAAATTGFMDMNVGPRR 112
>At3g44340 putative protein
Length = 1122
Score = 43.9 bits (102), Expect = 1e-04
Identities = 84/345 (24%), Positives = 113/345 (32%), Gaps = 70/345 (20%)
Query: 1 MSSTSPSQSPPSHHHHHQ-PLLSSLVVRPS-LTENSPPGAAAHSNDYEPGELRRDP--PP 56
M + P SPP P RPS + PP AA + P ++ + PP
Sbjct: 52 MPGSGPRPSPPFGQSPQSFPQQQQQQPRPSPMARPGPPPPAAMARPGGPPQVSQPGGFPP 111
Query: 57 VNYSRSDRYSDDAGYRFRAGSSSPVHHRRDADHRFPSDYNHFPRNRGF------------ 104
V G S+ P R + + FP+ GF
Sbjct: 112 V------------GRPVAPPSNQPPFGGRPSTGPLVGGGSSFPQPGGFPASGPPGGVPSG 159
Query: 105 --GGGRDFGRFRDPPHPYARGR----PGGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVR 158
G R G F PP P G P G P G +GP HG H N P
Sbjct: 160 PPSGARPIG-FGSPP-PMGPGMSMPPPSGMPGGPLSNGPPPSGMHGG----HLSNGPPPS 213
Query: 159 PREGDWMC---PDALCGNLNFAR-RDFCNQCKRPRPAAAGSPPRRG---------SPPLH 205
G + P + G F R F + P G PP G SPP H
Sbjct: 214 GMPGGPLSNGPPPPMMGPGAFPRGSQFTSGPMMAPPPPYGQPPNAGPFTGNSPLSSPPAH 273
Query: 206 A--PPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDG----------LRDYGPAAAALPPL 253
+ PP +PG P+ R P Y +PP +L G +++ + + P
Sbjct: 274 SIPPPTNFPGVPYGRPPMPGGFPYGAPPQQLPSAPGTPGSIYGMGPMQNQSMTSVSSPSK 333
Query: 254 RHEGRFPDPHLHRERMDYMDDAYRGRGKFDRPPP--LDWDNRDRG 296
+ P P + Y R K + PPP +D+ RD G
Sbjct: 334 IDLNQIPRPGSSSSPIVY---ETRVENKANPPPPTTVDYITRDTG 375
Score = 35.8 bits (81), Expect = 0.040
Identities = 30/90 (33%), Positives = 35/90 (38%), Gaps = 11/90 (12%)
Query: 172 GNLNFARRDFCN-QCKRPRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSP 230
GN N + N RP P GS PR SPP P+ +P + PM P
Sbjct: 31 GNPNSLAANMQNLNINRPPPPMPGSGPRP-SPPFGQSPQSFPQQQQQQPRPSPMARPGPP 89
Query: 231 PPRLMGRDGLRDYGPAAAALPPLRHEGRFP 260
PP M R G GP P + G FP
Sbjct: 90 PPAAMARPG----GP-----PQVSQPGGFP 110
>At2g17975 putative protein
Length = 268
Score = 42.4 bits (98), Expect = 4e-04
Identities = 18/30 (60%), Positives = 21/30 (70%), Gaps = 2/30 (6%)
Query: 160 REGDWMCPDALCGNLNFARRDFCNQCKRPR 189
REGDW C C N N+A R FCN+CK+PR
Sbjct: 5 REGDWECLG--CRNRNYAFRSFCNRCKQPR 32
Score = 37.7 bits (86), Expect = 0.010
Identities = 16/30 (53%), Positives = 20/30 (66%), Gaps = 2/30 (6%)
Query: 160 REGDWMCPDALCGNLNFARRDFCNQCKRPR 189
R+GDWMC + C N N+A R CN+CK R
Sbjct: 228 RDGDWMCTN--CKNHNYASRAECNRCKTTR 255
Score = 35.8 bits (81), Expect = 0.040
Identities = 17/47 (36%), Positives = 26/47 (55%), Gaps = 4/47 (8%)
Query: 149 LHGRNNPNVR--PREGDWMCPDALCGNLNFARRDFCNQCKRPRPAAA 193
