

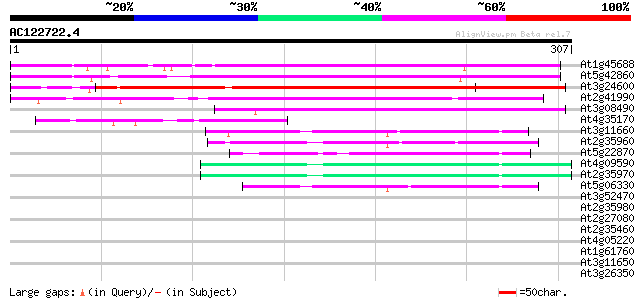
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122722.4 + phase: 0
(307 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g45688 unknown protein 214 6e-56
At5g42860 unknown protein 196 1e-50
At3g24600 hypothetical protein 192 2e-49
At2g41990 unknown protein 162 2e-40
At3g08490 hypothetical protein 73 2e-13
At4g35170 putative protein 50 1e-06
At3g11660 unknown protein 49 3e-06
At2g35960 putative harpin-induced protein 45 5e-05
At5g22870 putative protein 45 6e-05
At4g09590 putative protein 44 8e-05
At2g35970 putative harpin-induced protein 42 4e-04
At5g06330 harpin-induced protein-like 41 7e-04
At3g52470 unknown protein 40 0.001
At2g35980 similar to harpin-induced protein hin1 from tobacco 39 0.003
At2g27080 Unknown protein (At2g27080; T20P8.13) 39 0.004
At2g35460 similar to harpin-induced protein hin1 from tobacco 39 0.005
At4g05220 putative protein 37 0.010
At1g61760 hypothetical protein 37 0.010
At3g11650 unknown protein 37 0.013
At3g26350 hypothetical protein 35 0.039
>At1g45688 unknown protein
Length = 342
Score = 214 bits (544), Expect = 6e-56
Identities = 140/341 (41%), Positives = 192/341 (56%), Gaps = 45/341 (13%)
Query: 1 MHIKTDSDVTSFDPSSP-RSQKQTPYYVQSPSRESSHDGDKS-----STMQPSPMDSP-- 52
MH KTDS+VTS SSP RS ++ YYVQSPSR+S HDG+K+ ST SPM SP
Sbjct: 1 MHAKTDSEVTSLAASSPARSPRRPVYYVQSPSRDS-HDGEKTATSFHSTPVLSPMGSPPH 59
Query: 53 SHQSYGHHSRASSSSRISGGYNSSFLGRKVN------RKVN--DKGWIGSKVIEEENGGY 104
SH S G HSR SSSSR SG RKVN RK + +K W VIEEE G
Sbjct: 60 SHSSMGRHSRESSSSRFSGSLKPG--SRKVNPNDGSKRKGHGGEKQWKECAVIEEE-GLL 116
Query: 105 GDFDGDGNGVSRRVQFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEG 164
D D DG GV RR +VGF ++F F I++ A+ KP+++VKS+T G
Sbjct: 117 DDGDRDG-GVPRRCYVLAFIVGFFILFGFFSLILYGAAKPMKPKITVKSITFETLKIQAG 175
Query: 165 SDVTGVPTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKS 224
D GV T M+T+N ++RML N TFFG++V+S ++L +S + + +G +KK++Q RKS
Sbjct: 176 QDAGGVGTDMITMNATLRMLYRNTGTFFGVHVTSTPIDLSFSQIKIGSGSVKKFYQGRKS 235
Query: 225 RRTVSVNIQGSKVPLYGAGADVA-----------------------SSVDNGKISLTLVF 261
RTV V++ G K+PLYG+G+ + + +TL F
Sbjct: 236 ERTVLVHVIGEKIPLYGSGSTLLPPAPPAPLPKPKKKKGAPVPIPDPPAPPAPVPMTLSF 295
Query: 262 EVKSRGNVVGKLVRTKHTQRVSCSIAVDPHN-NKYIKLKEN 301
V+SR V+GKLV+ K +++ C I + N NK+I + +N
Sbjct: 296 VVRSRAYVLGKLVQPKFYKKIECDINFEHKNLNKHIVITKN 336
>At5g42860 unknown protein
Length = 320
Score = 196 bits (499), Expect = 1e-50
