

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
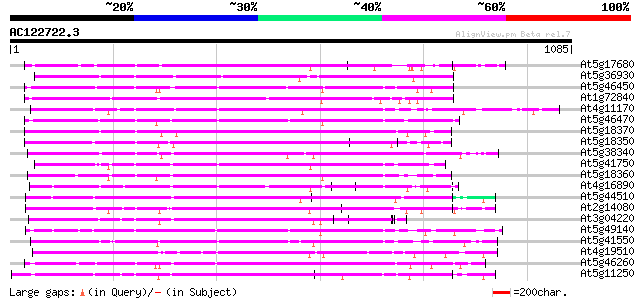
Query= AC122722.3 - phase: 0
(1085 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g17680 disease resistance protein RPP1-WsB - like protein 505 e-143
At5g36930 disease resistance like protein 443 e-124
At5g46450 disease resistance protein-like 437 e-122
At1g72840 hypothetical protein 427 e-119
At4g11170 RPP1-WsA-like disease resistance protein 422 e-118
At5g46470 disease resistance protein-like 412 e-115
At5g18370 disease resistance protein -like 410 e-114
At5g18350 disease resistance protein -like 408 e-114
At5g38340 disease resistance - like protein 408 e-113
At5g41750 disease resistance protein-like 406 e-113
At5g18360 disease resistance protein -like 406 e-113
At4g16890 disease resistance RPP5 like protein 404 e-112
At5g44510 disease resistance protein-like 403 e-112
At2g14080 putative disease resistance protein 403 e-112
At3g04220 putative disease resistance protein 402 e-112
At5g49140 disease resistance protein-like 399 e-111
At5g41550 disease resistance protein-like 399 e-111
At4g19510 TMV resistance protein N-like 394 e-109
At5g46260 disease resistance protein-like 394 e-109
At5g11250 RPP1 disease resistance protein - like 392 e-109
>At5g17680 disease resistance protein RPP1-WsB - like protein
Length = 1294
Score = 505 bits (1301), Expect = e-143
Identities = 316/845 (37%), Positives = 475/845 (55%), Gaps = 86/845 (10%)
Query: 29 MASSSKSSSALVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQKG 88
+ SSS SSS+ V + DVFV+FRGED R F LF + GI FRD ++LQ+G
Sbjct: 4 LPSSSSSSSSTVWKT-----DVFVSFRGEDVRKTFVSHLFCEFDRMGIKAFRDDLDLQRG 58
Query: 89 ECIGPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPSE 148
+ I PEL AI+ S+ + + S+NYA+S+WCL EL KI EC K + ++P+FY+VDPS+
Sbjct: 59 KSISPELIDAIKGSRFAIVVVSRNYAASSWCLDELLKIMECNKDT---IVPIFYEVDPSD 115
Query: 149 VRKQSGIYSEAFVKHEQRFQQDSMKVSRWREALEQVGSISGWDLRDEPLAREIKEIVQKI 208
VR+Q G + E H D KV +W+EAL+++ +ISG D R+ ++ IK+IV+ I
Sbjct: 116 VRRQRGSFGEDVESH-----SDKEKVGKWKEALKKLAAISGEDSRNWDDSKLIKKIVKDI 170
Query: 209 INILECKYSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLYGQ 268
+ L SK L+G+ S + LQ+ + + D VR +GI GMGG+GKTT+A LY Q
Sbjct: 171 SDKLVSTSWDDSKGLIGMSSHMDFLQSMISIVDKD-VRMLGIWGMGGVGKTTIAKYLYNQ 229
Query: 269 ISHQFSASCFIDDVTKI---YGLHDDPLDVQKQILFQTLGIEHQQICNRYHATTLIQRKL 325
+S QF CF+++V ++ YG+ +Q + L + ++ + +I+ +
Sbjct: 230 LSGQFQVHCFMENVKEVCNRYGVR----RLQVEFLCRMFQERDKEAWSSVSCCNIIKERF 285
Query: 326 CHERTLMILDNVDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVYKVSLLDW 385
H+ ++LD+VD+ EQL ++ W GPGSRII+ +RD H+L ++G+++VYKV L
Sbjct: 286 RHKMVFIVLDDVDRSEQLNELVKETGWFGPGSRIIVTTRDRHLLLSHGINLVYKVKCLPK 345
Query: 386 NEAHMLFCRKAFKDEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFGRNVTEWKSALT 445
EA LFC AF++E I+ ++ L Q ++YA GLPLA++VLGSFL+ R+ EW+S L
Sbjct: 346 KEALQLFCNYAFREEIILPHGFEELSVQAVNYASGLPLALRVLGSFLYRRSQIEWESTLA 405
Query: 446 RLRQSPVKDVMDVLQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNILNCCGFHADIGLR 505
RL+ P D+M+VL++S+DGL+E EK IFL+I+CF+N + V+ +L+ CG+ A+IG+
Sbjct: 406 RLKTYPHSDIMEVLRVSYDGLDEQEKAIFLYISCFYNMKQVDYVRKLLDLCGYAAEIGIT 465
Query: 506 VLIDKSLVSISYSIINMHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDVMLENM-EK 564
+L +KSL+ S + +H LLE++GR++V+ + P + LW E + ++ EN +
Sbjct: 466 ILTEKSLIVESNGCVKIHDLLEQMGRELVRQQAVNNPAQRLLLWDPEDICHLLSENSGTQ 525
Query: 565 HVEAIVLYYKEDEEA-----DFEHLSKMSNLRLLFIANYISTMLGFPSCLS---NKLRFV 616
VE I L E E FE LS + L ++ T + P+ LS KLR++
Sbjct: 526 LVEGISLNLSEISEVFASDRAFEGLSNLKLLNFYDLSFDGETRVHLPNGLSYLPRKLRYL 585
Query: 617 HWFRYPSKYLPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRHSRNLEKIIDFG 676
W YP K +PS F P LVEL ++ SN+++LW + L NL+ +DL + L ++ D
Sbjct: 586 RWDGYPLKTMPSRFFPEFLVELCMSNSNLEKLWDGIQPLRNLKKMDLSRCKYLVEVPDLS 645
Query: 677 EFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCG 736
+ NLE L+L C +LVE+ PSI L+ L L +C L IP I L SL+ + M G
Sbjct: 646 KATNLEELNLSYCQSLVEVTPSIKNLKGLSCFYLTNCIQLKDIPIGII-LKSLETVGMSG 704
Query: 737 CSKVFNNPRRLMKSGISSEKKQQHDIRESASHHLPGLKW----IILAHDSSHMLP-SLHS 791
CS + H P + W + L+ LP S+
Sbjct: 705 CSSL---------------------------KHFPEISWNTRRLYLSSTKIEELPSSISR 737
Query: 792 LCCLRKVDISFCYLSHVPDAIECLHWLERLNLAGNDFVTLPS-LRKLSKLVYLNLEHCKL 850
L CL K+D+S C +RL TLPS L L L LNL+ C+
Sbjct: 738 LSCLVKLDMSDC---------------QRLR-------TLPSYLGHLVSLKSLNLDGCRR 775
Query: 851 LESLP 855
LE+LP
Sbjct: 776 LENLP 780
Score = 79.0 bits (193), Expect = 1e-14
Identities = 99/374 (26%), Positives = 169/374 (44%), Gaps = 83/374 (22%)
Query: 654 YLPNLRTLDLRHSRNLEKIID-FGEFPNLERLDLEGCINLVE---LDPSIGLLR------ 703
+L +L++L+L R LE + D +LE L++ GC+N+ E + SI +LR
Sbjct: 761 HLVSLKSLNLDGCRRLENLPDTLQNLTSLETLEVSGCLNVNEFPRVSTSIEVLRISETSI 820
Query: 704 -----------KLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCGCSKVFNNPRRLMKSGI 752
+L L++ + K L S+P +I L SL+ L + GCS + + P + ++
Sbjct: 821 EEIPARICNLSQLRSLDISENKRLASLPVSISELRSLEKLKLSGCSVLESFPLEICQT-- 878
Query: 753 SSEKKQQHDIRESASHHLPG--------------------LKWII--------LAHDSSH 784
+ D+ ++ LP W I LA +S
Sbjct: 879 -MSCLRWFDLDRTSIKELPENIGNLVALEVLQASRTVIRRAPWSIARLTRLQVLAIGNSF 937
Query: 785 MLPS--LHSLCC-------LRKVDISFCYLSHVPDAIECLHWLERLNLAGNDFVTLP-SL 834
P LHSLC LR + +S ++ +P++I L L L+L+GN+F +P S+
Sbjct: 938 FTPEGLLHSLCPPLSRFDDLRALSLSNMNMTEIPNSIGNLWNLLELDLSGNNFEFIPASI 997
Query: 835 RKLSKLVYLNLEHCKLLESLP-QLP---------FPTNTGEVHREYDDYFCGAGLLIFNC 884
++L++L LNL +C+ L++LP +LP T+ + ++ Y C L+ NC
Sbjct: 998 KRLTRLNRLNLNNCQRLQALPDELPRGLLYIYIHSCTSLVSISGCFNQY-CLRKLVASNC 1056
Query: 885 PKLGEREHCRSMTLLWMKQFIKANPRSSSEIQIVNPGSEIPSWINNQRMGYSIAIDRSPI 944
KL + L ++ A P S PGS+IP+ N+Q MG S+ I
Sbjct: 1057 YKLDQAAQILIHRNLKLE---SAKPEHS-----YFPGSDIPTCFNHQVMGPSLNIQLP-- 1106
Query: 945 RHDNDNNIIGIVCC 958
+ ++ ++I+G C
Sbjct: 1107 QSESSSDILGFSAC 1120
>At5g36930 disease resistance like protein
Length = 1164
Score = 443 bits (1139), Expect = e-124
Identities = 303/849 (35%), Positives = 454/849 (52%), Gaps = 52/849 (6%)
Query: 48 YDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQKGECIGPELFRAIEISQVYVA 107
YDVFV+FRG D R NF L+D+L GI F D + LQ+GE I PEL AIE S++ +
Sbjct: 14 YDVFVSFRGADVRKNFLSHLYDSLRRCGISTFMDDVELQRGEYISPELLNAIETSKILIV 73
Query: 108 IFSKNYASSTWCLQELEKICECIKGSGKH-VLPVFYDVDPSEVRKQSGIYSEAFVKHEQR 166
+ +K+YASS WCL EL I + K + H V P+F VDPS++R Q G Y+++F KH+
Sbjct: 74 VLTKDYASSAWCLDELVHIMKSHKNNPSHMVFPIFLYVDPSDIRWQQGSYAKSFSKHKNS 133
Query: 167 FQQDSMKVSRWREALEQVGSISGWDLRDEPLAREIKEIVQKIINILECKYSCVSKDLVGI 226
+ +K WREAL +V +ISGWD+++ A I +I ++I+ L C+Y V VG+
Sbjct: 134 HPLNKLK--DWREALTKVANISGWDIKNRNEAECIADITREILKRLPCQYLHVPSYAVGL 191
Query: 227 DSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLYGQISHQFSASCFIDDVTKIY 286
S +Q + + L + S DGVR I I GMGGIGKTTLA + + SH F S F+++ +
Sbjct: 192 RSRLQHISSLLSIGS-DGVRVIVIYGMGGIGKTTLAKVAFNEFSHLFEGSSFLENFREYS 250
Query: 287 GLHDDPLDVQKQILFQTLGIEHQQICNRYHATTLIQRKLCHERTLMILDNVDQVEQLEKI 346
+ +Q Q+L L + HA ++ + +R L+++D+VD V QL
Sbjct: 251 KKPEGRTHLQHQLLSDILRRNDIEFKGLDHA---VKERFRSKRVLLVVDDVDDVHQLNSA 307
Query: 347 AVHREWLGPGSRIIIISRDEHVLKAYGVDVVYKVSLLDWNEAHMLFCRKAFKDEKIIMSN 406
A+ R+ G GSRIII +R+ H+LK + Y LD +E+ LF AF+ +
Sbjct: 308 AIDRDCFGHGSRIIITTRNMHLLKQLRAEGSYSPKELDGDESLELFSWHAFRTSEPPKEF 367
Query: 407 YQNLVDQILHYAKGLPLAIKVLGSFLFGRNVTEWKSALTRLRQSPVKDVMDVLQLSFDGL 466
Q+ ++++ Y GLPLA++VLG+FL R++ EW+S L L++ P ++ LQ+SF+ L
Sbjct: 368 LQHS-EEVVTYCAGLPLAVEVLGAFLIERSIREWESTLKLLKRIPNDNIQAKLQISFNAL 426
Query: 467 NETEKDIFLHIACFFNNDSEEDVKNILNCCGFHADIGLRVLIDKSLVSISYSIINMHSLL 526
+KD+FL IACFF V IL+ C + DI L +L+++ L++IS + I MH LL
Sbjct: 427 TIEQKDVFLDIACFFIGVDSYYVACILDGCNLYPDIVLSLLMERCLITISGNNIMMHDLL 486
Query: 527 EELGRKIVQNSSSKEPRKWSRLWSTEQLYDVM--------LENMEKHVEAIVLYYKEDEE 578
++GR+IV+ S K+ + SRLWS + V+ +E + + + Y E
Sbjct: 487 RDMGRQIVREISPKKCGERSRLWSHNDVVGVLKKKSGTNAIEGLSLKADVMDFQYFE--- 543
Query: 579 ADFEHLSKMSNLRLLFIANYISTMLGFPSCLSNKLRFVHWFRYPSKYLPSNFHPNELVEL 638
E +KM LRLL + Y+ + G LR++ W + + P N L L
Sbjct: 544 --VEAFAKMQELRLLEL-RYVD-LNGSYEHFPKDLRWLCWHGFSLECFPINLSLESLAAL 599
Query: 639 ILTESNIKQLWKNK---KYLPNLRTLDLRHSRNLEKIIDFGEFPNLERLDLEGCINLVEL 695
L SN+K+ WK + + ++ LDL HS L + DF FPN+E+L L C +LV +
Sbjct: 600 DLQYSNLKRFWKAQSPPQPANMVKYLDLSHSVYLRETPDFSYFPNVEKLILINCKSLVLV 659
Query: 696 DPSIGLL-RKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCGCSKVFNNPRRLMK-SGIS 753
SIG+L +KLV LNL C L +P I+ L SL+ L + CSK+ L + ++
Sbjct: 660 HKSIGILDKKLVLLNLSSCIELDVLPEEIYKLKSLESLFLSNCSKLERLDDALGELESLT 719
Query: 754 SEKKQQHDIRE--SASHHLPGLKWIIL------------------AHDSSHMLP-SLHSL 792
+ +RE S + L LK + L +H S + P SL L
Sbjct: 720 TLLADFTALREIPSTINQLKKLKRLSLNGCKGLLSDDIDNLYSEKSHSVSLLRPVSLSGL 779
Query: 793 CCLRKVDISFCYLSH--VPDAIECLHWLERLNLAGNDFVTLPS-LRKLSKLVYLNLEHCK 849
+R + + +C LS +P+ I L +L L+L GN F LP+ L L L L C
Sbjct: 780 TYMRILSLGYCNLSDELIPEDIGSLSFLRDLDLRGNSFCNLPTDFATLPNLGELLLSDCS 839
Query: 850 LLESLPQLP 858
L+S+ LP
Sbjct: 840 KLQSILSLP 848
Score = 37.0 bits (84), Expect = 0.061
Identities = 29/87 (33%), Positives = 40/87 (45%), Gaps = 4/87 (4%)
Query: 658 LRTLDLRHSRNLEKIIDFGEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLV 717
LR LDLR + DF PNL L L C L + + L R L++L++ C L
Sbjct: 807 LRDLDLRGNSFCNLPTDFATLPNLGELLLSDCSKLQSI---LSLPRSLLFLDVGKCIMLK 863
Query: 718 SIPNNIFGLSSLQYLNMCGCSKVFNNP 744
P +I S+L L + C +F P
Sbjct: 864 RTP-DISKCSALFKLQLNDCISLFEIP 889
>At5g46450 disease resistance protein-like
Length = 1123
Score = 437 bits (1123), Expect = e-122
Identities = 303/863 (35%), Positives = 467/863 (54%), Gaps = 59/863 (6%)
Query: 29 MASSSKSSSALVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQKG 88
MASSS +SS+ YDVF +F GED R F LE K I+ F+D +++
Sbjct: 1 MASSS-------SSSRNWSYDVFPSFSGEDVRKTFLSHFLRELERKSIITFKDN-EMERS 52
Query: 89 ECIGPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPSE 148
+ I PEL AI+ S++ V +FSKNYASS+WCL EL +I C K G+ V+PVFY +DPS
Sbjct: 53 QSIAPELVEAIKDSRIAVIVFSKNYASSSWCLNELLEIMRCNKYLGQQVIPVFYYLDPSH 112
Query: 149 VRKQSGIYSEAFVKHEQRFQQDSMKVSRWREALEQVGSISGWDLRD-EPLAREIKEIVQK 207
+RKQSG + EAF K Q Q + +K ++W++AL V +I G+ ++ A I+EI
Sbjct: 113 LRKQSGEFGEAFKKTCQN-QTEEVK-NQWKQALTDVSNILGYHSKNCNSEATMIEEISSH 170
Query: 208 IINILECKYSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLYG 267
