

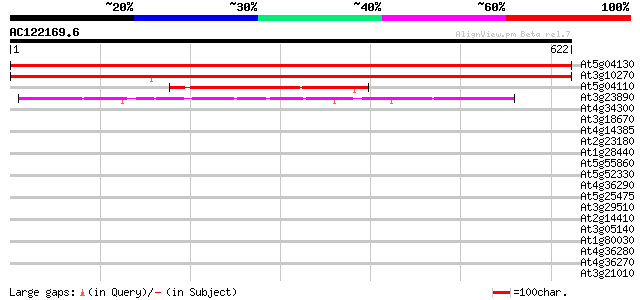
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122169.6 + phase: 0
(622 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g04130 DNA gyrase subunit B - like protein 1049 0.0
At3g10270 putative DNA gyrase subunit B 1035 0.0
At5g04110 DNA gyrase subunit B - like protein 265 4e-71
At3g23890 topoisomerase II 142 6e-34
At4g34300 putative protein 36 0.073
At3g18670 hypothetical protein 34 0.28
At4g14385 unknown protein 32 0.81
At2g23180 putative cytochrome P450 31 2.4
At1g28440 unknown protein 31 2.4
At5g55860 myosin heavy chain-like 30 3.1
At5g52330 putative protein 30 3.1
At4g36290 unknown protein 30 3.1
At5g25475 unknown protein 30 4.0
At3g29510 hypothetical protein 30 4.0
At2g14410 hypothetical protein 30 4.0
At3g05140 putative serine/threonine protein kinase 30 5.2
At1g80030 unknown protein 30 5.2
At4g36280 unknown protein 29 6.8
At4g36270 putative protein 29 6.8
At3g21010 unknown protein 29 6.8
>At5g04130 DNA gyrase subunit B - like protein
Length = 732
Score = 1049 bits (2712), Expect = 0.0
Identities = 510/622 (81%), Positives = 576/622 (91%)
Query: 1 MYIGSTGPRGLHHLVYEILDNAVDEAQAGFASKIDVVLHADDSVSIADNGRGIPTDMHPT 60
MYIGSTG RGLHHLVYEILDNA+DEAQAG+ASK+DVVLHAD SVS+ DNGRGIPTD+HP
Sbjct: 111 MYIGSTGSRGLHHLVYEILDNAIDEAQAGYASKVDVVLHADGSVSVVDNGRGIPTDLHPA 170
Query: 61 TKKSALETVLTILHAGGKFGGINSGYSVSGGLHGVGLSVVNALSEALEVTVWREGLEYTQ 120
TKKS+LETVLT+LHAGGKFGG +SGYSVSGGLHGVGLSVVNALSEALEV+VWR+G+E+ Q
Sbjct: 171 TKKSSLETVLTVLHAGGKFGGTSSGYSVSGGLHGVGLSVVNALSEALEVSVWRDGMEHKQ 230
Query: 121 TYSRGKPVTTLNCVMLPFENKDRQGTRIKFWPDKEVFTTAIEFEYNTIAGRIRELAFLNP 180
YSRGKP+TTL C +LP E+K +GT I+FWPDKEVFTTAIEF++NTIAGRIRELAFLNP
Sbjct: 231 NYSRGKPITTLTCRVLPLESKGTKGTSIRFWPDKEVFTTAIEFDHNTIAGRIRELAFLNP 290
Query: 181 KLTIALRKEDNDPEKVQSNEYFYAGGLAEYVQFLNTDKKAVHDVLSFRKEIDGITVDIAL 240
K+TI+L+KED+DPEK Q +EY +AGGL EYV +LNTDK +HDVL FR+EI+G TVD+AL
Sbjct: 291 KVTISLKKEDDDPEKTQYSEYSFAGGLTEYVSWLNTDKNPIHDVLGFRREINGATVDVAL 350
Query: 241 QWCEDAYSDTILGYANSIRTVDGGTHIDGMKASLTRTLNSLGKKSKVIKEKDITLSGEHV 300
QWC DAYSDT+LGYANSIRT+DGGTHI+G+KASLTRTLN+L KKSK +KEKDI+LSGEHV