Identities = 121/331 (36%), Positives = 178/331 (53%), Gaps = 47/331 (14%)
Query: 1 MHIKTDSDVTSFDPSSP-RSQKQTPYYVQSPSRESSHDGDKSST------MQPSPMDSPS 53
MH KTDS+VTS SSP RS ++ Y+VQSPSR+S HDG+K++T + SPM SP
Sbjct: 1 MHAKTDSEVTSLSASSPTRSPRRPAYFVQSPSRDS-HDGEKTATSFHSTPVLTSPMGSPP 59
Query: 54 HQSYGHHSRASSSSRISGGYNSSFLGRKVNRKVNDKGWIGSKVIEEENGGYGDFDGDGNG 113
H HS +S S+I+G G K + EE G D D +
Sbjct: 60 HS----HSSSSRFSKINGSKRKGHAGEKQFAMI------------EEEGLLDDGDREQEA 103
Query: 114 VSRRVQFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGVPTK 173
+ RR +VGF L+F+ F I++AA+ KP++SVKS+T G D G+ T
Sbjct: 104 LPRRCYVLAFIVGFSLLFAFFSLILYAAAKPQKPKISVKSITFEQLKVQAGQDAGGIGTD 163
Query: 174 MLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTVSVNIQ 233
M+T+N ++RML N TFFG++V+S ++L +S +T+ +G +KK++Q RKS+RTV VN+
Sbjct: 164 MITMNATLRMLYRNTGTFFGVHVTSSPIDLSFSQITIGSGSIKKFYQSRKSQRTVVVNVL 223
Query: 234 GSKVPLYGAGAD----------------------VASSVDNGKISLTLVFEVKSRGNVVG 271
G K+PLYG+G+ V + + L F V+SR V+G
Sbjct: 224 GDKIPLYGSGSTLVPPPPPAPIPKPKKKKGPIVIVEPPAPPAPVPMRLNFTVRSRAYVLG 283
Query: 272 KLVRTKHTQRVSCSIAVDPHN-NKYIKLKEN 301
KLV+ K +R+ C I + +K+I + N
Sbjct: 284 KLVQPKFYKRIVCLINFEHKKLSKHIPITNN 314
>At3g24600 hypothetical protein
Length = 506
Score = 192 bits (488), Expect = 2e-49
Identities = 105/258 (40%), Positives = 160/258 (61%), Gaps = 5/258 (1%)
Query: 48 PMDSPSHQSYGHHSRASSSSRISGGYNSSFLGRKVNRKVNDKGWIGSKVIEEENGGYGDF 107
P+ +P++ SR SSSSR S G + K + + ++ W E+ Y D
Sbjct: 249 PVHTPNYTILSE-SRLSSSSRTSNGTSGMGFRWKGSSRRSNMYWPEKPYTINEDEVYDDN 307
Query: 108 DGDGNGVSRRVQFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDV 167
G G R V + ++G V+VFS+FC ++W AS + P +SVKS+ +H+FY+GEG D
Sbjct: 308 RGLSVGQCRAV---LVILGTVVVFSVFCSVLWGASHPFSPIVSVKSVDIHSFYYGEGIDR 364
Query: 168 TGVPTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRT 227
TGV TK+L+ N S+++ + +PA +FGI+VSS + L +S LT+ATG+LK Y+Q RKS+
Sbjct: 365 TGVATKILSFNSSVKVTIDSPAPYFGIHVSSSTFKLTFSALTLATGQLKSYYQPRKSKHI 424
Query: 228 VSVNIQGSKVPLYGAGADVASSVDNGKISLTLVFEVKSRGNVVGKLVRTKHTQRVSCSIA 287
V + G++VPLYGAG +A+S GK+ + L FE++SRGN++GKLV++KH VSCS
Sbjct: 425 SIVKLTGAEVPLYGAGPHLAASDKKGKVPVKLEFEIRSRGNLLGKLVKSKHENHVSCSFF 484
Query: 288 V-DPHNNKYIKLKENECR 304
+ +K I+ C+
Sbjct: 485 ISSSKTSKPIEFTHKTCK 502
Score = 185 bits (469), Expect = 3e-47
Identities = 114/262 (43%), Positives = 157/262 (59%), Gaps = 23/262 (8%)
Query: 1 MHIKTDSDVTSFDPSSPRSQKQTPYYVQSPSRESSHDGDKSS-----TMQPSPMDSPSHQ 55
M+ K+DSDVTS D SSP K+ YYVQSPSR+S DKSS T Q +P +SPSH