I+ L S ++ VGI I+ ++ L L S D VR +GI G GIGKTT+A L+
Sbjct: 171 ILGKLSLTPSNDFEEFVGIKDHIEKVRLLLHLES-DEVRMVGIWGTSGIGKTTIARALFS 229
Query: 268 QISHQFSASCFIDDV-----TKIYGL-----HDDPLDVQKQILFQTLGIEHQQICNRYHA 317
+S QF +S +ID + YG ++ L +++ LF+ LG ++ +I
Sbjct: 230 NLSSQFQSSVYIDRAFISKSMEGYGRANPDDYNMKLRLRENFLFEILGKKNMKI------ 283
Query: 318 TTLIQRKLCHERTLMILDNVDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVV 377
++ +L H++ L+I+D++D + L+ + +W G GSRII++++++H L+A+G+D V
Sbjct: 284 -GAMEERLKHQKVLIIIDDLDDQDVLDALVGRTQWFGSGSRIIVVTKNKHFLRAHGIDHV 342
Query: 378 YKVSLLDWNEAHMLFCRKAFKDEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFGRNV 437
Y+ L A +FCR AF+ + L ++ A LPL +KVLGS+L GR++
Sbjct: 343 YEACLPSEELALEMFCRYAFRKNSP-PDGFMELSSEVALRAGNLPLGLKVLGSYLRGRDI 401
Query: 438 TEWKSALTRLRQSPVKDVMDVLQLSFDGLNETEKD-IFLHIACFFNNDSEEDVKNILNCC 496
+W + RL+ + L++S+DGLN + + IF HIAC FN + D+K +L
Sbjct: 402 EDWMDMMPRLQNDLDGKIEKTLRVSYDGLNNKKDEAIFRHIACLFNGEKVNDIKLLLAES 461
Query: 497 GFHADIGLRVLIDKSLVSISYSIINMHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYD 556
+IGL+ L+DKSL+ + I MH LL+++G++IV+ + S EP + L ++ +YD
Sbjct: 462 DLDVNIGLKNLVDKSLIFVREDTIEMHRLLQDMGKEIVR-AQSNEPGEREFLVDSKHIYD 520
Query: 557 VMLENM-EKHVEAIVLYYKEDEEADFEHLSKMSNLRLLFIANYISTML---------GFP 606
V+ +N K V I L E + + H S +R L N+ + GF
Sbjct: 521 VLEDNTGTKKVLGIALDINETDGL-YIHESAFKGMRNLLFLNFYTKQKKDVTWHLSEGFD 579
Query: 607 SCLSNKLRFVHWFRYPSKYLPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRHS 666
L KLR + W +YP + +PSNF P LV+L + ES +++LW L LR +DLR S
Sbjct: 580 H-LPPKLRLLSWEKYPLRCMPSNFRPENLVKLQMCESKLEKLWDGVHSLTGLRNMDLRGS 638
Query: 667 RNLEKIIDFGEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGL 726
NL++I D NL++LD+ C +LVEL +I L +L L ++ C++L ++P I L
Sbjct: 639 ENLKEIPDLSLATNLKKLDVSNCTSLVELSSTIQNLNQLEELQMERCENLENLPIGI-NL 697
Query: 727 SSLQYLNMCGCSKVFNNPRRLMKSGISSEKKQQHDIRESASH-HLPGLKWIILAHDSSHM 785
SL LN+ GCSK+ + P + + IS + I E + HL L ++ L S
Sbjct: 698 ESLYCLNLNGCSKLRSFPD--ISTTISELYLSETAIEEFPTELHLENLYYLGLYDMKSEK 755
Query: 786 L----PSLHSLCCLRKVDISFCYLSHVPDAIEC------LHWLERLNLAG-NDFVTLPSL 834
L L L + ++ +LS +P +E LH LE LN+A + TLP+
Sbjct: 756 LWKRVQPLTPLMTMLSPSLTKLFLSDIPSLVELPSSFQNLHNLEHLNIARCTNLETLPTG 815
Query: 835 RKLSKLVYLNLEHCKLLESLPQL 857
L L L+ C L S P +
Sbjct: 816 VNLELLEQLDFSGCSRLRSFPDI 838
>At1g72840 hypothetical protein
Length = 1183
Score = 427 bits (1099), Expect = e-119
Identities = 312/882 (35%), Positives = 461/882 (51%), Gaps = 76/882 (8%)
Query: 29 MASSSKSSSALVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQKG 88
MASSS SS+ ++ HYDVF++FRG DTR L+ AL G++ F+D L+ G
Sbjct: 1 MASSSSSSA-----TRLRHYDVFLSFRGVDTRQTIVSHLYVALRNNGVLTFKDDRKLEIG 55
Query: 89 ECIGPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPSE 148
+ I L +AI+ S V I S+NYA+STWCL+EL I + VLP+FY V PS+
Sbjct: 56 DTIADGLVKAIQTSWFAVVILSENYATSTWCLEELRLIMQLHSEEQIKVLPIFYGVKPSD 115
Query: 149 VRKQSGIYSEAFVKHEQRFQQDSMKVSRWREALEQVGSISGWDLR---DEPLAREIKEIV 205
VR Q G ++ AF ++E + + KVS+WR AL QV ++SG R DE A I E+V
Sbjct: 116 VRYQEGSFATAFQRYEADPEMEE-KVSKWRRALTQVANLSGKHSRNCVDE--ADMIAEVV 172
Query: 206 QKIINILECKYSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTL 265
I + L S +LVG+++ + + L + D V IGI GMGGIGK+T+A L
Sbjct: 173 GGISSRLPRMKSTDLINLVGMEAHMMKMTLLLNIGCEDEVHMIGIWGMGGIGKSTIAKCL 232
Query: 266 YGQISHQFSASCFIDDVTKIYGLHDDPLDVQKQILFQTLGIEHQQICNRYHATTLIQRKL 325
Y + S QF A CF+++V+K Y D +QK++L L E ++ + + I+ +L
Sbjct: 233 YDRFSRQFPAHCFLENVSKGY----DIKHLQKELLSHILYDEDVELWSMEAGSQEIKERL 288
Query: 326 CHERTLMILDNVDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVYKVSLLDW 385
H++ ++LDNVD+VEQL +A W GPGSRIII +RD+ +L + GV+ +Y+V LD
Sbjct: 289 GHQKVFVVLDNVDKVEQLHGLAKDPSWFGPGSRIIITTRDKGLLNSCGVNNIYEVKCLDD 348
Query: 386 NEAHMLFCRKAFKDEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFG-RNVTEWKSAL 444
+A +F + AF + ++ L + A GLP A+ S L + EW+ L
Sbjct: 349 KDALQVFKKLAF-GGRPPSDGFEQLFIRASRLAHGLPSALVAFASHLSAIVAIDEWEDEL 407
Query: 445 TRLRQSPVKDVMDVLQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNILNCCGFHADIGL 504
L P K+V ++L+ S+DGL++ +K +FLH+ACFFN ++ L C D +
Sbjct: 408 ALLETFPQKNVQEILRASYDGLDQYDKTVFLHVACFFNGGHLRYIRAFLKNC----DARI 463
Query: 505 RVLIDKSLVSISY-SIINMHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDVMLENM- 562
L K LV+IS I+MH LL + GR+IV+ S P K LW +++ V+ N
Sbjct: 464 NHLAAKCLVNISIDGCISMHILLVQTGREIVRQESDWRPSKQRFLWDPTEIHYVLDSNTG 523
Query: 563 EKHVEAIVLYYKEDEEADFEHLS---KMSNLRLLFIANYIS------TMLGFPSCLSNKL 613
+ VE + L+ E + S M NL L ++ ++ LS L
Sbjct: 524 TRRVEGLSLHLCEMADTLLLRNSVFGPMHNLTFLKFFQHLGGNVSNLQLISDDYVLSRNL 583
Query: 614 RFVHWFRYPSKYLPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRHSRNLEKII 673
+ +HW YP LP F P+ ++EL L S + LW K LPNLR LD+ SRNL ++
Sbjct: 584 KLLHWDAYPLTILPPIFRPHTIIELSLRYSKLNSLWDGTKLLPNLRILDVTGSRNLRELP 643
Query: 674 DFGEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSL----------------- 716
+ NLE L LE C +LV++ SI L L LN+ C L
Sbjct: 644 ELSTAVNLEELILESCTSLVQIPESINRL-YLRKLNMMYCDGLEGVILVNDLQEASLSRW 702
Query: 717 ------VSIPNNIFGLSSLQYLNMCGCSKVFNNPRRLMKSG----ISSEKKQQHDIRESA 766
+++P++ LSSL L + G K+F L +G SS +K H +S
Sbjct: 703 GLKRIILNLPHSGATLSSLTDLAIQG--KIFIKLSGLSGTGDHLSFSSVQKTAH---QSV 757
Query: 767 SHHLP----GLKWIILAHDSSHMLP------SLHSLCCLRKVDISFCYLSHVPDAIECLH 816
+H L GLK + + S + P S CL ++ + + +P+ I L
Sbjct: 758 THLLNSGFFGLKSLDIKRFSYRLDPVNFSCLSFADFPCLTELKLINLNIEDIPEDICQLQ 817
Query: 817 WLERLNLAGNDFVTLP-SLRKLSKLVYLNLEHCKLLESLPQL 857
LE L+L GNDFV LP S+ +L+ L YL+L +C+ L++LPQL
Sbjct: 818 LLETLDLGGNDFVYLPTSMGQLAMLKYLSLSNCRRLKALPQL 859
Score = 36.6 bits (83), Expect = 0.080
Identities = 22/62 (35%), Positives = 33/62 (52%), Gaps = 1/62 (1%)
Query: 681 LERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCGCSKV 740
L L LE C +LV L + KL YL+L + IP +I LS ++ L + C+K+
Sbjct: 917 LLELSLENCKSLVSLSEELSHFTKLTYLDLSSLE-FRRIPTSIRELSFMRTLYLNNCNKI 975
Query: 741 FN 742
F+
Sbjct: 976 FS 977
Score = 32.3 bits (72), Expect = 1.5
Identities = 30/111 (27%), Positives = 48/111 (43%), Gaps = 34/111 (30%)
Query: 655 LPNLRTLDLRHSRNLEKIIDFGEFPNLERLDLEGCINL------------------VELD 696
L L+ L L + R L+ + + +ERL L GC+ L VE
Sbjct: 839 LAMLKYLSLSNCRRLKALPQLSQ---VERLVLSGCVKLGSLMGILGAGRYNLLDFCVEKC 895
Query: 697 PSIGLL-------------RKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNM 734
S+G L +L+ L+L++CKSLVS+ + + L YL++
Sbjct: 896 KSLGSLMGILSVEKSAPGRNELLELSLENCKSLVSLSEELSHFTKLTYLDL 946
>At4g11170 RPP1-WsA-like disease resistance protein
Length = 1174
Score = 422 bits (1086), Expect = e-118
Identities = 336/1064 (31%), Positives = 533/1064 (49%), Gaps = 99/1064 (9%)
Query: 41 TSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQKGECIGPELFRAIE 100
+SS YDVF +FRGED RNNF L E+KGI+ FRD ++++ IG EL AI
Sbjct: 4 SSSNSWRYDVFPSFRGEDVRNNFLSHLLKEFESKGIVTFRDD-HIKRSHTIGHELRAAIR 62
Query: 101 ISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPSEVRKQSGIYSEAF 160
S++ V +FS+NYASS+WCL EL +I +C + G V+PVFY VDPS++RKQ+G + +F
Sbjct: 63 ESKISVVLFSENYASSSWCLDELIEIMKCKEEQGLKVMPVFYKVDPSDIRKQTGKFGMSF 122
Query: 161 VKHEQRFQQDSMKVSRWREALEQVGSISG-----WDLRDEPLAREIKEIVQKIINILECK 215
+ E + + WR AL +I G WD + K++++K L
Sbjct: 123 L--ETCCGKTEERQHNWRRALTDAANILGDHPQNWDNEAYKITTISKDVLEK----LNAT 176
Query: 216 YSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLYGQISHQFSA 275
S DLVG+++ I +++ L L S GVR +GI G G+GKTT+A LY Q F+
Sbjct: 177 PSRDFNDLVGMEAHIAKMESLLCLES-QGVRIVGIWGPAGVGKTTIARALYNQYHENFNL 235
Query: 276 SCFIDDVTKIYG---LHDDPLDVQKQILFQTLGIEHQQICNRYHATTLIQRKLCHERTLM 332
S F+++V + YG L D L + Q F + ++ + + R+ I+ +L ++ L+
Sbjct: 236 SIFMENVRESYGEAGLDDYGLKLHLQQRFLSKLLDQKDLRVRHLGA--IEERLKSQKVLI 293
Query: 333 ILDNVDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVYKVSLLDWNEAHMLF 392
ILD+VD +EQL+ +A +W G SRI++ ++++ +L ++ ++ +Y+V+ EA +F
Sbjct: 294 ILDDVDNIEQLKALAKENQWFGNKSRIVVTTQNKQLLVSHDINHMYQVAYPSKQEALTIF 353
Query: 393 CRKAFKDEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFGRNVTEWKSALTRLRQSPV 452
C+ AFK + ++L + A LPLA++VLGSF+ G+ EW+ +L L+
Sbjct: 354 CQHAFKQSSP-SDDLKHLAIEFTTLAGHLPLALRVLGSFMRGKGKEEWEFSLPTLKSRLD 412
Query: 453 KDVMDVLQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNILNCCG-FHADIGLRVLIDKS 511
+V VL++ +DGL++ EKD+FLHIAC F+ E +K ++ + GL+VL DKS
Sbjct: 413 GEVEKVLKVGYDGLHDHEKDLFLHIACIFSGQHENYLKQMIIANNDTYVSFGLQVLADKS 472
Query: 512 LVS-ISYSIINMHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDVMLENMEKH----V 566
L+ I MHSLL +LG+++V+ S EP K L + ++ V+ N +
Sbjct: 473 LIQKFENGRIEMHSLLRQLGKEVVRKQSIYEPGKRQFLMNAKETCGVLSNNTGTGTVLGI 532
Query: 567 EAIVLYYKED---EEADFEHLSKMSNLRLLF---IANYISTMLGFPS---CLSNKLRFVH 617
+ KE+ E FE + + L+ I + + L P +LR +H
Sbjct: 533 SLDMCEIKEELYISEKTFEEMRNLVYLKFYMSSPIDDKMKVKLQLPEEGLSYLPQLRLLH 592
Query: 618 WFRYPSKYLPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRHSRNLEKIIDFGE 677
W YP ++ PS+F P LVEL ++ S +K+LW + L NLRT++L SRNLE + + E
Sbjct: 593 WDAYPLEFFPSSFRPECLVELNMSHSKLKKLWSGVQPLRNLRTMNLNSSRNLEILPNLME 652
Query: 678 FPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCGC 737
L RLDL C +LVEL SI L+ L+ L + CK L IP NI L SL+ L+ C
Sbjct: 653 ATKLNRLDLGWCESLVELPSSIKNLQHLILLEMSCCKKLEIIPTNI-NLPSLEVLHFRYC 711
Query: 738 SKVFNNPRRLMKSGISSEKKQQHDIRESASHHLPGLKWIILAHDSSHMLPSLHSLCCLRK 797
+++ P IS+ + + I + + P +K+ S C+ +
Sbjct: 712 TRLQTFPE------ISTNIRLLNLIGTAITEVPPSVKYW-----------SKIDEICMER 754
Query: 798 VDISFCYLSHVPDAIECLHWLERLNLAGN-DFVTLPS-LRKLSKLVYLNLEHCKLLESLP 855
+ L HVP + LE+L L N + T+P L+ L +L +++ +C + SLP
Sbjct: 755 AKVK--RLVHVP------YVLEKLCLRENKELETIPRYLKYLPRLQMIDISYCINIISLP 806
Query: 856 QLPFPTNT-GEVHREYDDYFCG------AGLLIFNCPKLGEREHCRSMTLLWMKQFIKAN 908
+LP + V+ E G L NC KLG+R + +++ Q
Sbjct: 807 KLPGSVSALTAVNCESLQILHGHFRNKSIHLNFINCLKLGQRAQEKIHRSVYIHQ----- 861
Query: 909 PRSSSEIQIVNPGSEIPSWINNQRMGYSIAIDRSPIRHDNDNNIIGIVCCAAFTMAPYRE 968
SS I V PG +P++ + + G SI I + + N + A +
Sbjct: 862 ---SSYIADVLPGEHVPAYFSYRSTGSSIMIHSNKVDLSKFNRFKVCLVLGAGKRFEGCD 918
Query: 969 IFYSSELMNLAFKRIDSNERLLKMRVPVKLSLVTTKSSHLWII-------YLPREYPGYS 1021
I + +K+ R + VP L KS HL + + P E+
Sbjct: 919 IKF--------YKQFFCKPR--EYYVPKHLDSPLLKSDHLCMCEFELMPPHPPTEWELLH 968
Query: 1022 CHEFGKIELKFFEVEGL---EVESCGYRWVCKQDIQEFNLIMNH 1062
+EF +E+ F GL EV+ CG +++ + EF + H