+ +PN + PR GDW+C C N N+A R+ C +C + + AA
Sbjct: 35 MDNNTSPNSKWLPRIGDWICTG--CTNNNYASREKCKKCGQSKEVAA 79
>At1g26150 Pto kinase interactor, putative
Length = 760
Score = 42.4 bits (98), Expect = 4e-04
Identities = 38/139 (27%), Positives = 49/139 (34%), Gaps = 33/139 (23%)
Query: 188 PRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDGLRDYGPAA 247
P P + PP + PP H+PPR P PP P PPPR + +
Sbjct: 160 PDPPSNPLPPPKLVPPSHSPPRHLPSPPASEIP---------PPPRHLPSPPASE----R 206
Query: 248 AALPPLRHEGRFPDPHLHRERMDYMDDAYRGRGKFDRPPPLDWDNRDRGRDGFSNERKGF 307
+ PP E P P H +R + R P P D
Sbjct: 207 PSTPPSDSEHPSPPPPGHPKRREQPPPPGSKRPTPSPPSPSD------------------ 248
Query: 308 ERRPLSPSAPLLP--SLPP 324
+RP+ PS P P +LPP
Sbjct: 249 SKRPVHPSPPSPPEETLPP 267
Score = 34.7 bits (78), Expect = 0.089
Identities = 17/45 (37%), Positives = 20/45 (43%), Gaps = 2/45 (4%)
Query: 188 PRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPP 232
P P+ G PP + P+ PP P PP P N SPPP
Sbjct: 78 PSPSLTGPPPT--TIPVSPPPEPSPPPPLPTEAPPPANPVSSPPP 120
Score = 33.5 bits (75), Expect = 0.20
Identities = 16/47 (34%), Positives = 20/47 (42%)
Query: 186 KRPRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPP 232
+ P P G P RR PP R P PP +RP++ PP
Sbjct: 215 EHPSPPPPGHPKRREQPPPPGSKRPTPSPPSPSDSKRPVHPSPPSPP 261
Score = 28.1 bits (61), Expect = 8.3
Identities = 22/70 (31%), Positives = 27/70 (38%), Gaps = 8/70 (11%)
Query: 188 PRPAAAGSP--PRRGSPPLHAPPRRYPGPPFDRSPERPMNG---YRSPPPRLMGRDGLRD 242
P P A P P PP +PP P PP + P P+ +PPP L
Sbjct: 103 PLPTEAPPPANPVSSPPPESSPP---PPPPTEAPPTTPITSPSPPTNPPPPPESPPSLPA 159
Query: 243 YGPAAAALPP 252
P + LPP
Sbjct: 160 PDPPSNPLPP 169
Score = 28.1 bits (61), Expect = 8.3
Identities = 14/45 (31%), Positives = 20/45 (44%), Gaps = 2/45 (4%)
Query: 188 PRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPP 232
P+P+ P SPP PP PP P + ++G +P P
Sbjct: 268 PKPSPDPLPSNSSSPPTLLPPSSVVSPP--SPPRKSVSGPDNPSP 310
>At3g07100 putative Sec24-like COPII protein
Length = 1054
Score = 41.2 bits (95), Expect = 0.001
Identities = 38/157 (24%), Positives = 55/157 (34%), Gaps = 19/157 (12%)
Query: 188 PRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRS--------PERPMNGYRSPPPRLMGRDG 239
P P++ P PP AP +R+P PPF + P + + G+ SPP L +
Sbjct: 90 PPPSSNSFPSPAYGPPGGAPFQRFPSPPFPTTQNPPQGPPPPQTLAGHLSPPMSLRPQQP 149
Query: 240 LRDYGPAAAALPPLRHEGRFPDPHLHRERMDYMDDAYRGRGKFDRPPPLDWDNRDRGRDG 299
+ P A PP P + + DY A G + P + +