Sbjct: 3 MYPKSDSDVTSLDLSSP---KRPTYYVQSPSRDS----DKSSSVALTTHQTTPTESPSHP 55
Query: 56 SYGHHSRASSSSRISGGYNSSFLGRKVNRKVNDKGWIGSKVIEEENGGYGDFDGDGNGVS 115
S +SR+S G F K RK + W + E +G Y D D GVS
Sbjct: 56 SI--------ASRVSNGGGGGFRW-KGRRKYHGGIWWPADKEEGGDGRYEDLYEDNRGVS 106
Query: 116 R-RVQFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGVPTKM 174
+ + VV + +F L C +++ AS P + +K + V +FY+GEGSD TGVPTK+
Sbjct: 107 IVTCRLILGVVATLSIFFLLCSVLFGASQSSPPIVYIKGVNVRSFYYGEGSDNTGVPTKI 166
Query: 175 LTVNCSMRMLVHNPATFFGIYVSSKSVNLMYS-DLTVATGELKKYHQQRKSRRTVSVNIQ 233
+ V CS+ + HNP+T FGI+VSS +V+L+YS T+A LK YHQ ++S T +N+
Sbjct: 167 MNVKCSVVITTHNPSTLFGIHVSSTAVSLIYSRQFTLANARLKSYHQPKQSNHTSRINLI 226
Query: 234 GSKVPLYGAGADVASSVDNGKI 255
GSKVPLYGAGA++ +S ++G +
Sbjct: 227 GSKVPLYGAGAELVASDNSGGV 248
>At2g41990 unknown protein
Length = 297
Score = 162 bits (410), Expect = 2e-40
Identities = 103/302 (34%), Positives = 167/302 (55%), Gaps = 28/302 (9%)
Query: 1 MHIKTDSDVTSFDP---SSPRSQKQTPYYVQSPSRESSHDGDKSSTMQP-SPMDSPSHQS 56
MH KTDS+ TS D S PRS + YYVQSPS +HD +K S S M SP+H
Sbjct: 1 MHAKTDSEATSIDAAALSPPRSAIRPLYYVQSPS---NHDVEKMSFGSGCSLMGSPTHPH 57
Query: 57 YGH-----HSRASSSSRISGGYNSSFLGRKVNRKVNDKGWIGSKVIEEENGGYGDFDGDG 111
Y H HSR SS+SR S S+ + R+ + G +++ G GD
Sbjct: 58 YYHCSPIHHSRESSTSRFSDRALLSYKSIRERRRYINDG-------DDKTDG-----GDD 105
Query: 112 NGVSRRVQFFV-AVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGV 170
+ R V+ +V ++ + +F++F I+W AS Y P+++VK + V + G+D++GV
Sbjct: 106 DDPFRNVRLYVWLLLSVIFLFTVFSLILWGASKSYPPKVTVKGMLVRDLNLQAGNDLSGV 165
Query: 171 PTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTVSV 230
PT ML++N ++R+ NP+TFF ++V++ + L YS+L +++GE+ K+ R V
Sbjct: 166 PTDMLSLNSTVRIYYRNPSTFFAVHVTASPLLLHYSNLLLSSGEMNKFTVGRNGETNVVT 225
Query: 231 NIQGSKVPLYGAGADVASSVDNGKISLTLVFEVKSRGNVVGKLVRTKHTQRVSCSIAVDP 290
+QG ++PLYG V+ +D + L L + S+ ++G+LV +K R+ CS +D
Sbjct: 226 VVQGHQIPLYGG---VSFHLDTLSLPLNLTIVLHSKAYILGRLVTSKFYTRIICSFTLDA 282
Query: 291 HN 292
++
Sbjct: 283 NH 284
>At3g08490 hypothetical protein
Length = 271
Score = 72.8 bits (177), Expect = 2e-13
Identities = 48/196 (24%), Positives = 88/196 (44%), Gaps = 4/196 (2%)
Query: 113 GVSRRVQFFVAVVGFVLVFSL--FCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGV 170
G S + V VG+ +FSL + + A+ P +S + + F EG D GV
Sbjct: 73 GTSNSSWWIVLQVGWRFLFSLGVALLVFYIATQPPHPNISFRIGRFNQFMLEEGVDSHGV 132
Query: 171 PTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTVSV 230
TK LT NCS ++++ N + FG+++ S+ + L A + K + T +