Sbjct: 969 PNEF--LEVSFESRGGLYKCEVKECGLQFLEPHETSEFRYLSPH 1010
>At5g46470 disease resistance protein-like
Length = 1160
Score = 412 bits (1059), Expect = e-115
Identities = 301/869 (34%), Positives = 455/869 (51%), Gaps = 52/869 (5%)
Query: 29 MASSSKSSSALVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQKG 88
MASSS SSS + Y VF +F GED RN F L+ K I+ F+D +++
Sbjct: 1 MASSSSSSS------RNWSYHVFPSFSGEDVRNTFLSHFLKELDRKLIISFKDN-EIERS 53
Query: 89 ECIGPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPSE 148
+ + PEL I S++ V +FSK YASS+WCL EL +I +C K G+ V+P+FY++DPS
Sbjct: 54 QSLDPELKHGIRNSRIAVVVFSKTYASSSWCLNELLEIVKCKKEFGQLVIPIFYNLDPSH 113
Query: 149 VRKQSGIYSEAFVKHEQRFQQDSMKVSRWREALEQVGSISGWDLRD-EPLAREIKEIVQK 207
VRKQ+G + + F K + D RW+EAL V +I G+ + + A I+EI
Sbjct: 114 VRKQTGDFGKIFEKTCRNKTVDEK--IRWKEALTDVANILGYHIVTWDNEASMIEEIAND 171
Query: 208 IINILECKYSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLYG 267
I+ + S +DLVGI+ I + + L L S + VR +GI G GIGKTT+A L+
Sbjct: 172 ILGKMNISPSNDFEDLVGIEDHITKMSSLLHLESEE-VRMVGIWGPSGIGKTTIARALFS 230
Query: 268 QISHQFSASCFIDDV-----TKIYG---LHDDPLDVQKQILFQTLGIEHQQICNRYHATT 319
++S QF +S FID V ++Y L D + + Q F + + I + H
Sbjct: 231 RLSCQFQSSVFIDKVFISKSMEVYSGANLVDYNMKLHLQRAFLAEIFDKKDI--KIHVGA 288
Query: 320 LIQRKLCHERTLMILDNVDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVYK 379
+ ++ + H + L+++D++D + L+ +A +W G GSRII+++ ++H L+A +D +YK
Sbjct: 289 M-EKMVKHRKALIVIDDLDDQDVLDALADQTQWFGSGSRIIVVTENKHFLRANRIDHIYK 347
Query: 380 VSLLDWNEAHMLFCRKAFKDEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFGRNVTE 439
V L A +FCR AFK ++ L ++ A LPL + VLGS L G N
Sbjct: 348 VCLPSNALALEMFCRSAFKKNSP-PDDFLELSSEVALRAGNLPLGLNVLGSNLRGINKGY 406
Query: 440 WKSALTRLRQSPVKDVMDVLQLSFDGLNETEKD-IFLHIACFFNNDSEEDVKNILNCCGF 498
W L RL+ K + L++S+DGLN + + IF HIAC FN + D+K +L
Sbjct: 407 WIDMLPRLQGLDGK-IGKTLRVSYDGLNNRKDEAIFRHIACIFNGEKVSDIKLLLANSNL 465
Query: 499 HADIGLRVLIDKSLVSISYSIINMHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDVM 558
+IGL+ L+D+SL+ ++ + MHSLL+ELG++IV+ S+ +P + L + + DV+
Sbjct: 466 DVNIGLKNLVDRSLICERFNTLEMHSLLQELGKEIVRTQSN-QPGEREFLVDLKDICDVL 524
Query: 559 LENM-EKHVEAIVLYYKEDEEADFEHLSKMSNLRLLFIANYISTM-------LGFPS--- 607
N K V I L E +E S LLF+ Y + P
Sbjct: 525 EHNTGTKKVLGITLDIDETDELHIHESSFKGMHNLLFLKIYTKKLDQKKKVRWHLPERFD 584
Query: 608 CLSNKLRFVHWFRYPSKYLPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRHSR 667
L ++LR + + RYPSK LPSNFHP LV+L + +S +++LW L LR +DLR SR
Sbjct: 585 YLPSRLRLLRFDRYPSKCLPSNFHPENLVKLQMQQSKLEKLWDGVHSLAGLRNMDLRGSR 644
Query: 668 NLEKIIDFGEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLS 727
NL++I D NLE L L C +LVEL SI L KL L++ C L +IP+ + L
Sbjct: 645 NLKEIPDLSMATNLETLKLSSCSSLVELPSSIQYLNKLNDLDMSYCDHLETIPSGV-NLK 703
Query: 728 SLQYLNMCGCSKV---FNNPRRLMKSGISSEKKQQHDIRESASHHLPGLKWIILAHDSSH 784
SL LN+ GCS++ + P + I ++R L L +IL
Sbjct: 704 SLDRLNLSGCSRLKSFLDIPTNISWLDIGQTADIPSNLR------LQNLDELILCERVQL 757
Query: 785 MLP--SLHSLCCLRKVDISFCYLSHVPDAIECLHWLERLNLAG-NDFVTLPSLRKLSKLV 841
P ++ S R + VP +I+ L+ LE L + + VTLP+ L L+
Sbjct: 758 RTPLMTMLSPTLTRLTFSNNPSFVEVPSSIQNLYQLEHLEIMNCRNLVTLPTGINLDSLI 817
Query: 842 YLNLEHCKLLESLPQLPFPTNTGEVHREY 870
L+L HC L++ P + TN +++ Y
Sbjct: 818 SLDLSHCSQLKTFPDI--STNISDLNLSY 844
Score = 37.0 bits (84), Expect = 0.061
Identities = 38/136 (27%), Positives = 54/136 (38%), Gaps = 22/136 (16%)
Query: 626 LPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRHSRNLEKI-IDFGEFPNLERL 684
+PSN L ELIL E + P L L ++ + ++ LE L
Sbjct: 737 IPSNLRLQNLDELILCERVQLRTPLMTMLSPTLTRLTFSNNPSFVEVPSSIQNLYQLEHL 796
Query: 685 DLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPN--------------------NIF 724
++ C NLV L I L L+ L+L C L + P+ +I
Sbjct: 797 EIMNCRNLVTLPTGINL-DSLISLDLSHCSQLKTFPDISTNISDLNLSYTAIEEVPLSIE 855
Query: 725 GLSSLQYLNMCGCSKV 740
LS L YL+M GCS +
Sbjct: 856 KLSLLCYLDMNGCSNL 871
Score = 35.4 bits (80), Expect = 0.18
Identities = 26/91 (28%), Positives = 43/91 (46%), Gaps = 2/91 (2%)
Query: 626 LPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRHSRNLEKIIDFGEFPNLERLD 685
LP+ + + L+ L L S+ QL N+ L+L ++ E + + L LD
Sbjct: 807 LPTGINLDSLISLDL--SHCSQLKTFPDISTNISDLNLSYTAIEEVPLSIEKLSLLCYLD 864
Query: 686 LEGCINLVELDPSIGLLRKLVYLNLKDCKSL 716
+ GC NL+ + P+I L+ L + DC L
Sbjct: 865 MNGCSNLLCVSPNISKLKHLERADFSDCVEL 895
>At5g18370 disease resistance protein -like
Length = 1210
Score = 410 bits (1053), Expect = e-114
Identities = 293/861 (34%), Positives = 456/861 (52%), Gaps = 49/861 (5%)
Query: 30 ASSSKSSSALVTS--SKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQK 87
+SSS SS ++S S+ + VF +FRGED R F + ++KGI+ F D +++
Sbjct: 41 SSSSLSSPPSLSSPISRTWTHQVFPSFRGEDVRKGFLSHIQKEFKSKGIVPFIDD-EMKR 99
Query: 88 GECIGPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPS 147
GE IGP LF+AI S++ + + SKNYASS+WCL EL +I C + G+ V+ VFY VDPS
Sbjct: 100 GESIGPGLFQAIRESKIAIVLLSKNYASSSWCLNELVEIMNCREEIGQTVMTVFYQVDPS 159
Query: 148 EVRKQSGIYSEAFVKHEQRFQQDSMKVSRWREALEQVGSISGWDLRD-EPLAREIKEIVQ 206
+VRKQ+G + +AF K Q+ + RW AL V +I G D R + A I ++ +
Sbjct: 160 DVRKQTGDFGKAFKKTCVGKTQEVKQ--RWSRALMDVANILGQDSRKWDKEADMIVKVAK 217
Query: 207 KIINILECKYSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLY 266
+ ++L S D VGI I + + L L S D VR IGI G GIGKTT+A LY
Sbjct: 218 DVSDVLSYTPSRDFDDYVGIRPHITRINSLLCLESSD-VRMIGILGPPGIGKTTIARVLY 276
Query: 267 GQISHQFSASCFIDDVTKIY--GLHDD-----PLDVQKQILFQTLGIEHQQICNRYHATT 319
QIS +F S FI+++ Y G HD+ P+++ + L ++ + + ++
Sbjct: 277 DQISEKFQFSAFIENIRLSYWKGWHDEGNLDFPVEIMTGDRQRKLNLQRRLLSELFNQKD 336
Query: 320 L-------IQRKLCHERTLMILDNVDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAY 372
+ +Q +L + L+ILD VDQ+EQL +A +W G GSRIII ++D+ +L+A+
Sbjct: 337 IQVRHLGAVQERLRDHKVLVILDGVDQLEQLTALAKETQWFGYGSRIIITTQDQRLLRAH 396
Query: 373 GVDVVYKVSLLDWNEAHMLFCRKAFKDEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFL 432
++ VYKV L +EA +FC AF +K ++ L + A LPL ++VLGS+L
Sbjct: 397 EINHVYKVDLPATDEALQIFCLYAF-GQKFPYDGFKKLAREFTALAGELPLGLRVLGSYL 455
Query: 433 FGRNVTEWKSALTRLRQSPVKDVMDVLQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNI 492
G ++ EWK+AL RLR S ++ L+ +++ L++ +K +FLHIAC FN VK
Sbjct: 456 RGMSLEEWKNALPRLRTSLDGEIEKTLRFAYNVLSDKDKSLFLHIACLFNGCQVNHVKQW 515
Query: 493 LNCCGFHADIGLRVLIDKSLVSISYSIINMHSLLEELGRKIVQNSSSKEPRKWSRLWSTE 552
L + G VL +KSL+S ++ MHSLL++LG IV+ S EP K L
Sbjct: 516 LANSSLDVNHGFEVLSNKSLISTDMGLVRMHSLLQQLGVDIVRKQSIGEPEKRQFLVDVN 575
Query: 553 QLYDVMLENM-EKHVEAIVLYYKEDEEA---DFEHLSKMSNLRLLFIANYISTMLGFP-- 606
++ DV+ +N + I+L+ + E+ + +M+NL+ L + + L P
Sbjct: 576 EISDVITDNTGTGTILGIMLHVSKIEDVLVIEETVFDRMTNLQFLILDECLRDKLNLPLG 635
Query: 607 -SCLSNKLRFVHWFRYPSKYLPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRH 665
+CL K+R + W P PS F LVELI+ + ++LW+ + L NL+ ++L
Sbjct: 636 LNCLPRKIRLLRWDYCPLSIWPSKFSAKFLVELIMRANKFEKLWEGIQPLKNLKRMELGD 695
Query: 666 SRNLEKIIDFGEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFG 725
+RNL++I D NLE L L C +L+E+ SI L L+L C SLV + + I
Sbjct: 696 ARNLKEIPDLSNATNLESLLLSFCTSLLEIPSSIRGTTNLKELDLGGCASLVKLSSCICN 755
Query: 726 LSSLQYLNMCGCSKVFNNPRRLMKSGISSEKKQQHDIRESAS------HHLPGLKWIILA 779
+SL+ LN+ CS + P L G S+ + + +S ++ + L+
Sbjct: 756 ATSLEELNLSACSNLVELPCAL--PGDSNMRSLSKLLLNGSSRLKTFPEISTNIQELNLS 813
Query: 780 HDSSHMLPSLHSLCC-LRKVDISFC----YLSHVPDAIECLHWLERLNLAGNDFVTLPS- 833
+ +PS L L K+D+S C VPD I LNL+ + +P
Sbjct: 814 GTAIEEVPSSIRLWSRLDKLDMSRCKNLKMFPPVPDGISV------LNLSETEIEDIPPW 867
Query: 834 LRKLSKLVYLNLEHCKLLESL 854
+ LS+L + + CK L+++
Sbjct: 868 VENLSQLRHFVMIRCKKLDNI 888
>At5g18350 disease resistance protein -like
Length = 1193
Score = 408 bits (1049), Expect = e-114
Identities = 277/746 (37%), Positives = 399/746 (53%), Gaps = 33/746 (4%)
Query: 29 MASSSKSSSALVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQKG 88
M + SSS T ++ + VF++FRGED R F + E KGI F D +++G
Sbjct: 1 MQPNRASSSLSSTPTRTWTHHVFLSFRGEDVRKGFLSHIQKEFERKGIFPFVDT-KMKRG 59
Query: 89 ECIGPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPSE 148
IGP L AI +S++ + + SKNYASSTWCL EL I +C + G+ V+ VFY+VDPS+
Sbjct: 60 SSIGPVLSDAIIVSKIAIVLLSKNYASSTWCLNELVNIMKCREEFGQTVMTVFYEVDPSD 119
Query: 149 VRKQSGIYSEAFVKHEQRFQQDSMKVSR-WREALEQVGSISGWDLRDEPLARE-IKEIVQ 206
VRKQ+G + AF E + +V + WR+AL V +I G R + I +I +
Sbjct: 120 VRKQTGDFGIAF---ETTCVGKTEEVKQSWRQALIDVSNIVGEVYRIWSKESDLIDKIAE 176
Query: 207 KIINILECKYSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLY 266
+++ L S VGI ++ +++ L L S D VR IGI G GIGKTT+A L
Sbjct: 177 DVLDELNYTMSRDFDGYVGIGRHMRKMKSLLCLESGD-VRMIGIVGPPGIGKTTIARALR 235
Query: 267 GQISHQFSASCFIDDVTKIY--------GLHDDPL---DVQKQILFQTLGIEHQQICNR- 314
QIS F + FIDD+ Y GL D +++I+ QT + +I N+
Sbjct: 236 DQISENFQLTAFIDDIRLTYPRRCYGESGLKPPTAFMNDDRRKIVLQTNFLS--EILNQK 293
Query: 315 ---YHATTLIQRKLCHERTLMILDNVDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKA 371
H L + L+ILD+VD +EQL+ +A W G GSRIII ++D +LKA
Sbjct: 294 DIVIHNLNAAPNWLKDRKVLVILDDVDHLEQLDAMAKETGWFGYGSRIIITTQDRKLLKA 353
Query: 372 YGVDVVYKVSLLDWNEAHMLFCRKAFKDEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSF 431
+ +D +Y+V L ++A +FC AF + ++Q L ++ A LPL +KVLGS+
Sbjct: 354 HNIDYIYEVGLPRKDDALQIFCLSAF-GQNFPHDDFQYLACEVTQLAGELPLGLKVLGSY 412
Query: 432 LFGRNVTEWKSALTRLRQSPVKDVMDVLQLSFDGLNETEKDIFLHIACFFNNDSEEDVKN 491
L G ++ EWK+AL RL+ D+ L+ S+D L+ ++ +FLHIAC F VK
Sbjct: 413 LKGMSLEEWKNALPRLKTCLDGDIEKTLRYSYDALSRKDQALFLHIACLFRGYEVGHVKQ 472
Query: 492 ILNCCGFHADIGLRVLIDKSLVSISYSIINMHSLLEELGRKIVQNSSSKEPRKWSRLWST 551
L D GL VL KSL+SI +NMHSLL++LG +IV+N SS+EPR+ L
Sbjct: 473 WLGKSDLDVDHGLDVLRQKSLISIDMGFLNMHSLLQQLGVEIVRNQSSQEPRERQFLVDV 532
Query: 552 EQLYDVMLENM--EKHVEAIVLYYKEDEE---ADFEHLSKMSNLRLLFIANYISTMLGFP 606
+ DV N K + I L E EE D M+NL+ LF+ L P
Sbjct: 533 NDISDVFTYNTAGTKSILGIRLNVPEIEEKIVIDELVFDGMTNLQFLFVNEGFGDKLSLP 592
Query: 607 ---SCLSNKLRFVHWFRYPSKYLPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDL 663
+CL KLR +HW P + PS F N LVEL++ +N ++LW+ L +L+ +DL
Sbjct: 593 RGLNCLPGKLRVLHWNYCPLRLWPSKFSANFLVELVMRGNNFEKLWEKILPLKSLKRMDL 652
Query: 664 RHSRNLEKIIDFGEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNI 723
HS++L++I D NLE LDL C L+EL SIG L L L C L +P++I
Sbjct: 653 SHSKDLKEIPDLSNATNLEELDLSSCSGLLELTDSIGKATNLKRLKLACCSLLKKLPSSI 712
Query: 724 FGLSSLQYLNMCGCSKVFNNPRRLMK 749
++LQ L++ C P+ + K
Sbjct: 713 GDATNLQVLDLFHCESFEELPKSIGK 738