Sbjct: 351 QWCSDAYSDTMLGYANSIRTIDGGTHIEGVKASLTRTLNTLAKKSKTVKEKDISLSGEHV 410
Query: 301 REGLTCVVSVKVPNPEFEGQTKTRLGNPEVRKVVDQSIQEYLTEYLELHPDVLDSVLSKA 360
REGLTC+VSVKVPNPEFEGQTKTRLGNPEVRK+VDQS+QEYLTE+LELHPD+L+S++SK+
Sbjct: 411 REGLTCIVSVKVPNPEFEGQTKTRLGNPEVRKIVDQSVQEYLTEFLELHPDILESIISKS 470
Query: 361 LNAFKAALAAKRARELVRQKSVLRSSSLPGKLADCSSSNPEDCEIFIVEGDSAGGSAKQG 420
LNA+KAALAAKRARELVR KSVL+SSSLPGKLADCSS++PE EIFIVEGDSAGGSAKQG
Sbjct: 471 LNAYKAALAAKRARELVRSKSVLKSSSLPGKLADCSSTDPEVSEIFIVEGDSAGGSAKQG 530
Query: 421 RDRRFQAILPLRGKILNIERRDEAAMYKNEEIQNLILGLGLGVKGEDFKKDALRYHKIII 480
RDRRFQAILPLRGKILNIER+DEAAMYKNEEIQNLILGLGLGVKGEDFKK+ LRYHKIII
Sbjct: 531 RDRRFQAILPLRGKILNIERKDEAAMYKNEEIQNLILGLGLGVKGEDFKKENLRYHKIII 590
Query: 481 LTDADVDGAHIRTLLLTFFYRYQRALFDEGYIYVGVPPLYKVTRGKQVHYCYDEADLKKL 540
LTDADVDGAHIRTLLLTFF+RYQRALFD G IYVGVPPL+KV RGK YCYD+ADLKK+
Sbjct: 591 LTDADVDGAHIRTLLLTFFFRYQRALFDAGCIYVGVPPLFKVERGKNAQYCYDDADLKKI 650
Query: 541 KSSFAPNASYNMQRFKGLGEMMPLQLWETTMDPEQRLLKKLTVEDAAEANIVFSSLMGAR 600
S+F NASYN+QRFKGLGEMMP QLWETTM+PE R+LK+L V+D AEAN+ FSSLMGAR
Sbjct: 651 TSNFPANASYNIQRFKGLGEMMPEQLWETTMNPETRILKQLVVDDIAEANMTFSSLMGAR 710
Query: 601 VDVRKELIQNAANMIDLDQLDI 622
VDVRKELI+NAA I+L +LDI
Sbjct: 711 VDVRKELIKNAATRINLQRLDI 732
>At3g10270 putative DNA gyrase subunit B
Length = 657
Score = 1035 bits (2676), Expect = 0.0
Identities = 512/625 (81%), Positives = 577/625 (91%), Gaps = 3/625 (0%)
Query: 1 MYIGSTGPRGLHHLVYEILDNAVDEAQAGFASKIDVVLHADDSVSIADNGRGIPTDMHPT 60
MYIGSTG RGLHHLVYEILDNA+DEAQAGFASKIDVVLH+DDSVSI+DNGRGIPTD+HP
Sbjct: 33 MYIGSTGSRGLHHLVYEILDNAIDEAQAGFASKIDVVLHSDDSVSISDNGRGIPTDLHPA 92
Query: 61 TKKSALETVLTILHAGGKFGGINSGYSVSGGLHGVGLSVVNALSEALEVTVWREGLEYTQ 120
T KS+LETVLT+LHAGGKFGG +SGYSVSGGLHGVGLSVVNALSEALEV V R+G+E+ Q
Sbjct: 93 TGKSSLETVLTVLHAGGKFGGKSSGYSVSGGLHGVGLSVVNALSEALEVIVRRDGMEFQQ 152
Query: 121 TYSRGKPVTTLNCVMLPFENKDRQGTRIKFWPDKE---VFTTAIEFEYNTIAGRIRELAF 177
YSRGKPVTTL C +LP E++ QGT I+FWPDKE +FTTAI+F++NTIAGRIRELAF
Sbjct: 153 KYSRGKPVTTLTCHVLPPESRGTQGTCIRFWPDKEGFALFTTAIQFDHNTIAGRIRELAF 212
Query: 178 LNPKLTIALRKEDNDPEKVQSNEYFYAGGLAEYVQFLNTDKKAVHDVLSFRKEIDGITVD 237