Sbjct: 150 M---APVAMGPPPQSTTSGLPGANAYPPATDYHMPARPGFQQSMPPVTPSYPGVGGSQPS 206
Query: 300 FS--------NERKGFERRPLSPSAPLLPSLPPHRGG 328
F F+ P P + S PPH GG
Sbjct: 207 FPGYPSKQVLQAPTPFQTSQGPPGPPPVSSYPPHTGG 243
Score = 30.8 bits (68), Expect = 1.3
Identities = 20/57 (35%), Positives = 22/57 (38%), Gaps = 1/57 (1%)
Query: 74 RAGSSSPVHHRRDADHRFPSDYNHFPRNR-GFGGGRDFGRFRDPPHPYARGRPGGRP 129
R G SP + P N FP G GG F RF PP P + P G P
Sbjct: 73 RPGQPSPFVSQIPGSRPPPPSSNSFPSPAYGPPGGAPFQRFPSPPFPTTQNPPQGPP 129
>At1g76010 unknown protein
Length = 350
Score = 40.8 bits (94), Expect = 0.001
Identities = 51/196 (26%), Positives = 64/196 (32%), Gaps = 49/196 (25%)
Query: 123 GRPGGRPFGRAFDGPGYGHRHGRGEGLHGRNNPNVRPREGDWMCPDALCGNLNFARRDFC 182
G PGGR GR G G GRG G G NV +G W R+
Sbjct: 146 GSPGGRGRGR-----GRGRGRGRGRGGRGNAYVNVEHEDGGW-------------ERE-- 185
Query: 183 NQCKRPRPAAAGSPPR-RGSPPLHAPPRRYPGP---------PFDRSPERPMNGYRSPPP 232
R R G R RG + PP Y P P + GY +PP
Sbjct: 186 QSYGRGRGRGRGRSSRGRGRGGYNGPPNEYDAPQDGGYGYDAPHEHRGYDDRGGYDAPPQ 245
Query: 233 RLMGRDGLRDYGPAAAALPPLRHEGRFPDPHLHRERMDYMDDAYRGRGKFDRPPP--LDW 290
G DG + G + P R D +GRG +D P +
Sbjct: 246 GRGGYDGPQG-------------RGGYDGPQGRRG----YDGPPQGRGGYDGPSQGRGGY 288
Query: 291 DNRDRGRDGFSNERKG 306
D +GR G+ +G
Sbjct: 289 DGPSQGRGGYDGPSQG 304
Score = 32.7 bits (73), Expect = 0.34
Identities = 39/134 (29%), Positives = 48/134 (35%), Gaps = 11/134 (8%)
Query: 33 NSPPGAAAHSNDYEP---GELRRDPPPVNYSRSDRYSDDAGYRFRAGSSSPVHHRRDADH 89
N PP N+Y+ G D P + DR DA + R G P
Sbjct: 209 NGPP------NEYDAPQDGGYGYDAPHEHRGYDDRGGYDAPPQGRGGYDGPQGRGGYDGP 262
Query: 90 RFPSDYNHFPRNRGFGGGRDFGRFRDPPHPYARGRPGGRPFGRA-FDGPGYGHRHGRGEG 148
+ Y+ P+ RG G GR RG G GR +DGP G GRG G
Sbjct: 263 QGRRGYDGPPQGRGGYDGPSQGRGGYDGPSQGRGGYDGPSQGRGGYDGP-QGRGRGRGRG 321
Query: 149 LHGRNNPNVRPREG 162
GR R +G
Sbjct: 322 RGGRGRGGGRGGDG 335
Score = 28.9 bits (63), Expect = 4.9
Identities = 53/222 (23%), Positives = 63/222 (27%), Gaps = 70/222 (31%)
Query: 98 FPRNRGFGGGRDF-----GRFRDPPHPYARGRPGGRPFGRAFDGPGYGHRHGRGEGLHGR 152
+ R RG G GR G + PP+ Y + GG + + GY R G GR
Sbjct: 188 YGRGRGRGRGRSSRGRGRGGYNGPPNEYDAPQDGGYGYDAPHEHRGYDDRGGYDAPPQGR 247
Query: 153 NNPNVRPREGDWMCPDALCGNLNFARRDFCNQCKRPRPAAAGSPPRRGSPPLHAPPRRYP 212
G + P R G RRG Y
Sbjct: 248 ---------GGYDGPQG-------------------RGGYDGPQGRRG----------YD 269
Query: 213 GPPFDRSPERPMNGYRSPPPRLMGRDGLRDYGPAAAALPPLRHEGRFPDPHLHRERMDYM 272