Score = 51.2 bits (121), Expect = 3e-06
Identities = 58/214 (27%), Positives = 97/214 (45%), Gaps = 30/214 (14%)
Query: 657 NLRTLDLRHSRNLEKIIDFGEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSL 716
N++ LDLR++ + L RLD+ C NL E P++ + +V L+L + +
Sbjct: 800 NVKELDLRNTAIENVPSSICSWSCLYRLDMSECRNLKEF-PNVPV--SIVELDLSKTE-I 855
Query: 717 VSIPNNIFGLSSLQYLNMCGC----------SKVFN-NPRRLMKSGISSEKKQQHDIRES 765
+P+ I L L+ L M GC SK+ N L G+S + + E
Sbjct: 856 EEVPSWIENLLLLRTLTMVGCKRLNIISPNISKLKNLEDLELFTDGVSGDAASFYAFVEF 915
Query: 766 ASHHLPGLKWIILAHDSSHMLPSLHSLCCLRKVDISFCYLSH----VPDAIECLHWLERL 821
+ H W + + H + + CL K+ IS + S+ +PD I CL L L
Sbjct: 916 SDRH----DWTLESDFQVHYILPI----CLPKMAISLRFWSYDFETIPDCINCLPGLSEL 967
Query: 822 NLAG-NDFVTLPSLRKLSKLVYLNLEHCKLLESL 854
+++G + V+LP L L+ L+ +C+ LE +
Sbjct: 968 DVSGCRNLVSLPQLP--GSLLSLDANNCESLERI 999
Score = 32.3 bits (72), Expect = 1.5
Identities = 24/72 (33%), Positives = 35/72 (48%), Gaps = 5/72 (6%)
Query: 679 PNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCGCS 738
P L LD+ GC NLV L G L+ L+ +C+SL I N F + LN C
Sbjct: 962 PGLSELDVSGCRNLVSLPQLPG---SLLSLDANNCESLERI-NGSFQNPEI-CLNFANCI 1016
Query: 739 KVFNNPRRLMKS 750
+ R+L+++
Sbjct: 1017 NLNQEARKLIQT 1028
>At5g38340 disease resistance - like protein
Length = 1059
Score = 408 bits (1048), Expect = e-113
Identities = 303/956 (31%), Positives = 506/956 (52%), Gaps = 80/956 (8%)
Query: 35 SSSALVT--SSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQKGECIG 92
SS AL T SS YDVF +F G D R F + + KGI+ F D ++ + + IG
Sbjct: 42 SSLALPTIPSSLSRKYDVFPSFHGADVRKTFLSHMLKEFKRKGIVPFIDN-DIDRSKSIG 100
Query: 93 PELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPSEVRKQ 152
PEL AI S++ + + SKNYASS+WCL EL +I +C K + V+ +FY VDP++V+KQ
Sbjct: 101 PELDEAIRGSKIAIVMLSKNYASSSWCLNELVEITKCRKDLNQTVMTIFYGVDPTDVKKQ 160
Query: 153 SGIYSEAFVKHEQRFQQDSMKVSRWREALEQVGSISG--WDLRDEPLAREIKEIVQKIIN 210
+G + + F + + ++ +K WRE L+ +I+G W + D A I++I + N
Sbjct: 161 TGEFGKVFERTCESKTEEQVKT--WREVLDGAATIAGEHWHIWDNE-ASMIEKISIDVSN 217
Query: 211 ILE-CKYSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLYGQI 269
IL S DL+G+++ ++ +++ L L+S + V+ IGI G GIGKTT+A LY +
Sbjct: 218 ILNRSSPSRDFDDLIGMEAHMEKMKSLLSLHSNE-VKMIGIWGPSGIGKTTIARVLYNRF 276
Query: 270 SHQFSASCFIDDVTKIYGLHDDP---------LDVQKQILFQTLGIEHQQICNRYHATTL 320
S F S F+D++ ++ +H P L +Q Q++ + + +I + +
Sbjct: 277 SGDFGLSVFMDNIKEL--MHTRPVGSDDYSAKLHLQNQLMSEITNHKETKITH----LGV 330
Query: 321 IQRKLCHERTLMILDNVDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVYKV 380
+ +L + L++LD++DQ QL+ IA +W GPGSRIII ++D+ +L+A+ ++ +YKV
Sbjct: 331 VPDRLKDNKVLIVLDSIDQSIQLDAIAKETQWFGPGSRIIITTQDQKLLEAHDINNIYKV 390
Query: 381 SLLDWNEAHMLFCRKAFKDEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFGRNVTEW 440
EA +FC AF + ++ L ++ LPL ++V+GS + +W
Sbjct: 391 EFPSKYEAFQIFCTYAF-GQNFPKDGFEKLAWEVTDLLGELPLGLRVMGSHFRRMSKDDW 449
Query: 441 KSALTRLRQSPVKDVMDVLQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNILNCCGFHA 500
AL RL+ ++ +L+ S+D L+ +KD+FLHIAC FNN+ V++ L A
Sbjct: 450 VIALPRLKTRLDANIQSILKFSYDALSPEDKDLFLHIACLFNNEEIVKVEDYLALDFLDA 509
Query: 501 DIGLRVLIDKSLVS---ISYSIINMHSLLEELGRKIVQ----NSSSKEPRKWSRLWSTEQ 553
GL +L +KSL+ ++Y ++ MH+LLE+LG++IV+ + S +EP K L T+
Sbjct: 510 RHGLHLLAEKSLIDLEGVNYKVLKMHNLLEQLGKEIVRYHPAHHSIREPEKRQFLVDTKD 569
Query: 554 LYDVMLENM-EKHVEAIVLYYKEDEEADFEHLSK-----MSNLRLLFIANYISTMLGFPS 607
+ +V+ + K ++ I + D + ++S+ M+NL+ L + S L P
Sbjct: 570 ICEVLADGTGSKSIKGIC--FDLDNLSGRLNISERAFEGMTNLKFLRVLRDRSEKLYLPQ 627
Query: 608 CLS---NKLRFVHWFRYPSKYLPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDLR 664
L+ KLR + W +P K LPSNF LV L + +S +++LW+ K+ L NL+ ++L
Sbjct: 628 GLNYLPKKLRLIEWDYFPMKSLPSNFCTTYLVNLHMRKSKLEKLWEGKQPLGNLKWMNLS 687
Query: 665 HSRNLEKIIDFGEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIF 724
+SRNL+++ D L+ L+L C +LVE+ SIG L LNL C SLV +P++I
Sbjct: 688 NSRNLKELPDLSTATKLQDLNLTRCSSLVEIPFSIGNTTNLEKLNLVMCTSLVELPSSIG 747
Query: 725 GLSSLQYLNMCGCSKVFNNPRRLMKSGISSEKKQQHDIRESASHHLPGLKWIILAHDSSH 784
L L+ L + GCSK+ P + + + + +S +K + LA + +
Sbjct: 748 SLHKLRELRLRGCSKLEVLPTNISLESLDNLDITDCSLLKSFPDISTNIKHLSLARTAIN 807
Query: 785 MLPS-LHSLCCLRKVDISFCYLSHVPDAIECLHWLERLNLAGNDFVTLPS-LRKLSKLVY 842
+PS + S LR +S Y ++ ++ L + L+ LP ++K+S+L
Sbjct: 808 EVPSRIKSWSRLRYFVVS--YNENLKESPHALDTITMLSSNDTKMQELPRWVKKISRLET 865
Query: 843 LNLEHCKLLESLPQLPFP-TNTGEVHRE-----------YDDYFCGAGLLIFNCPKLGER 890
L LE CK L +LP+LP +N G ++ E + + F G NC KL +
Sbjct: 866 LMLEGCKNLVTLPELPDSLSNIGVINCESLERLDCSFYKHPNMFIG----FVNCLKLNKE 921
Query: 891 EHCRSMTLLWMKQFIKANPRSSSEIQIVNPGSEIPSWINNQRMGYSIAI--DRSPI 944
++ I+ SSS I+ PG +PS ++ G S+ + ++SP+
Sbjct: 922 ----------ARELIQT---SSSTCSIL-PGRRVPSNFTYRKTGGSVLVNLNQSPL 963
>At5g41750 disease resistance protein-like
Length = 1068
Score = 406 bits (1044), Expect = e-113
Identities = 280/818 (34%), Positives = 442/818 (53%), Gaps = 51/818 (6%)
Query: 48 YDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQKGECIGPELFRAIEISQVYVA 107
Y VF +F G D R F L +KGI F D + +G+ IGPEL + I ++V +
Sbjct: 13 YQVFSSFHGPDVRKGFLSHLHSVFASKGITTFNDQ-KIDRGQTIGPELIQGIREARVSIV 71
Query: 108 IFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPSEVRKQSGIYSEAFVKHEQRF 167
+ SK YASS+WCL EL +I +C + G+ V+ VFY+VDPS+V+KQSG++ EAF K Q
Sbjct: 72 VLSKKYASSSWCLDELVEILKCKEALGQIVMTVFYEVDPSDVKKQSGVFGEAFEKTCQG- 130
Query: 168 QQDSMKVSRWREALEQVGSISG-----WDLRDEPLAREIKEIVQKIINILECKYSCVSKD 222
+ + +K+ RWR AL V +I+G WD A+ I++IV + + L S +
Sbjct: 131 KNEEVKI-RWRNALAHVATIAGEHSLNWDNE----AKMIQKIVTDVSDKLNLTPSRDFEG 185
Query: 223 LVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLYGQISHQFSASCFIDDV 282
+VG+++ ++ L + L L S D V+ IGI G GIGKTT+A TL+ +IS F CF++++
Sbjct: 186 MVGMEAHLKRLNSLLCLES-DEVKMIGIWGPAGIGKTTIARTLFNKISSIFPFKCFMENL 244
Query: 283 T---KIYGLHDDPLDVQKQILFQTLGIEHQQICNRYHATTLIQRKLCHERTLMILDNVDQ 339
K H L +QKQ+L + L E+ +I H I++ L ++ L+ILD+VD
Sbjct: 245 KGSIKGGAEHYSKLSLQKQLLSEILKQENMKI----HHLGTIKQWLHDQKVLIILDDVDD 300
Query: 340 VEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVYKVSLLDWNEAHMLFCRKAFKD 399
+EQLE +A W G GSRII+ + D+++LKA+ + +Y V EA + C AFK
Sbjct: 301 LEQLEVLAEDPSWFGSGSRIIVTTEDKNILKAHRIQDIYHVDFPSEEEALEILCLSAFKQ 360
Query: 400 EKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFGRNVTEWKSALTRLRQSPVKDVMDVL 459
I ++ L +++ LPL + V+G+ L ++ EW+ L+R+ S K++ ++L
Sbjct: 361 SSI-PDGFEELANKVAELCGNLPLGLCVVGASLRRKSKNEWERLLSRIESSLDKNIDNIL 419
Query: 460 QLSFDGLNETEKDIFLHIACFFNNDSEEDVKNILNCCGFHADIGLRVLIDKSLVSISYS- 518
++ +D L+ ++ +FLHIACFFNN+ + + +L G +L D+SLV IS
Sbjct: 420 RIGYDRLSTEDQSLFLHIACFFNNEKVDYLTALLADRKLDVVNGFNILADRSLVRISTDG 479
Query: 519 -IINMHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDVMLENM-EKHVEAIVLYYKED 576
++ H LL++LGR+IV EP K L E++ DV+ + + V+ I
Sbjct: 480 HVVMHHYLLQKLGRRIVHEQWPNEPGKRQFLIEAEEIRDVLTKGTGTESVKGISFDTSNI 539
Query: 577 EEAD-----FEHLSKMSNLRLLFIANYISTMLGFPSCLSN--KLRFVHWFRYPSKYLPSN 629
EE FE + + LR+ + L P + +R +HW YP K LP
Sbjct: 540 EEVSVGKGAFEGMRNLQFLRIYRDSFNSEGTLQIPEDMEYIPPVRLLHWQNYPRKSLPQR 599
Query: 630 FHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRHSRNLEKIIDFGEFPNLERLDLEGC 689
F+P LV++ + S +K+LW + LPNL+++D+ S +L++I + + NLE L LE C
Sbjct: 600 FNPEHLVKIRMPSSKLKKLWGGIQPLPNLKSIDMSFSYSLKEIPNLSKATNLEILSLEFC 659
Query: 690 INLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCGCSKVFNNPRRLMK 749
+LVEL SI L KL LN+++C L IP NI L+SL+ L+M GCS++ P
Sbjct: 660 KSLVELPFSILNLHKLEILNVENCSMLKVIPTNI-NLASLERLDMTGCSELRTFP----- 713
Query: 750 SGISSEKKQQHDIRESASHHLPGLK--WIILAH--DSSHMLPSLHSLCCLRKVDISFCYL 805
ISS K+ ++ ++ +P W L H S L LH C+ + + +
Sbjct: 714 -DISSNIKKL-NLGDTMIEDVPPSVGCWSRLDHLYIGSRSLKRLHVPPCITSLVLWKSNI 771
Query: 806 SHVPDAIECLHWLERLNLAGNDFVTLPSLRKLSKLVYL 843
+P++I + L D++ + S RKL ++ L
Sbjct: 772 ESIPESI--------IGLTRLDWLNVNSCRKLKSILGL 801
Score = 33.5 bits (75), Expect = 0.67
Identities = 24/64 (37%), Positives = 38/64 (58%), Gaps = 4/64 (6%)
Query: 795 LRKVDISFCY-LSHVPDAIECLHWLERLNLAG-NDFVTLP-SLRKLSKLVYLNLEHCKLL 851
L+ +D+SF Y L +P+ + + LE L+L V LP S+ L KL LN+E+C +L
Sbjct: 628 LKSIDMSFSYSLKEIPNLSKATN-LEILSLEFCKSLVELPFSILNLHKLEILNVENCSML 686
Query: 852 ESLP 855
+ +P
Sbjct: 687 KVIP 690
>At5g18360 disease resistance protein -like
Length = 900
Score = 406 bits (1044), Expect = e-113
Identities = 292/845 (34%), Positives = 449/845 (52%), Gaps = 66/845 (7%)
Query: 34 KSSSALVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQKGECIGP 93
+ SS+L S + VF +F G+D R F L KGI F D ++++ + I
Sbjct: 2 EESSSLSLQSCNWRHHVFPSFSGKDVRRTFLSHLLKEFRRKGIRTFIDN-DIKRSQMISS 60
Query: 94 ELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPSEVRKQS 153
EL RAI S++ V + S+ YASS+WCL EL +I K + ++PVFY+VDPS+VRK++
Sbjct: 61 ELVRAIRESRIAVVVLSRTYASSSWCLNELVEI----KKVSQMIMPVFYEVDPSDVRKRT 116
Query: 154 GIYSEAFVKHEQRFQQDSMKVSRWREALEQVGSISG-----WDLRDEPLAREIKEIVQKI 208
G + +AF + +R + +K +WREAL + +I+G WD A I +I I
Sbjct: 117 GEFGKAFEEACERQPDEEVK-QKWREALVYIANIAGESSQNWDNE----ADLIDKIAMSI 171
Query: 209 INILECKYSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLYGQ 268
L S S +LVGID+ ++ L + L L S + V+ +GI G GIGKTT+A L+ +
Sbjct: 172 SYELNSTLSRDSYNLVGIDNHMRELDSLLCLESTE-VKMVGIWGPAGIGKTTIARALFNR 230
Query: 269 ISHQFSASCFIDDV-----TKIYGLHDDPLDVQKQILFQTLGIEHQQICNRYHATTLIQR 323
+S F + F+++V T + L +Q+Q L + + +H +I H L++
Sbjct: 231 LSENFQHTIFMENVKGSSRTSELDAYGFQLRLQEQFLSEVIDHKHMKI----HDLGLVKE 286
Query: 324 KLCHERTLMILDNVDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVYKVSLL 383
+L + L++LD+VD++EQL+ + +W G GSRII+ + ++ +L+A+G+ +Y++
Sbjct: 287 RLQDLKVLVVLDDVDKLEQLDALVKQSQWFGSGSRIIVTTENKQLLRAHGITCIYELGFP 346
Query: 384 DWNEAHMLFCRKAFKDEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFGRNVTEWKSA 443
+++ +FC+ AF E L +I A LPLA+KVLGS L G + E KSA
Sbjct: 347 SRSDSLQIFCQYAF-GESSAPDGCIELATEITKLAGYLPLALKVLGSSLRGMSKDEQKSA 405
Query: 444 LTRLRQSPVKDVMDVLQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNILNCCGFHADIG 503
L RLR S +D+ +VL++ +DG+++ +K IFLHIAC FN ++ + VK IL G G
Sbjct: 406 LPRLRTSLNEDIRNVLRVGYDGIHDKDKVIFLHIACLFNGENVDYVKQILASSGLDVTFG 465
Query: 504 LRVLIDKSLVSIS--YSIINMHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDVMLEN 561
L+VL +SL+ IS I MH+LLE+LGR+IV S EP K L ++YDV+ +N
Sbjct: 466 LQVLTSRSLIHISRCNRTITMHNLLEQLGREIVCEQSIAEPGKRQFLMDASEIYDVLADN 525