LNPK+TI+L+KED+DPE+ +EYFYAGGL EYV +LNTDKK +HDVL FRKEI+G TVD
Sbjct: 213 LNPKVTISLKKEDDDPERDVYSEYFYAGGLTEYVSWLNTDKKPLHDVLGFRKEINGSTVD 272
Query: 238 IALQWCEDAYSDTILGYANSIRTVDGGTHIDGMKASLTRTLNSLGKKSKVIKEKDITLSG 297
++LQWC DAYSDT+LGYANSIRT+DGGTHI+G+KASLTRTLNSL KK KVIKEKDI+LSG
Sbjct: 273 VSLQWCSDAYSDTMLGYANSIRTIDGGTHIEGVKASLTRTLNSLAKKLKVIKEKDISLSG 332
Query: 298 EHVREGLTCVVSVKVPNPEFEGQTKTRLGNPEVRKVVDQSIQEYLTEYLELHPDVLDSVL 357
EHVREGLTC+VSVKVPNPEFEGQTKTRLGNPEVRK+VDQS+QEYLTEYLELHPDVL+S++
Sbjct: 333 EHVREGLTCIVSVKVPNPEFEGQTKTRLGNPEVRKIVDQSVQEYLTEYLELHPDVLESII 392
Query: 358 SKALNAFKAALAAKRARELVRQKSVLRSSSLPGKLADCSSSNPEDCEIFIVEGDSAGGSA 417
SK+LNA+KAALAAKRARELVR KSVL+SSSLPGKLADCSS++P + EIFIVEGDSAGGSA
Sbjct: 393 SKSLNAYKAALAAKRARELVRSKSVLKSSSLPGKLADCSSTDPAESEIFIVEGDSAGGSA 452
Query: 418 KQGRDRRFQAILPLRGKILNIERRDEAAMYKNEEIQNLILGLGLGVKGEDFKKDALRYHK 477
KQGRDRRFQAILPLRGKILNIER+DEAAMYKNEEIQNLILGLGLGVKGEDF K+ LRYHK
Sbjct: 453 KQGRDRRFQAILPLRGKILNIERKDEAAMYKNEEIQNLILGLGLGVKGEDFNKENLRYHK 512
Query: 478 IIILTDADVDGAHIRTLLLTFFYRYQRALFDEGYIYVGVPPLYKVTRGKQVHYCYDEADL 537
IIILTDADVDGAHIRTLLLTFF+RYQRALFD G IYVGVPPL+KV RGKQ HYCYD+A L
Sbjct: 513 IIILTDADVDGAHIRTLLLTFFFRYQRALFDAGCIYVGVPPLFKVERGKQAHYCYDDAAL 572
Query: 538 KKLKSSFAPNASYNMQRFKGLGEMMPLQLWETTMDPEQRLLKKLTVEDAAEANIVFSSLM 597
KK+ +SF NASYN+QRFKGLGEMMP QLWETTM+P+ R+LK+L V+DAAE N+VFSSLM
Sbjct: 573 KKITASFPGNASYNIQRFKGLGEMMPAQLWETTMNPDTRILKQLVVDDAAETNMVFSSLM 632
Query: 598 GARVDVRKELIQNAANMIDLDQLDI 622
GARVDVRKELI++AA ++L+ LDI
Sbjct: 633 GARVDVRKELIKSAATRMNLENLDI 657
>At5g04110 DNA gyrase subunit B - like protein
Length = 257
Score = 265 bits (678), Expect = 4e-71
Identities = 135/223 (60%), Positives = 172/223 (76%), Gaps = 8/223 (3%)
Query: 178 LNPKLTIALRKEDNDPEKVQSNEYFYAGGLAEYVQFLNTDKKAVHDVLSFRKEIDGITVD 237
L ++T++L EDNDP K EY YA GL+E+V +LN DKK +HDVL FRKEI+G T++
Sbjct: 17 LRVQITVSLEMEDNDPNK----EYLYAKGLSEFVTWLNADKKPLHDVLGFRKEINGTTIN 72
Query: 238 IALQWCEDAYSDTILGYANSIRTVDGGTHIDGMKASLTRTLNSLGKKSKVIKEKDITLSG 297
IALQWC D YS+ ILGYAN IRT+DGGT+IDG+KAS+TRTLNSL +KSK++++KDI +
Sbjct: 73 IALQWCVDGYSNKILGYANGIRTMDGGTYIDGVKASITRTLNSLVEKSKLVEDKDIIFTE 132