GPP R GY P G DG P + G + P R D
Sbjct: 270 GPPQGR------GGYDGPSQGRGGYDG------------PSQGRGGYDGPSQGRGGYDGP 311
Query: 273 DDAYRGRGKFDRPPPLDWDNRDRGRDGFSNERKGFERRPLSP 314
RGRG+ RGR G GF R P
Sbjct: 312 QGRGRGRGR---------GRGGRGRGGGRGGDGGFNNRSDGP 344
>At5g17790 putative protein
Length = 758
Score = 39.7 bits (91), Expect = 0.003
Identities = 19/45 (42%), Positives = 25/45 (55%), Gaps = 3/45 (6%)
Query: 146 GEGLHGRNNPNVRPREGDWMCPDALCGNLNFARRDFCNQCKRPRP 190
G+G+ N NV + GDW+C + C +NFAR C QC RP
Sbjct: 263 GQGVRSFQN-NVEMKRGDWIC--SRCSGMNFARNVKCFQCDEARP 304
>At1g68690 protein kinase, putative
Length = 708
Score = 39.7 bits (91), Expect = 0.003
Identities = 18/44 (40%), Positives = 21/44 (46%)
Query: 189 RPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPP 232
RP + PP SPP P + P PP D P+ P SPPP
Sbjct: 191 RPTQSPPPPSPPSPPSDRPSQSPPPPPEDTKPQPPRRSPNSPPP 234
Score = 35.8 bits (81), Expect = 0.040
Identities = 24/66 (36%), Positives = 29/66 (43%), Gaps = 6/66 (9%)
Query: 188 PRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDGLRDYGPAA 247
P P A G+PP PPL PP PP P P + SPPP+ + P
Sbjct: 60 PPPVANGAPP----PPLPKPPESSSPPPQPVIPSPPPS--TSPPPQPVIPSPPPSASPPP 113
Query: 248 AALPPL 253
A +PPL
Sbjct: 114 ALVPPL 119
Score = 34.7 bits (78), Expect = 0.089
Identities = 28/89 (31%), Positives = 34/89 (37%), Gaps = 10/89 (11%)
Query: 188 PRPAAAGSPPRRGSPPLHA---------PPRRYPGPP-FDRSPERPMNGYRSPPPRLMGR 237
P P + SPP PPL + PPR P PP RSP + +SPPP R
Sbjct: 103 PSPPPSASPPPALVPPLPSSPPPPASVPPPRPSPSPPILVRSPPPSVRPIQSPPPPPSDR 162
Query: 238 DGLRDYGPAAAALPPLRHEGRFPDPHLHR 266
P+ + P R P P R
Sbjct: 163 PTQSPPPPSPPSPPSERPTQSPPSPPSER 191
Score = 32.3 bits (72), Expect = 0.44
Identities = 19/50 (38%), Positives = 20/50 (40%), Gaps = 5/50 (10%)
Query: 188 PRPAAAGSPPRRGSPPLHAPP-RRYPGPPFDRSPERPMNG----YRSPPP 232
P+P PP PP PP P PP P RP RSPPP
Sbjct: 98 PQPVIPSPPPSASPPPALVPPLPSSPPPPASVPPPRPSPSPPILVRSPPP 147
Score = 32.3 bits (72), Expect = 0.44
Identities = 16/44 (36%), Positives = 20/44 (45%), Gaps = 6/44 (13%)
Query: 189 RPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPP 232
RP + PP SPP P + P PP +R + P PPP
Sbjct: 162 RPTQSPPPPSPPSPPSERPTQSPPSPPSERPTQSP------PPP 199
Score = 29.3 bits (64), Expect = 3.7
Identities = 17/48 (35%), Positives = 19/48 (39%), Gaps = 3/48 (6%)
Query: 189 RPAAAGSPPRRGSPPLHAPPRRYPGPPFDR---SPERPMNGYRSPPPR 233
RP + P P PP P PP DR SP P + PPR
Sbjct: 179 RPTQSPPSPPSERPTQSPPPPSPPSPPSDRPSQSPPPPPEDTKPQPPR 226