Query: 562 M-EKHVEAIVLYYKEDEEADFEHLSKMSNLRLLFIANYISTM------LGFP---SCLSN 611
V I L + E + LLF+ Y S+ L P L
Sbjct: 526 TGTGAVLGISLDISKINELFLNERAFGGMHNLLFLRFYKSSSSKDQPELHLPRGLDYLPR 585
Query: 612 KLRFVHWFRYPSKYLPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRHSRNLEK 671
KLR +HW +P +P +F P LV + + ES +++LW+ + L +L+ +DL S NL++
Sbjct: 586 KLRLLHWDAFPMTSMPLSFCPQFLVVINIRESQLEKLWEGTQPLRSLKQMDLSKSENLKE 645
Query: 672 IIDFGEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQY 731
I D + N+E L L C +LV L SI L KLV L++K C L IP N+ L SL
Sbjct: 646 IPDLSKAVNIEELCLSYCGSLVMLPSSIKNLNKLVVLDMKYCSKLEIIPCNM-DLESLSI 704
Query: 732 LNMCGCSKVFNNPRRLMKSGISSEKKQQHDIRESASHHLPGLKWIILAHDSSHMLPSLHS 791
LN+ GCS++ + P K G S + E+A +P ++ S
Sbjct: 705 LNLDGCSRLESFPEISSKIGFLS-------LSETAIEEIP---------------TTVAS 742
Query: 792 LCCLRKVDISFC-YLSHVPDAIECLHWLERLNLAGNDFVTLPS-LRKLSKLVYLNLEHCK 849
CL +D+S C L P + + WL+ L+ + +P + KLSKL L + C
Sbjct: 743 WPCLAALDMSGCKNLKTFPCLPKTIEWLD---LSRTEIEEVPLWIDKLSKLNKLLMNSCM 799
Query: 850 LLESL 854
L S+
Sbjct: 800 KLRSI 804
Score = 33.1 bits (74), Expect = 0.88
Identities = 22/79 (27%), Positives = 34/79 (42%), Gaps = 4/79 (5%)
Query: 650 KNKKYLP----NLRTLDLRHSRNLEKIIDFGEFPNLERLDLEGCINLVELDPSIGLLRKL 705
KN K P + LDL + E + + L +L + C+ L + I L +
Sbjct: 755 KNLKTFPCLPKTIEWLDLSRTEIEEVPLWIDKLSKLNKLLMNSCMKLRSISSGISTLEHI 814
Query: 706 VYLNLKDCKSLVSIPNNIF 724
L+ CK++VS P IF
Sbjct: 815 KTLDFLGCKNIVSFPVEIF 833
Score = 32.3 bits (72), Expect = 1.5
Identities = 38/154 (24%), Positives = 65/154 (41%), Gaps = 24/154 (15%)
Query: 786 LPSLHSLCCLRKVDISFC-YLSHVPDAIECLHWLERLNLAG-NDFVTLPSLRKLSKLVYL 843
+P L + ++ +S+C L +P +I+ L+ L L++ + +P L L L
Sbjct: 646 IPDLSKAVNIEELCLSYCGSLVMLPSSIKNLNKLVVLDMKYCSKLEIIPCNMDLESLSIL 705
Query: 844 NLEHCKLLESLPQLP--------FPTNTGEVHREYDDYFCGAGLLIFNCPKLGEREHCRS 895
NL+ C LES P++ T E+ + C A L + C L + C
Sbjct: 706 NLDGCSRLESFPEISSKIGFLSLSETAIEEIPTTVASWPCLAALDMSGCKNL-KTFPCLP 764
Query: 896 MTLLWMKQFIKANPRSSSEIQIVNPGSEIPSWIN 929
T+ W+ S +EI+ E+P WI+
Sbjct: 765 KTIEWL-------DLSRTEIE------EVPLWID 785
Score = 30.8 bits (68), Expect = 4.4
Identities = 20/63 (31%), Positives = 36/63 (56%), Gaps = 2/63 (3%)
Query: 795 LRKVDISFCY-LSHVPDAIECLHWLERLNLAGNDFVTLPS-LRKLSKLVYLNLEHCKLLE 852
L+++D+S L +PD + ++ E V LPS ++ L+KLV L++++C LE
Sbjct: 632 LKQMDLSKSENLKEIPDLSKAVNIEELCLSYCGSLVMLPSSIKNLNKLVVLDMKYCSKLE 691
Query: 853 SLP 855
+P
Sbjct: 692 IIP 694
>At4g16890 disease resistance RPP5 like protein
Length = 1301
Score = 404 bits (1038), Expect = e-112
Identities = 291/852 (34%), Positives = 462/852 (54%), Gaps = 55/852 (6%)
Query: 39 LVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQKGECIGPELFRA 98
+ +SS YDVF +FRGED R++F L L K I D I ++ IGPEL A
Sbjct: 3 IASSSGSRRYDVFPSFRGEDVRDSFLSHLLKELRGKAITFIDDEI--ERSRSIGPELLSA 60
Query: 99 IEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPSEVRKQSGIYSE 158
I+ S++ + IFSKNYASSTWCL EL +I +C + V+P+F+ VD SEV+KQ+G + +
Sbjct: 61 IKESRIAIVIFSKNYASSTWCLNELVEIHKCYTNLNQMVIPIFFHVDASEVKKQTGEFGK 120
Query: 159 AFVKHEQRFQQDSMKVSRWREALEQVGSISGWDLRDEPL-AREIKEIVQKIINILECKYS 217
F + + +D + W++AL V ++G+DLR P A I+E+ + ++
Sbjct: 121 VFEETCKAKSEDEKQ--SWKQALAAVAVMAGYDLRKWPSEAAMIEELAEDVLRKTMTPSD 178
Query: 218 CVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLYGQISHQFSASC 277
DLVGI++ I+A+++ L L S + +GI G GIGK+T+ LY ++S QF
Sbjct: 179 DFG-DLVGIENHIEAIKSVLCLESKEARIMVGIWGQSGIGKSTIGRALYSKLSIQFHHRA 237
Query: 278 FID-DVTKIYGLHDDPLDVQKQILFQTLGIEHQQICNRYHATTLIQRKLCHERTLMILDN 336
FI T + L +K++L + LG + +I +++++L ++ L++LD+
Sbjct: 238 FITYKSTSGSDVSGMKLRWEKELLSEILGQKDIKI----EHFGVVEQRLKQQKVLILLDD 293
Query: 337 VDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVYKVSLLDWNEAHMLFCRKA 396
VD +E L+ + EW G GSRII+I++D +LKA+ +D++Y+V + A + CR A
Sbjct: 294 VDSLEFLKTLVGKAEWFGSGSRIIVITQDRQLLKAHEIDLIYEVEFPSEHLALTMLCRSA 353
Query: 397 F-KDEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFGRNVTEWKSALTRLRQSPVKDV 455
F KD +++ L ++ A LPL + VLGS L GR W + RLR D+
Sbjct: 354 FGKDSP--PDDFKELAFEVAKLAGNLPLGLSVLGSSLKGRTKEWWMEMMPRLRNGLNGDI 411
Query: 456 MDVLQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNILNCCGFHADIGLRVLIDKSLVSI 515
M L++S+D L++ ++D+FL+IAC FN VK++L ++G +L +KSL+ I
Sbjct: 412 MKTLRVSYDRLHQKDQDMFLYIACLFNGFEVSYVKDLLK-----DNVGFTMLTEKSLIRI 466
Query: 516 SYS-IINMHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDVMLENM-EKHVEAIVLYY 573
+ I MH+LLE+LGR+I + S P K L + E +++V+ E + + I L +
Sbjct: 467 TPDGYIEMHNLLEKLGREIDRAKSKGNPGKRRFLTNFEDIHEVVTEKTGTETLLGIRLPF 526
Query: 574 KEDEEA-----DFEHLSKMSNLRLLFIANY--ISTMLGFPSCLSNKLRFVHWFRYPSKYL 626
+E D E M NL+ L I Y + L + L KLR + W P K L
Sbjct: 527 EEYFSTRPLLIDKESFKGMRNLQYLEIGYYGDLPQSLVY---LPLKLRLLDWDDCPLKSL 583
Query: 627 PSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRHSRNLEKIIDFGEFPNLERLDL 686
PS F LV LI+ S +++LW+ L +L+ ++LR+S NL++I D NLE LDL
Sbjct: 584 PSTFKAEYLVNLIMKYSKLEKLWEGTLPLGSLKEMNLRYSNNLKEIPDLSLAINLEELDL 643
Query: 687 EGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCGCSKVFNNPR- 745
GC +LV L SI KL+YL++ DCK L S P ++ L SL+YLN+ GC + N P
Sbjct: 644 VGCKSLVTLPSSIQNATKLIYLDMSDCKKLESFPTDL-NLESLEYLNLTGCPNLRNFPAI 702
Query: 746 RLMKSGISSEKKQQHDIRESA--SHHLP-GLKWIILAHDSSHMLPSLHSLCCLRKVDISF 802
++ S + + + + E + +LP GL ++ + +P C R ++F
Sbjct: 703 KMGCSDVDFPEGRNEIVVEDCFWNKNLPAGLDYLDCL---TRCMP-----CEFRPEQLAF 754
Query: 803 CYL-----SHVPDAIECLHWLERLNLAGNDFVT-LPSLRKLSKLVYLNLEHCKLLESLPQ 856
+ + + I+ L LE ++L+ ++ +T +P L K +KL L L +CK L +L
Sbjct: 755 LNVRGYKHEKLWEGIQSLGSLEGMDLSESENLTEIPDLSKATKLESLILNNCKSLVTL-- 812
Query: 857 LPFPTNTGEVHR 868
P+ G +HR
Sbjct: 813 ---PSTIGNLHR 821
Score = 92.4 bits (228), Expect = 1e-18
Identities = 76/239 (31%), Positives = 118/239 (48%), Gaps = 14/239 (5%)
Query: 623 SKYLPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRHSRNLEKIIDFGEFPNLE 682
++ +P F P +L L + ++LW+ + L +L +DL S NL +I D + LE
Sbjct: 740 TRCMPCEFRPEQLAFLNVRGYKHEKLWEGIQSLGSLEGMDLSESENLTEIPDLSKATKLE 799
Query: 683 RLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCGCSKVFN 742
L L C +LV L +IG L +LV L +K+C L +P ++ LSSL+ L++ GCS + +
Sbjct: 800 SLILNNCKSLVTLPSTIGNLHRLVRLEMKECTGLEVLPTDV-NLSSLETLDLSGCSSLRS 858
Query: 743 NPRRLMKSGISSEKKQQHDIRESAS-----HHLPGLKWIILAHDSSHMLPSLHSLCCLRK 797
P L+ + I + I E S H L L+ + +LP+ +L L
Sbjct: 859 FP--LISTNIVWLYLENTAIEEIPSTIGNLHRLVRLE--MKKCTGLEVLPTDVNLSSLET 914
Query: 798 VDISFC-YLSHVPDAIECLHWLERLNLAGNDFVTLPSLRKLSKLVYLNLEHCKLLESLP 855
+D+S C L P E + WL N A + +P L K + L L L +CK L +LP
Sbjct: 915 LDLSGCSSLRSFPLISESIKWLYLENTAIEE---IPDLSKATNLKNLKLNNCKSLVTLP 970
Score = 54.3 bits (129), Expect = 4e-07
Identities = 50/188 (26%), Positives = 78/188 (40%), Gaps = 47/188 (25%)
Query: 669 LEKIIDFGEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSS 728
+E+I D + NL+ L L C +LV L +IG L+KLV +K+C L +P ++ LSS
Sbjct: 943 IEEIPDLSKATNLKNLKLNNCKSLVTLPTTIGNLQKLVSFEMKECTGLEVLPIDV-NLSS 1001
Query: 729 LQYLNMCGCSKVFNNPRRLMKSGISSEKKQQHDIRESASHHLPGLKWIILAHDSSHMLPS 788
L L++ GCS + P L+ + I W+ L + +
Sbjct: 1002 LMILDLSGCSSLRTFP--LISTNI---------------------VWLYLENTA------ 1032
Query: 789 LHSLCCLRKVDISFCYLSHVPDAIECLHWLERLNLAG-NDFVTLPSLRKLSKLVYLNLEH 847
+ +P I LH L +L + LP+ LS L+ L+L
Sbjct: 1033 ----------------IEEIPSTIGNLHRLVKLEMKECTGLEVLPTDVNLSSLMILDLSG 1076
Query: 848 CKLLESLP 855
C L + P
Sbjct: 1077 CSSLRTFP 1084
Score = 42.0 bits (97), Expect = 0.002
Identities = 46/181 (25%), Positives = 80/181 (43%), Gaps = 28/181 (15%)
Query: 587 MSNLRLLFIANYISTMLGFPSCLSNKLRFVHWFRYPSKYLPSNFHPNELVELILTESNIK 646
+S+L L ++ S++ FP +S +++++ + +P L L L +N K
Sbjct: 909 LSSLETLDLSG-CSSLRSFP-LISESIKWLYLENTAIEEIPDLSKATNLKNLKL--NNCK 964
Query: 647 QLWKNKKYLPNLRTL---DLRHSRNLEKIIDFGEFPNLERLDLEGCINL----------- 692
L + NL+ L +++ LE + +L LDL GC +L
Sbjct: 965 SLVTLPTTIGNLQKLVSFEMKECTGLEVLPIDVNLSSLMILDLSGCSSLRTFPLISTNIV 1024
Query: 693 ---------VELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCGCSKVFNN 743
E+ +IG L +LV L +K+C L +P ++ LSSL L++ GCS +
Sbjct: 1025 WLYLENTAIEEIPSTIGNLHRLVKLEMKECTGLEVLPTDV-NLSSLMILDLSGCSSLRTF 1083
Query: 744 P 744
P
Sbjct: 1084 P 1084
Score = 39.3 bits (90), Expect = 0.012
Identities = 31/85 (36%), Positives = 43/85 (50%), Gaps = 16/85 (18%)
Query: 805 LSHVPDAIECLHWLERLNLAG-NDFVTLPS-LRKLSKLVYLNLEHCKLLESLPQLPFPTN 862
L +PD ++ LE L+L G VTLPS ++ +KL+YL++ CK LES FPT+
Sbjct: 626 LKEIPDLSLAIN-LEELDLVGCKSLVTLPSSIQNATKLIYLDMSDCKKLES-----FPTD 679
Query: 863 TGEVHREYDDYFCGAGLLIFNCPKL 887
EY L + CP L
Sbjct: 680 LNLESLEY--------LNLTGCPNL 696
>At5g44510 disease resistance protein-like
Length = 1187
Score = 403 bits (1036), Expect = e-112
Identities = 289/857 (33%), Positives = 457/857 (52%), Gaps = 46/857 (5%)
Query: 31 SSSKSSSALVTSSKKNH-YDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQKGE 89
SSS SSS+ +S +N + VF++FRGED R + + GI F D +++G
Sbjct: 22 SSSLSSSSPPSSLSQNWLHPVFLSFRGEDVRKGLLSHIQKEFQRNGITPFID-NEMKRGG 80
Query: 90 CIGPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPSEV 149
IGPEL +AI S++ + + S+NY SS WCL EL +I +C + G+ V+ VFYDVDPS+V
Sbjct: 81 SIGPELLQAIRGSKIAIILLSRNYGSSKWCLDELVEIMKCREELGQTVMTVFYDVDPSDV 140
Query: 150 RKQSGIYSEAFVKHEQRFQQDSMKVSRWREALEQVGSISGWDLRD-EPLAREIKEIVQKI 208
RKQ G + + F K + V RW++AL +I G D R+ E A I +I + +
Sbjct: 141 RKQKGDFGKVFKK--TCVGRPEEMVQRWKQALTSAANILGEDSRNWENEADMIIKISKDV 198
Query: 209 INILECKYSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLYGQ 268
++L S + VGI++ + + L L+ ++ VR IGI G GIGKTT++ LY +
Sbjct: 199 SDVLSFTPSKDFDEFVGIEAHTTEITSLLQLD-LEEVRMIGIWGPAGIGKTTISRVLYNK 257
Query: 269 ISHQFSASCFIDDVTKIY--GLHDD---PLDVQKQILFQTLGIEHQQICNRYHATTLIQR 323
+ HQF ID++ Y HD+ L +QK++L Q + + + + + Q
Sbjct: 258 LFHQFQLGAIIDNIKVRYPRPCHDEYSAKLQLQKELLSQMINQKDMVVPH----LGVAQE 313
Query: 324 KLCHERTLMILDNVDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVYKVSLL 383
+L ++ L++LD+VD + QL+ +A +W G GSRII++++D +LKA+G+ +YKV
Sbjct: 314 RLKDKKVLLVLDDVDGLVQLDAMAKDVQWFGLGSRIIVVTQDLKLLKAHGIKYIYKVDFP 373
Query: 384 DWNEAHMLFCRKAFKDEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFGRNVTEWKSA 443
+EA +FC AF EK ++ + + A LPL ++V+GS+L + EW +
Sbjct: 374 TSDEALEIFCMYAF-GEKSPKVGFEQIARTVTTLAGKLPLGLRVMGSYLRRMSKQEWAKS 432
Query: 444 LTRLRQSPVKDVMDVLQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNILNCCGFHADIG 503