Sbjct: 133 STKFLTFNCSTKLIIDNKSNVFGLHIHPPSIKFFFGPLNFAKAQGPKLYGLSHESTTFQL 192
Query: 231 NIQGSKVPLYGAGADVASS-VDNGKISLTLVFEVKSRGNVVGKLVRTKHTQRVSCSIAV- 288
I + +YGAG ++ + + L L + S VV ++ K+ +V C + +
Sbjct: 193 YIATTNRAMYGAGTEMNDMLLSRAGLPLILRTSIISDYRVVWNIINPKYHHKVECLLLLA 252
Query: 289 DPHNNKYIKLKENECR 304
D + ++ + +CR
Sbjct: 253 DKERHSHVTMIREKCR 268
>At4g35170 putative protein
Length = 163
Score = 50.4 bits (119), Expect = 1e-06
Identities = 45/147 (30%), Positives = 70/147 (47%), Gaps = 21/147 (14%)
Query: 15 SSPRSQKQTPYYVQSPSRESSHDGDKSSTMQP-SPMDSPSHQ-----SYGHHSRASSSS- 67
SSP++ ++ Y V SP D DK ST SP SP + ++ HHS A SSS
Sbjct: 8 SSPQNTRKPVYVVHSPPNT---DVDKISTGSGFSPFGSPLNDQGQVSNFQHHSVAESSSY 64
Query: 68 -RISGGYNSSFLGRKVNRKVNDKGWIGSKVIEEENGGYGDFDGDGNGVSRRVQFFVAVV- 125
R SG + + +V+ + + E+E+ Y + DG R +F+ ++
Sbjct: 65 PRSSGPLRNEYSSVQVHD-------LDRRTHEDED--YDEMDGPDEKRRRITRFYSCLLF 115
Query: 126 GFVLVFSLFCFIIWAASLHYKPQLSVK 152
VL F+LFC I+W S + P ++K
Sbjct: 116 TLVLAFTLFCLILWGVSKSFAPIATLK 142
>At3g11660 unknown protein
Length = 209
Score = 49.3 bits (116), Expect = 3e-06
Identities = 47/183 (25%), Positives = 79/183 (42%), Gaps = 14/183 (7%)
Query: 108 DGDGNGVSRRV---QFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEG 164
D + +G SRR + F +++ + + L +IWA KP+ ++ TV+ F
Sbjct: 3 DCENHGHSRRKLIRRIFWSIIFVLFIIFLTILLIWAILQPSKPRFILQDATVYAF----- 57
Query: 165 SDVTGVPTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMY--SDLTVATGELKKYHQQR 222
+V+G P +LT N + + NP GIY V Y +T T + +Q
Sbjct: 58 -NVSGNPPNLLTSNFQITLSSRNPNNKIGIYYDRLDVYATYRSQQITFPT-SIPPTYQGH 115
Query: 223 KSRRTVSVNIQGSKVPLYG-AGADVASSVDNGKISLTLVFEVKSRGNVVGKLVRTKHTQR 281
K S + G+ VP+ G + + DNG + L + + + R VG + K+
Sbjct: 116 KDVDIWSPFVYGTSVPIAPFNGVSLDTDKDNGVVLLIIRADGRVRWK-VGTFITGKYHLH 174
Query: 282 VSC 284
V C
Sbjct: 175 VKC 177
>At2g35960 putative harpin-induced protein
Length = 210
Score = 45.1 bits (105), Expect = 5e-05
Identities = 42/184 (22%), Positives = 76/184 (40%), Gaps = 15/184 (8%)
Query: 109 GDGNGVSRRVQFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVT 168
G G G + R+ ++GF+++ + F++W KP+ ++ TV+ F +
Sbjct: 12 GGGGGTASRI--CGVIIGFIIIVLITIFLVWIILQPTKPRFILQDATVYAFNLSQ----- 64
Query: 169 GVPTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMY--SDLTVATGELKKYHQQRKSRR 226
+LT N + + N + GIY V Y +T+ T + +Q K
Sbjct: 65 ---PNLLTSNFQITIASRNRNSRIGIYYDRLHVYATYRNQQITLRTA-IPPTYQGHKEDN 120
Query: 227 TVSVNIQGSKVPLYGAGADVASSVDNGKISLTLVFEVKSRGN-VVGKLVRTKHTQRVSCS 285