Score = 28.5 bits (62), Expect = 6.4
Identities = 43/163 (26%), Positives = 49/163 (29%), Gaps = 24/163 (14%)
Query: 188 PRPAAAGSPPRR---GSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDGLRDYG 244
P P + SPP + SPP A P PP SP P + PPPR +
Sbjct: 88 PSPPPSTSPPPQPVIPSPPPSASPPPALVPPLPSSPPPPAS---VPPPRPSPSPPILVRS 144
Query: 245 PAAAALPPLRHEGRFPDPHLHRERMDYMDDAYRGRGKFDRPPPLDWDNRDRGRDGFSNER 304
P P +R P P R PPP + R S
Sbjct: 145 PP----PSVRPIQSPPPPPSDRPTQS--------------PPPPSPPSPPSERPTQSPPS 186
Query: 305 KGFERRPLSPSAPLLPSLPPHRGGDRWSRDVRDRSRSPIRGGP 347
ER SP P PS P R D P R P
Sbjct: 187 PPSERPTQSPPPPSPPSPPSDRPSQSPPPPPEDTKPQPPRRSP 229
>At1g61080 hypothetical protein
Length = 907
Score = 39.3 bits (90), Expect = 0.004
Identities = 57/203 (28%), Positives = 66/203 (32%), Gaps = 44/203 (21%)
Query: 188 PRPAAAGSPPRRGSPP--LHAPPRRYPGPPFDRSPERP-----MNGYRSPPPRLMGRDGL 240
P P AA +PP PP APP P P +P P N SPPP MG G
Sbjct: 528 PPPRAAVAPPPPPPPPGTAAAPPPPPPPPGTQAAPPPPPPPPMQNRAPSPPPMPMGNSG- 586
Query: 241 RDYGPAAAALPPLRHEGRFPDPHLHRERMDYMDDAYRGRGKFDRPPPLDWDNRDRG---- 296
GP P G P P PPP+ N G
Sbjct: 587 -SGGPPPPPPPMPLANGATPPP---------------------PPPPMAMANGAAGPPPP 624
Query: 297 --RDGFSNERKGFERRPLSPSAPLLPSLPPHRGGDRWSRDVRDRSRSPIRGGPPAKDYRR 354
R G +N G P P A SL P + + R + + I G K R
Sbjct: 625 PPRMGMANGAAG---PPPPPGA--ARSLRPKKAATKLKRSTQLGNLYRILKG---KVEGR 676
Query: 355 DPVMSRGGRDDRRGGVGRDRIGG 377
DP G R+ G G GG
Sbjct: 677 DPNAKTGSGSGRKAGAGSAPAGG 699
>At5g07750 putative protein
Length = 1289
Score = 38.1 bits (87), Expect = 0.008
Identities = 29/81 (35%), Positives = 31/81 (37%), Gaps = 15/81 (18%)
Query: 188 PRPAAAGSPP------RRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDGLR 241
P P G+PP R G+PP PP PP P PM G PPP G G
Sbjct: 1090 PPPMRGGAPPPPPPPMRGGAPPPPPPPMHGGAPP---PPPPPMRGGAPPPPPPPGGRG-- 1144
Query: 242 DYGPAAAALPPLRHEGRFPDP 262
A PP GR P P
Sbjct: 1145 ----PGAPPPPPPPGGRAPGP 1161
Score = 36.6 bits (83), Expect = 0.023
Identities = 31/84 (36%), Positives = 32/84 (37%), Gaps = 15/84 (17%)
Query: 188 PRPAAAGSPP-------RRGS-PPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDG 239
P P + GSPP GS PP PP Y PP P P GY SPPP
Sbjct: 935 PPPPSYGSPPPPPPPPPSYGSPPPPPPPPPSYGSPP---PPPPPPPGYGSPPP---PPPP 988
Query: 240 LRDYG-PAAAALPPLRHEGRFPDP 262
YG P PP H P P
Sbjct: 989 PPSYGSPPPPPPPPFSHVSSIPPP 1012
Score = 35.0 bits (79), Expect = 0.068