+ RLR S D+ VL+ S++ L E EKD+FLHI CFF + E ++ L G
Sbjct: 433 IPRLRTSLDDDIESVLKFSYNSLAEQEKDLFLHITCFFRRERIETLEVFLAKKSVDMRQG 492
Query: 504 LRVLIDKSLVSISYSIINMHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDVMLEN-- 561
L++L DKSL+S++ I MH+LL +LG IV+ S +P K L TE + +V+ ++
Sbjct: 493 LQILADKSLLSLNLGNIEMHNLLVQLGLDIVRKQSIHKPGKRQFLVDTEDICEVLTDDTG 552
Query: 562 ------MEKHVEAIVLYYKEDEEADFEHLSKMSNLRLLF-IANYISTMLGFP---SCLSN 611
++ + ++ E FE + + LR + +L P S +S
Sbjct: 553 TRTLIGIDLELSGVIEGVINISERAFERMCNLQFLRFHHPYGDRCHDILYLPQGLSHISR 612
Query: 612 KLRFVHWFRYPSKYLPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRHSRNLEK 671
KLR +HW RYP LP F+P LV++ + +S +++LW + + NL+ +DL NL++
Sbjct: 613 KLRLLHWERYPLTCLPPKFNPEFLVKINMRDSMLEKLWDGNEPIRNLKWMDLSFCVNLKE 672
Query: 672 IIDFGEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQY 731
+ DF NL+ L L C++LVEL SIG L+ L+L DC SLV +P++I L++L+
Sbjct: 673 LPDFSTATNLQELRLINCLSLVELPSSIGNATNLLELDLIDCSSLVKLPSSIGNLTNLKK 732
Query: 732 LNMCGCSKVFNNP---------RRLMKSGISSEKKQQHDIRESASHHLPGLKWIILAHDS 782
L + CS + P + L SG SS + S+ ++ LK + S
Sbjct: 733 LFLNRCSSLVKLPSSFGNVTSLKELNLSGCSSLLE-----IPSSIGNIVNLKKVYADGCS 787
Query: 783 S--HMLPSLHSLCCLRKVDISFC-YLSHVPDAIECLHWLERLNLAG-NDFVTLPSLRKLS 838
S + S+ + L+++ + C L P ++ L LE LNL+G V LPS+ +
Sbjct: 788 SLVQLPSSIGNNTNLKELHLLNCSSLMECPSSMLNLTRLEDLNLSGCLSLVKLPSIGNVI 847
Query: 839 KLVYLNLEHCKLLESLP 855
L L L C L LP
Sbjct: 848 NLQSLYLSDCSSLMELP 864
Score = 53.1 bits (126), Expect = 8e-07
Identities = 91/374 (24%), Positives = 150/374 (39%), Gaps = 72/374 (19%)
Query: 584 LSKMSNLRLLFIANYISTMLGFPSCLSN--KLRFVHWFRYPSKY-LPSNFHPNELVELIL 640
+ ++NL+ LF+ N S+++ PS N L+ ++ S +PS+ ++ +
Sbjct: 724 IGNLTNLKKLFL-NRCSSLVKLPSSFGNVTSLKELNLSGCSSLLEIPSSIGNIVNLKKVY 782
Query: 641 TE--SNIKQLWKNKKYLPNLRTLDLRHSRNL-EKIIDFGEFPNLERLDLEGCINLVELDP 697
+ S++ QL + NL+ L L + +L E LE L+L GC++LV+L P
Sbjct: 783 ADGCSSLVQLPSSIGNNTNLKELHLLNCSSLMECPSSMLNLTRLEDLNLSGCLSLVKL-P 841
Query: 698 SIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCGCSKVFNNPRRLMKSGISSEKK 757
SIG + L L L DC SL+ +P I ++L L + GCS +
Sbjct: 842 SIGNVINLQSLYLSDCSSLMELPFTIENATNLDTLYLDGCSNLL---------------- 885
Query: 758 QQHDIRESASHHLPGLKWIILAHDSSHMLPSLHSLCCLRKVDISFCYLSHVPDAIECLHW 817
LP W I L SL+ C L +P +E
Sbjct: 886 -----------ELPSSIWNIT------NLQSLYLNGC--------SSLKELPSLVENAIN 920
Query: 818 LERLNLAG-NDFVTLP-SLRKLSKLVYLNLEHCKLLESLPQLPFPTNTGEVHREYDDYFC 875
L+ L+L + V LP S+ ++S L YL++ +C L L + P D
Sbjct: 921 LQSLSLMKCSSLVELPSSIWRISNLSYLDVSNCSSLLELNLVSHPVVP-------DSLIL 973
Query: 876 GAGLLIFNCPKLGEREHC---RSMTLLWMKQFIKANPRSSSEI-------QIVNPGSEIP 925
AG +C L +R C +L K N + I + PG ++P
Sbjct: 974 DAG----DCESLVQRLDCFFQNPKIVLNFANCFKLNQEARDLIIQTSACRNAILPGEKVP 1029
Query: 926 SWINNQRMGYSIAI 939
++ + G S+ +
Sbjct: 1030 AYFTYRATGDSLTV 1043
Score = 29.6 bits (65), Expect = 9.7
Identities = 17/44 (38%), Positives = 27/44 (60%), Gaps = 2/44 (4%)
Query: 372 YGVDVVYKVSLLDWNEAHMLFCRKAFKDEKIIMSNYQNLVDQIL 415
+GV VV+ LD E+ C+ ++D I++SN QN++D IL
Sbjct: 1137 FGVHVVW--CHLDQYESKSPSCKPFWRDFPIVISNGQNILDAIL 1178
>At2g14080 putative disease resistance protein
Length = 1215
Score = 403 bits (1035), Expect = e-112
Identities = 281/866 (32%), Positives = 455/866 (52%), Gaps = 63/866 (7%)
Query: 31 SSSKSSSALVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQKGEC 90
SSS +S + S +DVF +F G D R +F + + KGI F D N+++ +
Sbjct: 39 SSSPPTSPQSSLSCNQKHDVFPSFHGADVRKSFLSHILKEFKRKGIDTFIDN-NIERSKS 97
Query: 91 IGPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPSEVR 150
IGPEL AI+ S++ V + SK+YASS+WCL EL +I +C K + V+ +FY+VDP++V+
Sbjct: 98 IGPELIEAIKGSKIAVVLLSKDYASSSWCLNELVEIMKCRKMLDQTVMTIFYEVDPTDVK 157
Query: 151 KQSGIYSEAFVKHEQRFQQDSMKVSR-WREALEQVGSISG-----WDLRDEPLAREIKEI 204
KQ+G + + F K + VSR W EAL +V +I+G WD + + +I
Sbjct: 158 KQTGDFGKVFKK---TCMGKTNAVSRKWIEALSEVATIAGEHSINWDTEAAMIEKISTDI 214
Query: 205 VQKIINILECKYSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATT 264
K+ N + LVG+ + ++ L+ L L+S + VR IGI G GIGKTT+
Sbjct: 215 SNKLNNSTPLRDF---DGLVGMGAHMEKLELLLCLDSCE-VRMIGIWGPPGIGKTTIVRF 270
Query: 265 LYGQISHQFSASCFIDDVTKIYGL------HDDPLDVQKQILFQTLGIEHQQICNRYHAT 318
LY Q+S F S F++++ ++ + + L +Q+Q L + L +H+ I
Sbjct: 271 LYNQLSSSFELSIFMENIKTMHTILASSDDYSAKLILQRQFLSKIL--DHKDI--EIPHL 326
Query: 319 TLIQRKLCHERTLMILDNVDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVY 378
++Q +L +++ L++LD+VDQ QL+ +A W GP SRI+I ++D +LKA+ ++ +Y
Sbjct: 327 RVLQERLYNKKVLVVLDDVDQSVQLDALAKETRWFGPRSRILITTQDRKLLKAHRINNIY 386
Query: 379 KVSLLDWNEAHMLFCRKAFKDEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFGRNVT 438
KV L + ++A +FC AF +K + L ++ PL ++V+GS+ +
Sbjct: 387 KVDLPNSDDALQIFCMYAF-GQKTPYDGFYKLARKVTWLVGNFPLGLRVVGSYFREMSKQ 445
Query: 439 EWKSALTRLRQSPVKDVMDVLQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNILNCCGF 498
EW+ + RLR + VL+ S+D L + +KD+FLHIACFFN++S E +++ L
Sbjct: 446 EWRKEIPRLRARLDGKIESVLKFSYDALCDEDKDLFLHIACFFNHESIEKLEDFLGKTFL 505
Query: 499 HADIGLRVLIDKSLVSISYSIINMHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDVM 558
VL +KSL+SI+ + + MH L +LG++IV+ S +EP + L + +V+
Sbjct: 506 DIAQRFHVLAEKSLISINSNFVEMHDSLAQLGKEIVRKQSVREPGQRQFLVDARDISEVL 565
Query: 559 LENMEKHVEAIVLY---YKEDE-----EADFEHLSKMSNLRLLFIANYISTMLGFPSCL- 609
++ I +Y ++ D+ E FE +S + LR+ N ++ P CL
Sbjct: 566 ADDTAGGRSVIGIYLDLHRNDDVFNISEKAFEGMSNLQFLRVKNFGNLFPAIVCLPHCLT 625
Query: 610 --SNKLRFVHWFRYPSKYLPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRHSR 667
S KLR + W +P PS F+P LVEL + S +++LW+ + L NL+ +DL S+
Sbjct: 626 YISRKLRLLDWMYFPMTCFPSKFNPEFLVELNMWGSKLEKLWEEIQPLRNLKRMDLFSSK 685
Query: 668 NLEKIIDFGEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLS 727
NL+++ D NLE L+L GC +LVEL SIG KL+ L L C SL+ +P++I
Sbjct: 686 NLKELPDLSSATNLEVLNLNGCSSLVELPFSIGNATKLLKLELSGCSSLLELPSSIGNAI 745
Query: 728 SLQYLNMCGCSKVFNNPRRLMKSGISSEKKQQHDIRESASHHLPGLKWIILAHDSSHMLP 787
+LQ ++ C + P S +++E LK + + + L
Sbjct: 746 NLQTIDFSHCENLVELP---------SSIGNATNLKELDLSCCSSLKELPSSIGNCTNLK 796
Query: 788 SLHSLCC---------------LRKVDISFC-YLSHVPDAIECLHWLERLNLAG-NDFVT 830
LH +CC L+++ ++ C L +P +I LE+L LAG V
Sbjct: 797 KLHLICCSSLKELPSSIGNCTNLKELHLTCCSSLIKLPSSIGNAINLEKLILAGCESLVE 856
Query: 831 LPS-LRKLSKLVYLNLEHCKLLESLP 855
LPS + K + L LNL + L LP
Sbjct: 857 LPSFIGKATNLKILNLGYLSCLVELP 882
Score = 60.1 bits (144), Expect = 7e-09
Identities = 88/319 (27%), Positives = 131/319 (40%), Gaps = 48/319 (15%)
Query: 643 SNIKQLWKNKKYLPNLRTLDLRHSRNLEKI-IDFGEFPNLERLDLEGCINLVELDPSIGL 701
S++K+L + NL+ L L +L K+ G NLE+L L GC +LVEL IG
Sbjct: 804 SSLKELPSSIGNCTNLKELHLTCCSSLIKLPSSIGNAINLEKLILAGCESLVELPSFIGK 863
Query: 702 LRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCGCSKVFNNPRRLMKSGISSEKKQQHD 761
L LNL LV +P+ I L L L + GC K+ +++ + I+ E + D
Sbjct: 864 ATNLKILNLGYLSCLVELPSFIGNLHKLSELRLRGCKKL-----QVLPTNINLEFLNELD 918
Query: 762 IRES-------------ASHHLPGLKWIILAHDSSHMLPSLHSLCCLRKVDISFCYLSHV 808
+ + HL G + I S P L L L ++S SHV
Sbjct: 919 LTDCILLKTFPVISTNIKRLHLRGTQ-IEEVPSSLRSWPRLEDLQMLYSENLS--EFSHV 975
Query: 809 PDAIECLHWLERLNLAGNDFVTLPSLRKLSKLVYLNLEHCKLLESLPQLP------FPTN 862
+ I L L +N+ P L ++++L L L C L SLPQL N
Sbjct: 976 LERITVLE-LSDINIR----EMTPWLNRITRLRRLKLSGCGKLVSLPQLSDSLIILDAEN 1030
Query: 863 TGEVHREYDDYFCGAGLLIFNCP--KLGEREHCRSMTLLWMKQFIKANPRSSSEIQIVNP 920
G + R C FN P K + +C + I+A R S + P
Sbjct: 1031 CGSLER----LGCS-----FNNPNIKCLDFTNCLKLDKEARDLIIQATARHYS----ILP 1077
Query: 921 GSEIPSWINNQRMGYSIAI 939
E+ +I N+ +G S+ +
Sbjct: 1078 SREVHEYITNRAIGSSLTV 1096
>At3g04220 putative disease resistance protein
Length = 896
Score = 402 bits (1032), Expect = e-112
Identities = 256/746 (34%), Positives = 416/746 (55%), Gaps = 48/746 (6%)
Query: 36 SSALVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQKGECIGPEL 95
S L + S YDVF +FRGED R +F + + +GI F D N+++GE IGPEL
Sbjct: 51 SPPLSSLSLTRKYDVFPSFRGEDVRKDFLSHIQKEFQRQGITPFVDN-NIKRGESIGPEL 109
Query: 96 FRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPSEVRKQSGI 155
RAI S++ + + SKNYASS+WCL EL +I +C + G+ V+ +FY VDPS V+K +G
Sbjct: 110 IRAIRGSKIAIILLSKNYASSSWCLDELVEIIKCKEEMGQTVIVIFYKVDPSLVKKLTGD 169
Query: 156 YSEAFVKHEQRFQQDSMKVSRWREALEQVGSISGWDLRD-EPLAREIKEIVQKIINILEC 214
+ + F + ++++ + RWREA ++V +I+G+D R + + I++IV I +L
Sbjct: 170 FGKVFRNTCKGKEREN--IERWREAFKKVATIAGYDSRKWDNESGMIEKIVSDISEMLN- 226
Query: 215 KYSCVSKD---LVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLYGQISH 271
+S S+D L+G+ ++ ++ L ++S D ++ IGI G G+GKTT+A +LY Q S
Sbjct: 227 -HSTPSRDFDDLIGMGDHMEKMKPLLDIDS-DEMKTIGIWGPPGVGKTTIARSLYNQHSD 284
Query: 272 QFSASCFIDDVTKIYGL------HDDPLDVQKQILFQTLGIEHQQICNRYHATTLIQRKL 325
+F S F++ + Y + + + L +Q++ L Q E+ QI + + Q +L
Sbjct: 285 KFQLSVFMESIKTAYTIPACSDDYYEKLQLQQRFLSQITNQENVQIPH----LGVAQERL 340
Query: 326 CHERTLMILDNVDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVYKVSLLDW 385
++ L+++D+V+Q Q++ +A +WLGPGSRIII ++D +L+A+G++ +Y+V ++
Sbjct: 341 NDKKVLVVIDDVNQSVQVDALAKENDWLGPGSRIIITTQDRGILRAHGIEHIYEVDYPNY 400
Query: 386 NEAHMLFCRKAFKDEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFGRNVTEWKSALT 445
EA +FC AF +K ++ L Q+ + LPL +KV+GS+ G EW AL
Sbjct: 401 EEALQIFCMHAF-GQKSPYDGFEELAQQVTTLSGRLPLGLKVMGSYFRGMTKQEWTMALP 459
Query: 446 RLRQSPVKDVMDVLQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNILNCCGFHADIGLR 505
R+R + +L+LS+D L + +K +FLH+AC F+ND E V+ L GL
Sbjct: 460 RVRTHLDGKIESILKLSYDALCDVDKSLFLHLACSFHNDDTELVEQQLGKKFSDLRQGLH 519
Query: 506 VLIDKSLVSISYSIINMHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDVMLENMEKH 565
VL +KSL+ + +I MH LL +LGR+IV+ S EP + L + +V+ ++
Sbjct: 520 VLAEKSLIHMDLRLIRMHVLLAQLGREIVRKQSIHEPGQRQFLVDATDIREVLTDDTGSR 579
Query: 566 VEAIVLYYKEDEEADFEHLSK----MSNLRLLFIANYI--------------------ST 601
+ + E + + K MSNL+ + I + +
Sbjct: 580 SVIGIDFDFNTMEKELDISEKAFRGMSNLQFIRIYGDLFSRHGVYYFGGRGHRVSLDYDS 639
Query: 602 MLGFP---SCLSNKLRFVHWFRYPSKYLPSNFHPNELVELILTESNIKQLWKNKKYLPNL 658
L FP L KLR +HW ++P LPS FH LV+L + S +++LW+ + L NL
Sbjct: 640 KLHFPRGLDYLPGKLRLLHWQQFPMTSLPSEFHAEFLVKLCMPYSKLEKLWEGIQPLRNL 699
Query: 659 RTLDLRHSRNLEKIIDFGEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVS 718
LDL SRNL+++ D NL+RL +E C +LV+L SIG L +NL++C SLV
Sbjct: 700 EWLDLTCSRNLKELPDLSTATNLQRLSIERCSSLVKLPSSIGEATNLKKINLRECLSLVE 759
Query: 719 IPNNIFGLSSLQYLNMCGCSKVFNNP 744