Query: 298 EHVREGLTCVVSVKVPNPEFEGQTKTRLGNPEVRKVVDQSIQEYLTEYLELHPDVLDSVL 357
EHV EGLTC+VSV VP PEFEGQT+ RLGNP VR++VDQS+QE L E ELHPDV +S++
Sbjct: 133 EHVMEGLTCIVSVIVPKPEFEGQTQ-RLGNPNVREIVDQSVQECLMESFELHPDVFESIM 191
Query: 358 SKALNAFKAALAAKRARELVRQKS---VLRSSSLPGKLADCSS 397
SK+ NA+K LA KRAR++ +S V S+P KL + SS
Sbjct: 192 SKSYNAYKTDLAVKRARDVYSSESVAMVCAIESIPMKLTNSSS 234
>At3g23890 topoisomerase II
Length = 1473
Score = 142 bits (358), Expect = 6e-34
Identities = 147/576 (25%), Positives = 257/576 (44%), Gaps = 63/576 (10%)
Query: 10 GLHHLVYEILDNAVDEAQAGFA-SKIDVVLHADDS-VSIADNGRGIPTDMHPTTKKSALE 67
GL+ + EIL NA D Q + VV+ + + +S+ ++G G+P ++H E
Sbjct: 80 GLYKIFDEILVNAADNKQRDAKMDSVQVVIDVEQNLISVCNSGAGVPVEIHQEEGIYVPE 139
Query: 68 TVLTILHAGGKFGGINSGYSVSGGLHGVGLSVVNALSEA--LEVTVWREGLEYTQTYS-- 123
+ L + ++ +GG +G G + N S +E + +Y Q +
Sbjct: 140 MIFGHLLTSSNYD--DNVKKTTGGRNGYGAKLTNIFSTEFIIETADGKRLKKYKQVFENN 197
Query: 124 ---RGKPVTTLNCVMLPFENKDRQGTRIKFWPDKEVFTTAIEFEYNTIAGRIRELAFLNP 180
+ +PV T C NK T++ F PD + F E E + +A + + +
Sbjct: 198 MGKKSEPVIT-KC------NKSENWTKVTFKPDLKKFNMT-ELEDDVVALMSKRVFDIAG 249
Query: 181 KLTIALRKEDNDPEKVQSNEYFYAGGLAEYVQFLNT--DKKAVHDVLSFRKEIDGITVDI 238
L +++ E N + +YV + +K D L E ++
Sbjct: 250 CLGKSVKVELNGKQ-------IPVKSFTDYVDLYLSAANKSRTEDPLPRLTEKVNDRWEV 302
Query: 239 ALQWCEDAYSDTILGYANSIRTVDGGTHIDGMKASLTRTLNSLGKKSKVIKEKDITLSGE 298
+ E + + + NSI T+ GGTH+D + + +T + + K K K+ +
Sbjct: 303 CVSLSEGQFQQ--VSFVNSIATIKGGTHVDYVTSQITNHIVAAVNK----KNKNANVKAH 356
Query: 299 HVREGLTCVVSVKVPNPEFEGQTKTRLGNPEVRKVVDQSIQEYLTEYLEL--HPDVLDSV 356
+V+ L V+ + NP F+ QTK L +R+ S E ++L+ V++++
Sbjct: 357 NVKNHLWVFVNALIDNPAFDSQTKETL---TLRQSSFGSKCELSEDFLKKVGKSGVVENL 413
Query: 357 LS----KALNAFKAALAAKRARELVRQKSVLRSSSLPGKLADCSSSNPEDCEIFIVEGDS 412
LS K K + AK R LV + A+ N C + + EGDS
Sbjct: 414 LSWADFKQNKDLKKSDGAKTGRVLVEKLD---------DAAEAGGKNSRLCTLILTEGDS 464
Query: 413 AGGSAKQGRD---RRFQAILPLRGKILNIERRDEAAMYKNEEIQNL--ILGLGLGVKGED 467
A A GR + + PLRGK+LN+ + N+EI+NL ILGL +K E+
Sbjct: 465 AKSLALAGRSVLGNNYCGVFPLRGKLLNVREASTTQITNNKEIENLKKILGLKQNMKYEN 524