Identities = 23/61 (37%), Positives = 27/61 (43%), Gaps = 11/61 (18%)
Query: 188 PRPAAAGSPPRR--------GSPPLHAPPR-RYPGPPFDRSPERPMNGYRSPPPRLMGRD 238
P P G+PP G+PP PP R PGPP P P G PPP ++G
Sbjct: 1126 PPPMRGGAPPPPPPPGGRGPGAPPPPPPPGGRAPGPPPPPGPRPPGGG--PPPPPMLGAR 1183
Query: 239 G 239
G
Sbjct: 1184 G 1184
Score = 34.3 bits (77), Expect = 0.12
Identities = 27/81 (33%), Positives = 33/81 (40%), Gaps = 11/81 (13%)
Query: 188 PRPAAAGSPPRRGSPPLH--APPRRYPGPPFDRSPERP----MNGYRSPPPRLMGRDGLR 241
P P G+PP PP+H APP P P +P P M+G PPP G +
Sbjct: 1015 PPPMHGGAPPPPPPPPMHGGAPPPPPPPPMHGGAPPPPPPPPMHGGAPPPPPPPMFGGAQ 1074
Query: 242 DYGPAAAALPPLRHEGRFPDP 262
P PP+R P P
Sbjct: 1075 PPPP-----PPMRGGAPPPPP 1090
Score = 33.1 bits (74), Expect = 0.26
Identities = 26/75 (34%), Positives = 29/75 (38%), Gaps = 9/75 (12%)
Query: 188 PRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDGLRDYGPAA 247
P P + GSPP PP + P PP P PM+G PPP G G
Sbjct: 987 PPPPSYGSPPPPPPPPF-SHVSSIPPPP----PPPPMHGGAPPPPPPPPMHG----GAPP 1037
Query: 248 AALPPLRHEGRFPDP 262
PP H G P P
Sbjct: 1038 PPPPPPMHGGAPPPP 1052
Score = 31.6 bits (70), Expect = 0.75
Identities = 33/106 (31%), Positives = 35/106 (32%), Gaps = 23/106 (21%)
Query: 188 PRPAAAGSPP------RRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGR---- 237
P P G+PP G+PP PP R PP P G PPP GR
Sbjct: 1102 PPPMRGGAPPPPPPPMHGGAPPPPPPPMRGGAPPPPPPPGGRGPGAPPPPPPPGGRAPGP 1161
Query: 238 ---DGLRDYGPAAAALPPLRHEGRFPDPHLHRERMDYMDDAYRGRG 280
G R G P L G DP A RGRG
Sbjct: 1162 PPPPGPRPPGGGPPPPPMLGARGAAVDPR----------GAGRGRG 1197
Score = 31.2 bits (69), Expect = 0.98
Identities = 21/51 (41%), Positives = 22/51 (42%), Gaps = 11/51 (21%)
Query: 191 AAAGSPPRRGSPPL---------HAPPRRYPGPPFDRSPERPMNGYRSPPP 232
A A PP PPL PP P PPF S ERP +G PPP
Sbjct: 643 AVASPPPPPPPPPLPTYSHYQTSQLPPPPPPPPPF--SSERPNSGTVLPPP 691
Score = 30.4 bits (67), Expect = 1.7
Identities = 19/52 (36%), Positives = 20/52 (37%), Gaps = 10/52 (19%)
Query: 188 PRPAAAGSPPRRGSPP-------LHAPPRRYPGPPFDRSPERPMNGYRSPPP 232
P P +PP PP L PP Y PP P P Y SPPP
Sbjct: 910 PSPPVKTAPPPPPPPPFSNAHSVLSPPPPSYGSPP---PPPPPPPSYGSPPP 958
Score = 30.0 bits (66), Expect = 2.2
Identities = 43/162 (26%), Positives = 49/162 (29%), Gaps = 41/162 (25%)
Query: 190 PAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPPRLMGRDGLRDYGPAAAA 249
P P G+PP PP + G P P PM+G PPP +G A
Sbjct: 1011 PPPPPPPMHGGAPPPPPPPPMHGGAP-PPPPPPPMHGGAPPPPPPPPM-----HGGAPPP 1064