+P++ L++LQ L++ CS + P
Sbjct: 760 LPSSFGNLTNLQELDLRECSSLVELP 785
Score = 63.2 bits (152), Expect = 8e-10
Identities = 38/114 (33%), Positives = 63/114 (54%), Gaps = 5/114 (4%)
Query: 655 LPNLRTLDLRHSRNLEKI-IDFGEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDC 713
L NL+ LDLR +L ++ FG N+E L+ C +LV+L + G L L L L++C
Sbjct: 767 LTNLQELDLRECSSLVELPTSFGNLANVESLEFYECSSLVKLPSTFGNLTNLRVLGLREC 826
Query: 714 KSLVSIPNNIFGLSSLQYLNMCGCSKVFNNPRRLMKSGISSEKKQQHDIRESAS 767
S+V +P++ L++LQ LN+ CS + P S ++ + D+R+ +S
Sbjct: 827 SSMVELPSSFGNLTNLQVLNLRKCSTLVELP----SSFVNLTNLENLDLRDCSS 876
Score = 60.5 bits (145), Expect = 5e-09
Identities = 42/118 (35%), Positives = 66/118 (55%), Gaps = 3/118 (2%)
Query: 626 LPSNF-HPNELVELILTE-SNIKQLWKNKKYLPNLRTLDLRHSRNLEKIID-FGEFPNLE 682
LPS+F + L EL L E S++ +L + L N+ +L+ +L K+ FG NL
Sbjct: 760 LPSSFGNLTNLQELDLRECSSLVELPTSFGNLANVESLEFYECSSLVKLPSTFGNLTNLR 819
Query: 683 RLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCGCSKV 740
L L C ++VEL S G L L LNL+ C +LV +P++ L++L+ L++ CS +
Sbjct: 820 VLGLRECSSMVELPSSFGNLTNLQVLNLRKCSTLVELPSSFVNLTNLENLDLRDCSSL 877
Score = 46.6 bits (109), Expect = 8e-05
Identities = 31/84 (36%), Positives = 45/84 (52%), Gaps = 3/84 (3%)
Query: 655 LPNLRTLDLRHSRNLEKI-IDFGEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDC 713
L NLR L LR ++ ++ FG NL+ L+L C LVEL S L L L+L+DC
Sbjct: 815 LTNLRVLGLRECSSMVELPSSFGNLTNLQVLNLRKCSTLVELPSSFVNLTNLENLDLRDC 874
Query: 714 KSLVSIPNNIFGLSSLQYLNMCGC 737
SL +P++ ++ L+ L C
Sbjct: 875 SSL--LPSSFGNVTYLKRLKFYKC 896
>At5g49140 disease resistance protein-like
Length = 980
Score = 399 bits (1026), Expect = e-111
Identities = 302/949 (31%), Positives = 494/949 (51%), Gaps = 89/949 (9%)
Query: 31 SSSKSSSALVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQKGEC 90
SSS SSS YDVF +FRGED R NF L E+KGI+ F+D + +++ +
Sbjct: 5 SSSGSSSCW-------KYDVFPSFRGEDVRGNFLSHLMKEFESKGIVTFKDDL-IERSQT 56
Query: 91 IGPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPSEVR 150
IG EL A+ S+++V IFSKNYASS+WCL EL +I +C + + ++P+FY V+PS+VR
Sbjct: 57 IGLELKEAVRQSKIFVVIFSKNYASSSWCLDELVEILKCKEE--RRLIPIFYKVNPSDVR 114
Query: 151 KQSGIYSEAFVKHEQRFQQDSMKVSRWREALEQVGSISGWDLRD-EPLAREIKEIVQKII 209
Q+G + F E ++ ++W+ AL + +I+G D + + A + +I + I+
Sbjct: 115 NQTGKFGRGF--RETCEGKNDETQNKWKAALTEAANIAGEDSQSWKNEADFLTKIAKDIL 172
Query: 210 NILECKYSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLYGQI 269
L S ++++GI+S ++ + L LN D VR +GI G GIGKTT+A L+ +
Sbjct: 173 AKLNGTPSNDFENIIGIESHMEKMVQLLCLND-DDVRMVGIWGPAGIGKTTIARVLHSRF 231
Query: 270 SHQFSASCFIDDVTKIYGLHDDP---LDVQKQILFQTLGIEHQQICNRYHATTLIQRKLC 326
S F + F+++V Y D ++Q ++ + L I Q + + I+ +L
Sbjct: 232 SGDFRFTVFMENVRGNYQRIVDSGGEYNLQARLQKEFLPIIFNQKDRKINHLWKIEERLK 291
Query: 327 HERTLMILDNVDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVYKVSLLDWN 386
++ L++L +VD+VEQLE +A W GPGSRII+ ++D+ +L + ++ +Y+V L
Sbjct: 292 KQKVLIVLGDVDKVEQLEALANETRWFGPGSRIIVTTKDKQILVGHEINHIYEVKLPCRK 351
Query: 387 EAHMLFCRKAFKDEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFGRNVTEWKSALTR 446
A + C AFK + + ++ ++V ++ + LPL ++VLGS + G++ WK L R
Sbjct: 352 TALEILCLYAFK-QNVAPDDFMDVVVEVAELSGHLPLGLRVLGSHMRGKSKDRWKLELGR 410
Query: 447 LRQSPVKDVMDVLQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNILNCCGFHADIGLRV 506
L S + V +L++S+D L+ +K +FLHIAC FN ++ + VK +L +GL++
Sbjct: 411 LTTSLDEKVEKILKISYDDLHIRDKALFLHIACMFNGENIDLVKQMLVNSDLDVSLGLQL 470
Query: 507 LIDKSLVSISYSI-INMHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDVMLENM-EK 564
L+DKSL+ I+ I MHSLL ++G+++V SS EP K L++T++ +++ N +
Sbjct: 471 LLDKSLIQINDDREIVMHSLLLKMGKEVVCQHSS-EPGKRQFLFNTKETCNILSNNTGSE 529
Query: 565 HVEAIVLYYKEDEEADFEH---LSKMSNLRLLFIANYI-----STMLGFPSCLSN--KLR 614
V I L E + F M NL+ L N S L P L+ +R
Sbjct: 530 AVLGISLDTSEIQNDVFMSERVFEDMRNLKFLRFYNKKIDENPSLKLHLPRGLNYLPAVR 589
Query: 615 FVHWFRYPSKYLPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRHSRNLEKIID 674
+HW YP KY+PS F P LVEL + S + +LW+ + L L+T+DL S NL ++ D
Sbjct: 590 LLHWDSYPMKYIPSQFRPECLVELRMMHSKVVKLWEGTQTLAYLKTIDLSFSNNLVEVPD 649
Query: 675 FGEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNM 734
+ +LE L LEGC +L EL S+ L +L +L L C+ L IP +I L+SL+ L+M
Sbjct: 650 LSKAISLETLCLEGCQSLAELPSSVLNLHRLKWLRLTMCEKLEVIPLHI-NLASLEVLDM 708
Query: 735 CGCSKVFNNPRRLMKSGISSEKKQQHDIRESASHHLPGLKWIILAHDSSHMLPSLHSLCC 794
GC K+ + P S+ ++ ++ + +P PS+
Sbjct: 709 EGCLKLKSFPD-------ISKNIERIFMKNTGIEEIP---------------PSISQWSR 746
Query: 795 LRKVDISFC----YLSHVPDAIECLHWLERLNLAGNDFVTLPS-LRKLSKLVYLNLEHCK 849
L +DIS C SHVP ++ ++ L + LP ++ L+ L YL +++C+
Sbjct: 747 LESLDISGCLNLKIFSHVPKSVVYIY------LTDSGIERLPDCIKDLTWLHYLYVDNCR 800
Query: 850 LLESLPQLP------FPTNTGEVHREYDDYFCGAGLLIFNCPKLGEREHCRSMTLLWMKQ 903
L SLP+LP N + R + C + F+ + E R +T W+ +
Sbjct: 801 KLVSLPELPSSIKILSAINCESLERISSSFDCPNAKVEFSKSMNFDGEARRVITQQWVYK 860
Query: 904 FIKANPRSSSEIQIVNPGSEIPSWINNQRMGYSIAIDRSPIRHDNDNNI 952
+ PG E+P +++ G S+ I H D N+
Sbjct: 861 ------------RACLPGKEVPLEFSHRARGGSLTI------HLEDENV 891
>At5g41550 disease resistance protein-like
Length = 1085
Score = 399 bits (1024), Expect = e-111
Identities = 310/932 (33%), Positives = 457/932 (48%), Gaps = 81/932 (8%)
Query: 41 TSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQKGECIGPELFRAIE 100
+SS Y VF +F G D R F L +KGI F+D ++KG IGPEL AI
Sbjct: 6 SSSNIRRYHVFPSFHGPDVRKGFLSHLHYHFASKGITTFKDQ-EIEKGNTIGPELVNAIR 64
Query: 101 ISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPSEVRKQSGIYSEAF 160
S+V + + SK YASS+WCL EL +I +C + G+ V+ +FYDVDPS VRKQ G + F
Sbjct: 65 ESRVSIVLLSKKYASSSWCLDELVEILKCKEDQGQIVMTIFYDVDPSSVRKQKGDFGSTF 124
Query: 161 VKHEQRFQQDSMKVSRWREALEQVGSISGWDLRDEPLAREIKEIVQKIINILECKYSCV- 219
+K + ++ + RW +AL V +I G + A E +++QKI + K S
Sbjct: 125 MKTCEGKSEEVKQ--RWTKALTHVANIKGEHSLN--WANEA-DMIQKIATDVSTKLSVTP 179
Query: 220 SKD---LVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLYGQISHQFSAS 276
S+D +VG+++ + L N LL D V+ IGI G GIGK+T+A LY Q+S F
Sbjct: 180 SRDFEGMVGLEAHLTKL-NSLLCFEGDDVKMIGIWGPAGIGKSTIARALYNQLSSSFQLK 238
Query: 277 CFIDDVT----KIYGLHDDPLDVQKQILFQTLGIEHQQICNRYHATTLIQRKLCHERTLM 332
CF+ ++ I G+ D + QK + L Q R H I+ L +R L+
Sbjct: 239 CFMGNLKGSLKSIVGV--DHYEFQKSLQKLLLAKILNQGDMRVHNLAAIKEWLQDQRVLI 296
Query: 333 ILDNVDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVYKVSLLDWNEAHMLF 392
ILD+VD +EQLE +A W G GSRII+ + D+ +LK +G++ +Y V EA +
Sbjct: 297 ILDDVDDLEQLEVLAKELSWFGSGSRIIVATEDKKILKEHGINDIYHVDFPSMEEALEIL 356
Query: 393 CRKAFKDEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFGRNVTEWKSALTRLRQSPV 452
C AFK + ++ L +++H LPL + ++GS L G + EW+ L R+ S
Sbjct: 357 CLSAFKQSSV-PDGFEELAKKVVHLCGNLPLGLSIVGSSLRGESKHEWELQLPRIEASLD 415
Query: 453 KDVMDVLQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNILNCCGFHADIGLRVLIDKSL 512
+ +L++ ++ L++ + +FLHIACFFN S + V +L GL+ L DK
Sbjct: 416 GKIESILKVGYERLSKKNQSLFLHIACFFNYRSVDYVTVMLADSNLDVRNGLKTLADKCF 475
Query: 513 VSISYS--IINMHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDVMLENMEKHVEAIV 570
V IS + I+ H LL++LGR+IV S EP K L E++ V+ + E +++
Sbjct: 476 VHISINGWIVMHHHLLQQLGRQIVLEQSD-EPGKRQFLIEAEEIRAVLTD--ETGTGSVI 532
Query: 571 LYYKEDEEADFEHLSK-----MSNLRLLFIANYIST---MLGFPSCLSN--KLRFVHWFR 620
+SK M NLR L I NY+ + L P + LR +HW R
Sbjct: 533 GISYNTSNIGEVSVSKGAFEGMRNLRFLRIFNYLFSGKCTLQIPEDMEYLPPLRLLHWDR 592
Query: 621 YPSKYLPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRHSRNLEKIIDFGEFPN 680
YP K LP+ F P L+EL + SN+++LW + LPN++++DL S L++I + N
Sbjct: 593 YPRKSLPTKFQPERLLELHMPHSNLEKLWGGIQPLPNIKSIDLSFSIRLKEIPNLSNATN 652
Query: 681 LERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCGCSKV 740
LE L+L C LVEL SI L KL L + C+ L IP NI L+SL+ + M CS++
Sbjct: 653 LETLNLTHCKTLVELPSSISNLHKLKKLKMSGCEKLRVIPTNI-NLASLEVVRMNYCSRL 711
Query: 741 FNNPRRLMKSGISSEKKQQHDIRESASHHLPGL--KWIILAHDSSHMLPSLHSLCCLRKV 798
P ISS K + P + W LA L
Sbjct: 712 RRFP------DISSNIKTLSVGNTKIENFPPSVAGSWSRLAR--------------LEIG 751
Query: 799 DISFCYLSHVPDAIECLHWLERLNLAGNDFVTLPS-LRKLSKLVYLNLEHCKLLESLPQL 857
S L+H P +I LNL+ +D +P + L LV L +E+C+ L ++P L
Sbjct: 752 SRSLKILTHAPQSI------ISLNLSNSDIRRIPDCVISLPYLVELIVENCRKLVTIPAL 805
Query: 858 P--FPTNTGEVHREYDDYFCGAG----LLIFNCPKLGEREHCRSMTLLWMKQFIKANPRS 911
P + C G L +NC KL E + I P
Sbjct: 806 PPWLESLNANKCASLKRVCCSFGNPTILTFYNCLKLDEEAR---------RGIIMQQP-- 854
Query: 912 SSEIQIVNPGSEIPSWINNQRMGYSIAIDRSP 943
+ I PG EIP+ +++ +G SI I +P
Sbjct: 855 -VDEYICLPGKEIPAEFSHKAVGNSITIPLAP 885
>At4g19510 TMV resistance protein N-like
Length = 1164
Score = 394 bits (1013), Expect = e-109
Identities = 307/950 (32%), Positives = 483/950 (50%), Gaps = 79/950 (8%)
Query: 45 KNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQKGECIGPELFRAIEISQV 104
K +DVFV+FRG DTR++FT L L KGI VF D L+ GE I LF IE S++
Sbjct: 21 KCEFDVFVSFRGADTRHDFTSHLVKYLRGKGIDVFSDA-KLRGGEYISL-LFDRIEQSKM 78
Query: 105 YVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPSEVRKQSGIYSEAFVKHE 164
+ +FS++YA+S WCL+E+ KI + K VLP+FY V S+V Q+G + F
Sbjct: 79 SIVVFSEDYANSWWCLEEVGKIMQRRKEFNHGVLPIFYKVSKSDVSNQTGSFEAVFQSPT 138
Query: 165 QRFQQDSMKVSRWREALEQVGSISGWDLRDEPLARE-IKEIVQKIINILECKYSCV-SKD 222
+ F D K+ + AL+ +I G+ + + + EIV+ +L CV D
Sbjct: 139 KIFNGDEQKIEELKVALKTASNIRGFVYPENSSEPDFLDEIVKNTFRMLNELSPCVIPDD 198
Query: 223 LVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLYGQISHQFSASCFIDDV 282
L GI+S + L+ L+ ++ + VR +G+ GM GIGKTT+A +Y Q +F F++D+
Sbjct: 199 LPGIESRSKELEKLLMFDNDECVRVVGVLGMTGIGKTTVADIVYKQNFQRFDGYEFLEDI 258
Query: 283 ---TKIYGLHDDPLDVQKQILFQTLGIEHQQICNRYHATTLIQRKLCHERTLMILDNVDQ 339
+K YGL P QK +L + L E+ + + ++ K + ++LDNV +
Sbjct: 259 EDNSKRYGL---PYLYQK-LLHKLLDGENVDVRAQGRPENFLRNK----KLFIVLDNVTE 310
Query: 340 VEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVVYKVSLLDWNEAHMLFCRKAFKD 399
+Q+E + + GSRI+II+RD+ +L+ D Y V L+ EA LFC + F +
Sbjct: 311 EKQIEYLIGKKNVYRQGSRIVIITRDKKLLQK-NADATYVVPRLNDREAMELFCLQVFGN 369
Query: 400 EKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFGRNVTEWKSALTRLRQSPVKDVMDVL 459
+ +L + + YAKGLPLA+K+LG L ++ WK L L+ +P K++ L
Sbjct: 370 H-YPTEEFVDLSNDFVCYAKGLPLALKLLGKGLLTHDINYWKKKLEFLQVNPDKELQKEL 428
Query: 460 QLSFDGLNETEKDIFLHIACFFNNDSEEDVKNILNCCGFHADIGLRVLIDKSLVSISYSI 519
+ S+ L++ +K +FL IACFF ++ + V +IL A +R L +K LV+ISY
Sbjct: 429 KSSYKALDDDQKSVFLDIACFFRSEKADFVSSILKSDDIDAKDVMRELEEKCLVTISYDR 488