S + G+ VP+ A VA + + +TL+ R VG L+ K+ V C
Sbjct: 121 VWSPFVYGNSVPIAPFNA-VALGDEQNRGFVTLIIRADGRVRWKVGTLITGKYHLHVRCQ 179
Query: 286 IAVD 289
++
Sbjct: 180 AFIN 183
>At5g22870 putative protein
Length = 207
Score = 44.7 bits (104), Expect = 6e-05
Identities = 39/166 (23%), Positives = 72/166 (42%), Gaps = 18/166 (10%)
Query: 121 FVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGVPTKMLTVNCS 180
F+A VGF+ I W + K + +V++ +V NF + ++ T T+
Sbjct: 38 FMAAVGFL--------ITWLETKPKKLRYTVENASVQNFNLTNDNHMSA--TFQFTIQS- 86
Query: 181 MRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTVSVNIQGSKVPLY 240
HNP +Y SS + + + D T+A ++ +HQ R + + + + V +
Sbjct: 87 -----HNPNHRISVYYSSVEIFVKFKDQTLAFDTVEPFHQPRMNVKQIDETLIAENVAVS 141
Query: 241 GA-GADVASSVDNGKISLTLVFEVKSRGNVVGKLVRTKHTQRVSCS 285
+ G D+ S GKI + + + R VG + T ++ CS
Sbjct: 142 KSNGKDLRSQNSLGKIGFEVFVKARVRFK-VGIWKSSHRTAKIKCS 186
>At4g09590 putative protein
Length = 211
Score = 44.3 bits (103), Expect = 8e-05
Identities = 41/206 (19%), Positives = 81/206 (38%), Gaps = 12/206 (5%)
Query: 105 GDFDGDGNGVSRRVQFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEG 164
G+ G G G + A++GF+++ + F++W P+ ++ TV+ F +
Sbjct: 7 GNHGGGGGGGGTACRICGAIIGFIIIVLMTIFLVWIILQPKNPEFILQDTTVYAFNLSQ- 65
Query: 165 SDVTGVPTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVA-TGELKKYHQQRK 223
+LT + + N + GIY Y + + +L +Q+ K
Sbjct: 66 -------PNLLTSKFQITIASRNRNSNIGIYYDHLHAYASYRNQQITLASDLPPTYQRHK 118
Query: 224 SRRTVSVNIQGSKVPLYGAGADVASSVDN-GKISLTLVFEVKSRGNVVGKLVRTKHTQRV 282
S + G++VP+ A N G +LT+ + + R VG L + V
Sbjct: 119 EDSVWSPLLYGNQVPIAPFNAVALGDEQNSGVFTLTICVDGQVRWK-VGTLTIGNYHLHV 177
Query: 283 SCSIAVDPHNNKY-IKLKENECRYNI 307
C ++ + + + EN +Y +
Sbjct: 178 RCQAFINQADKAAGVHVGENTVKYTL 203
>At2g35970 putative harpin-induced protein
Length = 211
Score = 42.0 bits (97), Expect = 4e-04
Identities = 41/206 (19%), Positives = 81/206 (38%), Gaps = 12/206 (5%)
Query: 105 GDFDGDGNGVSRRVQFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEG 164
G+ G G G + A++GF+++ + F++ KP+ ++ TV+ F +
Sbjct: 7 GNHGGGGGGGGTACRICGAIIGFIIIVLMTIFLVSIILQPKKPEFILQDTTVYAFNLSQ- 65
Query: 165 SDVTGVPTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVA-TGELKKYHQQRK 223
+LT + + N + GIY Y + + +L +Q+ K
Sbjct: 66 -------PNLLTSKFQITIASRNRNSNIGIYYDHLHAYASYRNQQITLASDLPPTYQRHK 118
Query: 224 SRRTVSVNIQGSKVPLYGAGADVASSVDN-GKISLTLVFEVKSRGNVVGKLVRTKHTQRV 282
S + G++VP+ A N G +LT+ + + R VG L + V
Sbjct: 119 ENSVWSPLLYGNQVPIAPFNAVALGDEQNSGVFTLTICVDGRVRWK-VGTLTIGNYHLHV 177