Query: 250 LPPLRHEGRFPDPHLHRERMDYMDDAYRGRGKFDRPPPLDWDNRDRGRDGFSNERKGFER 309
PP G P P RG PPP+ R G
Sbjct: 1065 PPPPMFGGAQPPP----------PPPMRGGAPPPPPPPM---------------RGGAPP 1099
Query: 310 RPLSP---SAPLLPSLPPHRGGDRWSRDVRDRSRSPIRGGPP 348
P P AP P P H G P+RGG P
Sbjct: 1100 PPPPPMRGGAPPPPPPPMHGGAP-------PPPPPPMRGGAP 1134
Score = 28.9 bits (63), Expect = 4.9
Identities = 18/45 (40%), Positives = 20/45 (44%), Gaps = 3/45 (6%)
Query: 188 PRPAAAGSPPRRGSPPLHAPPRRYPGPPFDRSPERPMNGYRSPPP 232
P P S R S + PP P PP S ERP +G PPP
Sbjct: 692 PPPPPPFSSERPNSGTVLPPP---PPPPLPFSSERPNSGTVLPPP 733
>At1g79730 unknown protein
Length = 589
Score = 37.7 bits (86), Expect = 0.010
Identities = 33/113 (29%), Positives = 44/113 (38%), Gaps = 25/113 (22%)
Query: 183 NQCKRPRPAAAGSPPRRGSPPLHAPPRRYPGPP-------FDRSPERP-MNGYRSPPPRL 234
N P P + PP PP H P YP PP + + P P N ++PPP
Sbjct: 18 NSLAPPPPPPSLPPPVPPPPPSHQP-YSYPPPPPPPPHAYYQQGPHYPQFNQLQAPPP-- 74
Query: 235 MGRDGLRDYGPAAAALPPLRHEGRFPDPHLHRERMDYMDDAYRGRGKFDRPPP 287
P +A PPL PDP H+ D+ A + G+ +R P
Sbjct: 75 ---------PPPPSAPPPL-----VPDPPRHQGPNDHEKGASKQVGRRERAKP 113
>At1g55040 putative protein
Length = 849
Score = 37.7 bits (86), Expect = 0.010
Identities = 15/35 (42%), Positives = 21/35 (59%), Gaps = 2/35 (5%)
Query: 156 NVRPREGDWMCPDALCGNLNFARRDFCNQCKRPRP 190
+V ++GDW+CP C +NFA+ C QC RP
Sbjct: 367 DVEMKKGDWLCPK--CDFMNFAKNTICLQCDAKRP 399
Score = 29.6 bits (65), Expect = 2.9
Identities = 15/39 (38%), Positives = 20/39 (50%), Gaps = 4/39 (10%)
Query: 162 GDWMCPDALCGNLNFARRDFCNQC--KRPRPAAAGSPPR 198
G+W CP+ C LN+ R C C KRP A + P+
Sbjct: 406 GEWECPE--CNFLNYRRNMACFHCDCKRPADAILENKPQ 442
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.143 0.479
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,000,131
Number of Sequences: 26719
Number of extensions: 708669
Number of successful extensions: 4324
Number of sequences better than 10.0: 238
Number of HSP's better than 10.0 without gapping: 80
Number of HSP's successfully gapped in prelim test: 165
Number of HSP's that attempted gapping in prelim test: 2384
Number of HSP's gapped (non-prelim): 1339
length of query: 379
length of database: 11,318,596
effective HSP length: 101
effective length of query: 278
effective length of database: 8,619,977
effective search space: 2396353606
effective search space used: 2396353606
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC122722.5