Query: 520 INMHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDVMLENM-EKHVEAIVLYYKEDEE 578
I MH LL +G++I + S ++ + RLW+ + + D++ N + V I L E
Sbjct: 489 IEMHDLLHAMGKEIGKEKSIRKAGERRRLWNHKDIRDILEHNTGTECVRGIFLNMSEVRR 548
Query: 579 -----ADFEHLSKMSNLRLLFIANYISTMLG---------FPSCLSNKLRFVHWFRYPSK 624
A F LSK+ L+ F +++ S P ++L ++HW YP
Sbjct: 549 IKLFPAAFTMLSKLKFLK--FHSSHCSQWCDNDHIFQCSKVPDHFPDELVYLHWQGYPYD 606
Query: 625 YLPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRHSRNLEKIIDFGEFPNLERL 684
LPS+F P ELV+L L S+IKQLW+++K +LR +DL S++L + NLERL
Sbjct: 607 CLPSDFDPKELVDLSLRYSHIKQLWEDEKNTESLRWVDLGQSKDLLNLSGLSRAKNLERL 666
Query: 685 DLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCGCSKVFNNP 744
DLEGC +L +L S+ + +L+YLNL+DC SL S+P F + SL+ L + GC K+
Sbjct: 667 DLEGCTSL-DLLGSVKQMNELIYLNLRDCTSLESLPKG-FKIKSLKTLILSGCLKL--KD 722
Query: 745 RRLMKSGISSEKKQQHDIRESASHHLPGLKWIILAH----DSSHMLPS-LHSLCCLRKVD 799
++ I S + I E H+ L +IL + + LP+ L+ L L+++
Sbjct: 723 FHIISESIESLHLEGTAI-ERVVEHIESLHSLILLNLKNCEKLKYLPNDLYKLKSLQELV 781
Query: 800 ISFC-YLSHVPDAIECLHWLERLNLAGNDFVTLPSLRKLSKL---------------VYL 843
+S C L +P E + LE L + G P + LS L +YL
Sbjct: 782 LSGCSALESLPPIKEKMECLEILLMDGTSIKQTPEMSCLSNLKICSFCRPVIDDSTGLYL 841
Query: 844 NLEHCKLLESLPQ-LPFPTNTGEVHREYDDYFCGAGLLIFNCPKLGEREHCRSMTLLWMK 902
+ C LE++ + L P T +H + + +C KL + E + +K
Sbjct: 842 DAHGCGSLENVSKPLTIPLVTERMHTTF---------IFTDCFKLNQAEKEDIVAQAQLK 892
Query: 903 -QFIKANPRSSSE--------IQIVNPGSEIPSWINNQRMGYSIAIDRSP 943
Q + R + + + PG +IPSW ++Q+MG I D P
Sbjct: 893 SQLLARTSRHHNHKGLLLDPLVAVCFPGHDIPSWFSHQKMGSLIETDLLP 942
>At5g46260 disease resistance protein-like
Length = 1205
Score = 394 bits (1011), Expect = e-109
Identities = 303/935 (32%), Positives = 479/935 (50%), Gaps = 73/935 (7%)
Query: 29 MASSSKSSSALVTSSKKNHYDVFVTFRGEDTRNNFTDFLFDALETKGIMVFRDVINLQKG 88
MASSS S + L YDVF++FRG D R F + K I FRD +++
Sbjct: 1 MASSSSSRNWL--------YDVFLSFRGGDVRVTFRSHFLKEFDRKLITAFRDN-EIERS 51
Query: 89 ECIGPELFRAIEISQVYVAIFSKNYASSTWCLQELEKICECIKGSGKHVLPVFYDVDPSE 148
+ P+L +AI+ S++ V +FSKNYASS+WCL EL +I C + K ++PVFY VDPS+
Sbjct: 52 HSLWPDLEQAIKESRIAVVVFSKNYASSSWCLNELLEIVNC---NDKIIIPVFYGVDPSQ 108
Query: 149 VRKQSGIYSEAFVKHEQRFQQDSMKVSRWREALEQVGSISGWDLRD-EPLAREIKEIVQK 207
VR Q G + + F K +R Q + +K ++W++AL V ++ G+D + A+ I+EI
Sbjct: 109 VRYQIGEFGKIFEKTCKR-QTEEVK-NQWKKALTHVANMLGFDSSKWDDEAKMIEEIAND 166
Query: 208 IINILECKYSCVSKDLVGIDSPIQALQNHLLLNSVDGVRAIGICGMGGIGKTTLATTLYG 267
++ L S +D VG++ I + L L S + V+ +GI G GIGKTT+A L+
Sbjct: 167 VLRKLLLTTSKDFEDFVGLEDHIANMSALLDLESKE-VKMVGIWGSSGIGKTTIARALFN 225
Query: 268 QISHQFSASCFID-----DVTKIYGL-----HDDPLDVQKQILFQTLGIEHQQICNRYHA 317
+ F FID +I+ H+ L +Q+ L + L + + +I +
Sbjct: 226 NLFRHFQVRKFIDRSFAYKSREIHSSANPDDHNMKLHLQESFLSEILRMPNIKIDH---- 281
Query: 318 TTLIQRKLCHERTLMILDNVDQVEQLEKIAVHREWLGPGSRIIIISRDEHVLKAYGVDVV 377
++ +L H++ L+I+D+VD L+ + +W G GSRII+++ ++H L A+G+D +
Sbjct: 282 LGVLGERLQHQKVLIIIDDVDDQVILDSLVGKTQWFGNGSRIIVVTNNKHFLTAHGIDRM 341
Query: 378 YKVSLLDWNEAHMLFCRKAFKDEKIIMSNYQNLVDQILHYAKGLPLAIKVLGSFLFGRNV 437
Y+VSL A + C+ AFK +K ++ LV Q+ YA LPL +KVLGS+L G++
Sbjct: 342 YEVSLPTEEHALAMLCQSAFK-KKSPPEGFEMLVVQVARYAGSLPLVLKVLGSYLSGKDK 400
Query: 438 TEWKSALTRLRQSPVKDVMDVLQLSFDGLNETEKDIFLHIACFFNNDSEEDVKNILNCCG 497
W L RL+ + +L++S+DGL ++ IF HIAC FN+ +K++L
Sbjct: 401 EYWIDMLPRLQNGLNDKIERILRISYDGLESEDQAIFRHIACIFNHMEVTTIKSLLANSI 460
Query: 498 FHADIGLRVLIDKSLVSISYSIINMHSLLEELGRKIVQNSSSKEPRKWSRLWSTEQLYDV 557
+ A++GL+ L+DKS++ + + + MH LL+E+GRKIV+ S +PRK L + DV
Sbjct: 461 YGANVGLQNLVDKSIIHVRWGHVEMHPLLQEMGRKIVRTQSIGKPRKREFLVDPNDICDV 520
Query: 558 MLENME-KHVEAIVLYYKEDEEADFEH--LSKMSNLRLLFIANYI---STMLGFPSC--- 608
+ E ++ + V I L + +E +M NLR L I I L P
Sbjct: 521 LSEGIDTQKVLGISLETSKIDELCVHESAFKRMRNLRFLKIGTDIFGEENRLHLPESFDY 580
Query: 609 LSNKLRFVHWFRYPSKYLPSNFHPNELVELILTESNIKQLWKNKKYLPNLRTLDLRHSRN 668
L L+ + W +P + +PSNF P LV L +T S + +LW+ L L+ +DL S N
Sbjct: 581 LPPTLKLLCWSEFPMRCMPSNFCPKNLVTLKMTNSKLHKLWEGAVPLTCLKEMDLDGSVN 640
Query: 669 LEKIIDFGEFPNLERLDLEGCINLVELDPSIGLLRKLVYLNLKDCKSLVSIPNNIFGLSS 728
L++I D NLE L+ E C +LVEL I L KL+ LN+ C SL ++P F L S
Sbjct: 641 LKEIPDLSMATNLETLNFENCKSLVELPSFIQNLNKLLKLNMAFCNSLETLPTG-FNLKS 699
Query: 729 LQYLNMCGCSKVFNNPRRLMKSGISSEKKQQHDIRESASH-HLPGL------KWIILAHD 781
L ++ CSK+ P + IS +I E S+ HL L K I
Sbjct: 700 LNRIDFTKCSKLRTFPD--FSTNISDLYLTGTNIEELPSNLHLENLIDLRISKKEIDGKQ 757
Query: 782 SSHMLPSLHSLCCLRKVDISFCYLSHVPDAIEC------LHWLERLNLAG-NDFVTLPSL 834
++ L L + ++ L ++P+ +E L LE L++ + TLP+
Sbjct: 758 WEGVMKPLKPLLAMLSPTLTSLQLQNIPNLVELPCSFQNLIQLEVLDITNCRNLETLPTG 817
Query: 835 RKLSKLVYLNLEHCKLLESLPQLPFPTNTGEVHRE---------YDDYFCGAGLLIFN-C 884
L L L+ + C L S P++ TN ++ E + D F GLL + C
Sbjct: 818 INLQSLDSLSFKGCSRLRSFPEI--STNISSLNLEETGIEEVPWWIDKFSNLGLLSMDRC 875
Query: 885 PKLGEREHCRSMTLLWMKQFIKANPRSSSEIQIVN 919
+L C S+ + +K+ K + + + IV+
Sbjct: 876 SRL----KCVSLHISKLKRLGKVDFKDCGALTIVD 906
>At5g11250 RPP1 disease resistance protein - like
Length = 1189
Score = 392 bits (1006), Expect = e-109
Identities = 298/880 (33%), Positives = 466/880 (52%), Gaps = 48/880 (5%)
Query: 4 RCHILTKKKKKSRYHIQSSKHLYIQMASSSKSSSALVTSSKKNHYDVFVTFRGEDTRNNF 63
R H K+KK + SS + SSS + T + VF +F GED R +F
Sbjct: 24 RFHHDNKEKKSTSLSPSSSPSSLSPSSVPPPSSSCIWT------HQVFPSFSGEDVRRDF 77
Query: 64 TDFLFDALETKGIMVFRDVINLQKGECIGPELFRAIEISQVYVAIFSKNYASSTWCLQEL 123
+ + GI F D +++GE IGPEL RAI S++ + + S+NYASS WCL EL
Sbjct: 78 LSHIQMEFQRMGITPFVDN-EIKRGESIGPELLRAIRGSKIAIILLSRNYASSKWCLDEL 136
Query: 124 EKICECIKGSGKHVLPVFYDVDPSEVRKQSGIYSEAFVKHEQRFQQDSMKVSRWREALEQ 183
+I +C + G+ V+ +FY VDPS+V+ +G + + F K + + RWR+A E+
Sbjct: 137 VEIMKCREEYGQTVMAIFYKVDPSDVKNLTGDFGKVFRK--TCAGKPKKDIGRWRQAWEK 194
Query: 184 VGSISGW-DLRDEPLAREIKEIVQKIINILECKYSCVSKD---LVGIDSPIQALQNHLLL 239
V +++G+ + + A IK+I I NIL S S+D LVG+ + ++ ++ L L
Sbjct: 195 VATVAGYHSINWDNEAAMIKKIATDISNILI--NSTPSRDFDGLVGMRAHLEKMKPLLCL 252
Query: 240 NSVDGVRAIGICGMGGIGKTTLATTLYGQISHQFSASCFIDDVTKIY----GLHDDPLDV 295
++ D VR IGI G GIGKTT+A +Y Q+SH F S F++++ Y G D +
Sbjct: 253 DT-DEVRIIGIWGPPGIGKTTIARVVYNQLSHSFQLSVFMENIKANYTRPTGSDDYSAKL 311
Query: 296 QKQILFQTLGIEHQQICNRYHATTLIQRKLCHERTLMILDNVDQVEQLEKIAVHREWLGP 355
Q Q +F + + + I + Q +L ++ L++LD V+Q QL+ +A W GP
Sbjct: 312 QLQQMFMSQITKQKDI--EIPHLGVAQDRLKDKKVLVVLDGVNQSVQLDAMAKEAWWFGP 369
Query: 356 GSRIIIISRDEHVLKAYGVDVVYKVSLLDWNEAHMLFCRKAFKDEKIIMSNYQNLVDQIL 415
GSRIII ++D+ + +A+G++ +YKV EA +FC AF + +QNL +++
Sbjct: 370 GSRIIITTQDQKLFRAHGINHIYKVDFPPTEEALQIFCMYAF-GQNSPKDGFQNLAWKVI 428
Query: 416 HYAKGLPLAIKVLGSFLFGRNVTEWKSALTRLRQSPVKDVMDVLQLSFDGLNETEKDIFL 475
+ A LPL ++++GS+ G + EWK +L RL S D+ +L+ S+D L++ +K++FL
Sbjct: 429 NLAGNLPLGLRIMGSYFRGMSREEWKKSLPRLESSLDADIQSILKFSYDALDDEDKNLFL 488
Query: 476 HIACFFNNDSEEDVKNILNCCGFHADIGLRVLIDKSLVSIS-YSIINMHSLLEELGRKIV 534
HIACFFN + ++ L L VL +KSL+S S + I MH LL +LG +IV
Sbjct: 489 HIACFFNGKEIKILEEHLAKKFVEVRQRLNVLAEKSLISFSNWGTIEMHKLLAKLGGEIV 548
Query: 535 QNSSSKEPRKWSRLWSTEQLYDVMLENM--EKHVEAIVLYYKEDEEADFEH--LSKMSNL 590
+N S EP + L+ E++ DV+ + K V I +Y +EE D MSNL
Sbjct: 549 RNQSIHEPGQRQFLFDGEEICDVLNGDAAGSKSVIGIDFHYIIEEEFDMNERVFEGMSNL 608
Query: 591 RLL-FIANYISTMLGFP-SCLSNKLRFVHWFRYPSKYLPSNFHPNELVELILTESNIKQL 648
+ L F ++ + L S LS KL+ + W +P LPS + L+EL LT S + L
Sbjct: 609 QFLRFDCDHDTLQLSRGLSYLSRKLQLLDWIYFPMTCLPSTVNVEFLIELNLTHSKLDML 668
Query: 649 WKNKKYLPNLRTLDLRHSRNLEKIIDFGEFPNLERLDLEGCINLVELDPSIGLLRKLVYL 708
W+ K L NLR +DL +S NL+++ D NL +L L C +L++L IG L L
Sbjct: 669 WEGVKPLHNLRQMDLSYSVNLKELPDLSTAINLRKLILSNCSSLIKLPSCIGNAINLEDL 728
Query: 709 NLKDCKSLVSIPNNIFG-LSSLQYLNMCGCSKVFNNPRRLMKSGISSEKKQQHDIRESAS 767
+L C SLV +P+ FG +LQ L + CS + P + G + ++ S+
Sbjct: 729 DLNGCSSLVELPS--FGDAINLQKLLLRYCSNLVELPSSI---GNAINLRELDLYYCSSL 783
Query: 768 HHLP-----GLKWIIL-AHDSSHML---PSLHSLCCLRKVDISFC-YLSHVPDAIECLHW 817
LP + +IL + S++L S+ + L+K+D+ C L +P +I
Sbjct: 784 IRLPSSIGNAINLLILDLNGCSNLLELPSSIGNAINLQKLDLRRCAKLLELPSSIGNAIN 843
Query: 818 LERLNLAG-NDFVTLP-SLRKLSKLVYLNLEHCKLLESLP 855
L+ L L + + LP S+ + LVY+NL +C L LP
Sbjct: 844 LQNLLLDDCSSLLELPSSIGNATNLVYMNLSNCSNLVELP 883
Score = 83.2 bits (204), Expect = 7e-16
Identities = 95/366 (25%), Positives = 164/366 (43%), Gaps = 32/366 (8%)
Query: 589 NLRLLFIANYISTMLGFPSCLSNKLRFVHWFRYPSKYL---PSNFHPNELVELILTE--- 642
NL+ L + Y S ++ PS + N + Y L PS+ + L++ +
Sbjct: 747 NLQKLLL-RYCSNLVELPSSIGNAINLRELDLYYCSSLIRLPSSI--GNAINLLILDLNG 803
Query: 643 -SNIKQLWKNKKYLPNLRTLDLRH-SRNLEKIIDFGEFPNLERLDLEGCINLVELDPSIG 700
SN+ +L + NL+ LDLR ++ LE G NL+ L L+ C +L+EL SIG
Sbjct: 804 CSNLLELPSSIGNAINLQKLDLRRCAKLLELPSSIGNAINLQNLLLDDCSSLLELPSSIG 863
Query: 701 LLRKLVYLNLKDCKSLVSIPNNIFGLSSLQYLNMCGCSKVFNNPRRLMKSGISSEKKQQH 760
LVY+NL +C +LV +P +I L LQ L + GCSK+ + P + +
Sbjct: 864 NATNLVYMNLSNCSNLVELPLSIGNLQKLQELILKGCSKLEDLPININLESLDILVLNDC 923
Query: 761 DIRESASHHLPGLKWIILAHDSSHMLPSLHSLCCLRKVDISFCYLSHVPDAIECLHWLER 820
+ + ++ + L + +P L R ++ Y ++ + L +
Sbjct: 924 SMLKRFPEISTNVRALYLCGTAIEEVP-LSIRSWPRLDELLMSYFDNLVEFPHVLDIITN 982
Query: 821 LNLAGNDFVTLPSL-RKLSKLVYLNLEHCKLLESLPQLPFPTNTGEVHREYDDYFCGAGL 879
L+L+G + +P L +++S+L L L+ + + SLPQ+P + + D C
Sbjct: 983 LDLSGKEIQEVPPLIKRISRLQTLILKGYRKVVSLPQIP------DSLKWIDAEDCE--- 1033
Query: 880 LIFNCPKLGEREHCRSMTLLWMKQFIKANPRSSSEI------QIVNPGSEIPSWINNQRM 933
+ +L H +TL + K F K N + I Q V PG E+P++ ++
Sbjct: 1034 ---SLERLDCSFHNPEITLFFGKCF-KLNQEARDLIIQTPTKQAVLPGREVPAYFTHRAS 1089
Query: 934 GYSIAI 939
G S+ I
Sbjct: 1090 GGSLTI 1095
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.138 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,295,907
Number of Sequences: 26719
Number of extensions: 1131252
Number of successful extensions: 5935
Number of sequences better than 10.0: 390
Number of HSP's better than 10.0 without gapping: 255
Number of HSP's successfully gapped in prelim test: 138
Number of HSP's that attempted gapping in prelim test: 3304
Number of HSP's gapped (non-prelim): 1232
length of query: 1085
length of database: 11,318,596
effective HSP length: 110
effective length of query: 975
effective length of database: 8,379,506
effective search space: 8170018350
effective search space used: 8170018350
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 65 (29.6 bits)
Medicago: description of AC122722.3