Query: 283 SCSIAVDPHNNKY-IKLKENECRYNI 307
C ++ + + + EN +Y +
Sbjct: 178 RCQAFINQADKAAGVHVGENTVKYTL 203
>At5g06330 harpin-induced protein-like
Length = 207
Score = 41.2 bits (95), Expect = 7e-04
Identities = 42/165 (25%), Positives = 67/165 (40%), Gaps = 11/165 (6%)
Query: 128 VLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGVPTKMLTVNCSMRMLVHN 187
+L+ + ++WA KP+ ++ TV NF +V+G P +LT N + N
Sbjct: 28 LLLILVVILLVWAILQPSKPRFVLQDATVFNF------NVSGNPPNLLTSNFQFTLSSRN 81
Query: 188 PATFFGIYVSSKSVNLMY--SDLTVATGELKKYHQQRKSRRTVSVNIQGSKVPLYGAGA- 244
P GIY V Y +T+ + L Y Q K S + G VP+ A
Sbjct: 82 PNDKIGIYYDRLDVYASYRSQQITLPSPMLTTY-QGHKEVNVWSPFVGGYSVPVAPYNAF 140
Query: 245 DVASSVDNGKISLTLVFEVKSRGNVVGKLVRTKHTQRVSCSIAVD 289
+ +G I L L + + R VG + K+ V C ++
Sbjct: 141 YLDQDHSSGAIMLMLHLDGRVRWK-VGSFITGKYHLHVRCHALIN 184
>At3g52470 unknown protein
Length = 208
Score = 40.4 bits (93), Expect = 0.001
Identities = 42/193 (21%), Positives = 81/193 (41%), Gaps = 22/193 (11%)
Query: 123 AVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGVPTKMLTVNCSMR 182
A++ F+++ + F++W KP+ ++ TV+ F + +LT N +
Sbjct: 22 AIIAFIVIVLITIFLVWVILRPTKPRFVLQDATVYAFNLSQ--------PNLLTSNFQVT 73
Query: 183 MLVHNPATFFGIYVSSKSVNLMY--SDLTVATGELKKYHQQRKSRRTVSVNIQGSKVPL- 239
+ NP + GIY V Y +T+ T + +Q K S + G+ VP+
Sbjct: 74 IASRNPNSKIGIYYDRLHVYATYMNQQITLRTA-IPPTYQGHKEVNVWSPFVYGTAVPIA 132
Query: 240 -YGAGADVASSVDNGKISLTLVFEVKSRGNV---VGKLVRTKHTQRVSCSIAVDPHNNKY 295
Y + A + D G + L +++ G V V L+ K+ V C ++ N
Sbjct: 133 PYNSVA-LGEEKDRGFVGLM----IRADGTVRWKVRTLITGKYHIHVRCQAFINLGNKAA 187
Query: 296 -IKLKENECRYNI 307
+ + +N +Y +
Sbjct: 188 GVLVGDNAVKYTL 200
>At2g35980 similar to harpin-induced protein hin1 from tobacco
Length = 227
Score = 39.3 bits (90), Expect = 0.003
Identities = 37/172 (21%), Positives = 67/172 (38%), Gaps = 9/172 (5%)
Query: 102 GGYGDFDGDGNGVSRRVQFFVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYF 161
G Y G G G F ++ +++ + I W + V ++ F
Sbjct: 23 GYYRRGHGRGCGCCLLSLFVKVIISLIVILGVAALIFWLIVRPRAIKFHVTDASLTRF-- 80
Query: 162 GEGSDVTGVPTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQ 221
D T P +L N ++ + V NP G+Y + Y +T L ++Q
Sbjct: 81 ----DHTS-PDNILRYNLALTVPVRNPNKRIGLYYDRIEAHAYYEGKRFSTITLTPFYQG 135
Query: 222 RKSRRTVSVNIQGSKVPLYGAGADVASSVDNGKISLTLVFEVKSRGNVVGKL 273
K+ ++ QG + ++ AG + +++ +IS E+K R V KL
Sbjct: 136 HKNTTVLTPTFQGQNLVIFNAGQ--SRTLNAERISGVYNIEIKFRLRVRFKL 185
>At2g27080 Unknown protein (At2g27080; T20P8.13)