Query: 468 FKKDALRYHKIIILTDADVDGAHIRTLLLTFFYRYQRALFD-EGYIYVGVPPLYKVTR-- 524
++LRY +++I+TD D DG+HI+ LL+ F + + +L ++ + P+ K TR
Sbjct: 525 V--NSLRYGQMMIMTDQDHDGSHIKGLLINFIHSFWPSLLQVPSFLVEFITPIVKATRKG 582
Query: 525 GKQVHYCYDEADLKKLKSSFAPNAS-YNMQRFKGLG 559
K+V Y + ++ K S NA+ ++++ +KGLG
Sbjct: 583 TKKVLSFYSMPEYEEWKESLKGNATGWDIKYYKGLG 618
>At4g34300 putative protein
Length = 313
Score = 35.8 bits (81), Expect = 0.073
Identities = 20/81 (24%), Positives = 37/81 (44%)
Query: 75 AGGKFGGINSGYSVSGGLHGVGLSVVNALSEALEVTVWREGLEYTQTYSRGKPVTTLNCV 134
AG G +SG + S + +L + VTVW+ G YT+ ++ P + +
Sbjct: 161 AGPTHNGHSSGSNHSSIIGSTHNHTAPSLGRKIAVTVWKNGYGYTEWTAKHAPFYVNDVL 220
Query: 135 MLPFENKDRQGTRIKFWPDKE 155
+ + N D+ ++ K DK+
Sbjct: 221 VFTYNNNDQTQSKTKHHHDKK 241
>At3g18670 hypothetical protein
Length = 598
Score = 33.9 bits (76), Expect = 0.28
Identities = 22/83 (26%), Positives = 40/83 (47%), Gaps = 4/83 (4%)
Query: 293 ITLSGEHVREGLTCVVSVKVPNPEFEGQTKTRLG-NPEVRKVVDQSIQEYLTEYLELHPD 351
+ L +E L C+ P+F+ + G N + K V+ I EY+ E + +PD
Sbjct: 258 LKLGHAQAKEILDCICQ---EIPKFDAAQQKNAGLNQALFKAVENGIVEYIEEMMRHYPD 314
Query: 352 VLDSVLSKALNAFKAALAAKRAR 374
++ S S LN F A++ ++ +
Sbjct: 315 IVWSKNSSGLNIFFYAVSQRQEK 337
>At4g14385 unknown protein
Length = 163
Score = 32.3 bits (72), Expect = 0.81
Identities = 26/105 (24%), Positives = 50/105 (46%), Gaps = 4/105 (3%)
Query: 334 VDQSIQEYLTEYLELHPDVLDSV--LSKALNAFKAALAAKRARELVRQKSVLRSSSLPGK 391
+++ + E T YL+ + +++ L++ K+ +AKR+R+ + V SS+
Sbjct: 32 IEKQVYELETSYLQESSHIGNALKGFEGFLSSSKSTASAKRSRKFQPEDRVFSLSSVTSP 91
Query: 392 LADCSSSNPEDCEIFIVEGDSAGGSAKQGRDR--RFQAILPLRGK 434
A+ ED + G S GG + QG+ + R Q+I+ K
Sbjct: 92 AAEELGVGREDGRAELGPGRSKGGLSTQGKPKKGRGQSIIAREAK 136
>At2g23180 putative cytochrome P450
Length = 516
Score = 30.8 bits (68), Expect = 2.4
Identities = 21/74 (28%), Positives = 33/74 (44%), Gaps = 3/74 (4%)
Query: 502 YQRALFDEGYIYV-GVPPLYKVTRGKQVHYC--YDEADLKKLKSSFAPNASYNMQRFKGL 558
+QR FD ++ GV P T Q+ + DEA+ P + MQRF G
Sbjct: 177 FQRFTFDTSFVLATGVDPGCLSTEMPQIEFARALDEAEEAIFFRHVKPEIVWKMQRFIGF 236
Query: 559 GEMMPLQLWETTMD 572
G+ + ++ +T D
Sbjct: 237 GDELKMKKAHSTFD 250