Length = 260
Score = 38.9 bits (89), Expect = 0.004
Identities = 39/176 (22%), Positives = 79/176 (44%), Gaps = 20/176 (11%)
Query: 121 FVAVVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGV---PTKMLTV 177
F+A V ++V + F + L Y+P+ S+ EG V+G+ T ++
Sbjct: 78 FLAAVFILIVLAGISFAV--LYLIYRPEAPKYSI--------EGFSVSGINLNSTSPISP 127
Query: 178 NCSMRMLVHNPATFFGIYVSSKS-VNLMYSDLTVATGELKKYHQQRKSRRTVSVNIQGSK 236
+ ++ + N G+Y +S V++ Y+D+ ++ G + ++Q K+ V + + GSK
Sbjct: 128 SFNVTVRSRNGNGKIGVYYEKESSVDVYYNDVDISNGVMPVFYQPAKNVTVVKLVLSGSK 187
Query: 237 VPLYGAGADVASSVDNGKISLTLVFEVKSRGNV---VGKLVRTKHTQRVSCSIAVD 289
+ L + + + N T+ F++K + V G + V C + VD
Sbjct: 188 IQL---TSGMRKEMRNEVSKKTVPFKLKIKAPVKIKFGSVKTWTMIVNVDCDVTVD 240
>At2g35460 similar to harpin-induced protein hin1 from tobacco
Length = 238
Score = 38.5 bits (88), Expect = 0.005
Identities = 26/120 (21%), Positives = 44/120 (36%), Gaps = 7/120 (5%)
Query: 124 VVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGVPTKMLTVNCSMRM 183
++G ++ + I+W + V + F F S L N S+
Sbjct: 58 LIGVLVCLGVVALILWFILRPNVVKFQVTEADLTRFEFDPRSH-------NLHYNISLNF 110
Query: 184 LVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTVSVNIQGSKVPLYGAG 243
+ NP GI+ V Y D + + ++Q K+ V + G K+ L GAG
Sbjct: 111 SIRNPNQRLGIHYDQLEVRGYYGDQRFSAANMTSFYQGHKNTTVVGTELNGQKLVLLGAG 170
>At4g05220 putative protein
Length = 226
Score = 37.4 bits (85), Expect = 0.010
Identities = 35/181 (19%), Positives = 75/181 (41%), Gaps = 18/181 (9%)
Query: 114 VSRRVQFFVAVVGFVLVF--SLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGVP 171
+S R+ F+ + +++F + FI+W + ++P+ ++ V +G D P
Sbjct: 38 LSTRISKFICAMFLLVLFFVGVIAFILWLSLRPHRPRFHIQDFVV------QGLDQ---P 88
Query: 172 TKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVA-TGELKKYHQQRKSRRTVSV 230
T + + + + NP G+Y S ++ Y D V L + QQ + V+
Sbjct: 89 TGVENARIAFNVTILNPNQHMGVYFDSMEGSIYYKDQRVGLIPLLNPFFQQPTNTTIVTG 148
Query: 231 NIQGSKVPLYGAGADVASSVDNGKISLTLVFEVKSRGNVVGKL---VRTKHTQRVSCSIA 287
+ G+ + + ++ + N + T+ F + + KL + H +C+I
Sbjct: 149 TLTGASLTV---NSNRWTEFSNDRAQGTVGFRLDIVSTIRFKLHRWISKHHRMHANCNIV 205
Query: 288 V 288
V
Sbjct: 206 V 206
>At1g61760 hypothetical protein
Length = 224
Score = 37.4 bits (85), Expect = 0.010
Identities = 31/183 (16%), Positives = 71/183 (37%), Gaps = 16/183 (8%)
Query: 125 VGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGVPTKMLTVNCSMRML 184
+ +L + FI+W + ++P++ ++ ++ +G + T + S ++
Sbjct: 49 LSLLLCLGIITFILWISLQPHRPRVHIRGFSISGLSRPDGFE---------TSHISFKIT 99
Query: 185 VHNPATFFGIYVSSKSVNLMYSDLTVATGEL-KKYHQQRKSRRTVSVNIQGSKVPLYGAG 243
HNP GIY S ++ Y + + + +L ++Q K+ ++ + P
Sbjct: 100 AHNPNQNVGIYYDSMEGSVYYKEKRIGSTKLTNPFYQDPKNTSSIDGALSR---PAMAVN 156
Query: 244 ADVASSVDNGKISLTLVFEVKSRGNVVGKLV---RTKHTQRVSCSIAVDPHNNKYIKLKE 300
D ++ + ++F +K R + K+ H SC I + K+
Sbjct: 157 KDRWMEMERDRNQGKIMFRLKVRSMIRFKVYTWHSKSHKMYASCYIEIGWDGMLLSATKD 216
Query: 301 NEC 303
C
Sbjct: 217 KRC 219
>At3g11650 unknown protein
Length = 240
Score = 37.0 bits (84), Expect = 0.013
Identities = 22/121 (18%), Positives = 45/121 (37%), Gaps = 8/121 (6%)
Query: 124 VVGFVLVFSLFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGVPTKMLTVNCSMRM 183
++ ++ + I+W + V ++ F F P L + +
Sbjct: 59 LIAVAVILGVAALILWLIFRPNAVKFYVADANLNRFSFD--------PNNNLHYSLDLNF 110
Query: 184 LVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTVSVNIQGSKVPLYGAG 243
+ NP G+Y SV+ Y D + + ++Q K+ + I+G + + G G
Sbjct: 111 TIRNPNQRVGVYYDEFSVSGYYGDQRFGSANVSSFYQGHKNTTVILTKIEGQNLVVLGDG 170
Query: 244 A 244
A
Sbjct: 171 A 171
>At3g26350 hypothetical protein
Length = 356
Score = 35.4 bits (80), Expect = 0.039
Identities = 34/182 (18%), Positives = 75/182 (40%), Gaps = 15/182 (8%)
Query: 112 NGVSRRVQFFVAVVGFVLVFS-LFCFIIWAASLHYKPQLSVKSLTVHNFYFGEGSDVTGV 170
N ++ F A+ +L+ L I++ P + + + ++ Y G + G
Sbjct: 170 NAMTWSAAFCCAIFWVILILGGLIILIVYLVYRPRSPYVDISAANLNAAYLDMGFLLNGD 229
Query: 171 PTKMLTVNCSMRMLVHNPATFFGIYVSSKSVNLMYSDLTVATGELKKYHQQRKSRRTVSV 230
T + V NP+ + S + L Y + +AT ++ + +K+ +V
Sbjct: 230 LTILANVT--------NPSKKSSVEFSYVTFELYYYNTLIATQYIEPFKVPKKTSMFANV 281
Query: 231 NIQGSKVPLYGA-GADVASSVDNGKISLTL--VFEVKSRGNVVGKLVRTKHTQRVSCSIA 287
++ S+V L ++ ++ G + L L +F +S +G L R + CS++
Sbjct: 282 HLVSSQVQLQATQSRELQRQIETGPVLLNLRGMFHARSH---IGPLFRYSYKLHTHCSVS 338
Query: 288 VD 289
++
Sbjct: 339 LN 340
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.132 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,293,016
Number of Sequences: 26719
Number of extensions: 330027
Number of successful extensions: 1339
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 33
Number of HSP's that attempted gapping in prelim test: 1277
Number of HSP's gapped (non-prelim): 77
length of query: 307
length of database: 11,318,596
effective HSP length: 99
effective length of query: 208
effective length of database: 8,673,415
effective search space: 1804070320
effective search space used: 1804070320
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC122722.4