>At1g28440 unknown protein
Length = 996
Score = 30.8 bits (68), Expect = 2.4
Identities = 26/92 (28%), Positives = 49/92 (53%), Gaps = 10/92 (10%)
Query: 279 NSLGKKSKVIKEKDITLSG--EHVRE---GLTCVVSVKVPNPEFEGQTKTRLGNPEVRKV 333
+SLG+ SK++ + D+ L+ H+ GLT VV +++ N G+ LGN + ++
Sbjct: 223 DSLGQLSKLV-DLDLALNDLVGHIPPSLGGLTNVVQIELYNNSLTGEIPPELGNLKSLRL 281
Query: 334 VDQSIQEYLTEYLELHPDVLDSVLSKALNAFK 365
+D S+ + + PD L V ++LN ++
Sbjct: 282 LDASMNQLTGKI----PDELCRVPLESLNLYE 309
>At5g55860 myosin heavy chain-like
Length = 649
Score = 30.4 bits (67), Expect = 3.1
Identities = 39/167 (23%), Positives = 69/167 (40%), Gaps = 12/167 (7%)
Query: 276 RTLNSLGKKSKVIKEKDITLSGEHVREGLTCVVSVKVPN-----PEFEGQTKTRLGNPEV 330
RT++ L +K + + E S E ++ P + QT+ EV
Sbjct: 101 RTVDELTRKLEAVNESRD--SANKATEAAKSLIEEAKPGNVSVASSSDAQTRDMEEYGEV 158
Query: 331 RKVVDQSIQEYLTEYLELHPDVLDS---VLSKALNAFKAALAAKRARELVRQKSVLRSSS 387
K +D + QE L + ++ ++L++ LSK A K + EL+R++ + S
Sbjct: 159 CKELDTAKQE-LRKIRQVSNEILETKTVALSKVEEAKKVSKVHSEKIELLRKEIAAVNES 217
Query: 388 LPGKLADCSSSNPEDCEIFIVEGDSAGGSAKQGRDRRFQAILPLRGK 434
+ CS + E EIF E + S K G + + L L+ +
Sbjct: 218 VEQTKLACSQARKEQSEIF-AEKEIQQKSYKAGMEESAKKSLALKNE 263
>At5g52330 putative protein
Length = 397
Score = 30.4 bits (67), Expect = 3.1
Identities = 14/29 (48%), Positives = 20/29 (68%)
Query: 352 VLDSVLSKALNAFKAALAAKRARELVRQK 380
VLD + SK A+K A+A KR EL+R++
Sbjct: 349 VLDHIFSKIEVAYKEAIALKRQEELIREE 377
>At4g36290 unknown protein
Length = 635
Score = 30.4 bits (67), Expect = 3.1
Identities = 18/57 (31%), Positives = 29/57 (50%), Gaps = 4/57 (7%)
Query: 1 MYIGSTGPRGLHHLVYEILDNAVDEAQAGFA----SKIDVVLHADDSVSIADNGRGI 53
++ +T + + E+LDNAVDE Q G KI++V ++ DNG G+
Sbjct: 92 LHSNATSHKWAFGAIAELLDNAVDEIQNGATVVKIDKINIVKDNTPALVFQDNGGGM 148
>At5g25475 unknown protein
Length = 282
Score = 30.0 bits (66), Expect = 4.0
Identities = 31/128 (24%), Positives = 51/128 (39%), Gaps = 16/128 (12%)
Query: 34 IDVVLHADDSVSIADNGRGIPTD---MHPTTKKSALETVLTILHAGGKFGGINSGYSVSG 90
++V + ++ V I D G+ + HPT KK ET A G SG S +G
Sbjct: 106 LEVQIFKNNGVEIIDVPLGVEPETEPFHPTPKKPHKETTPASSFASG------SGCSANG 159
Query: 91 GLHGVGLSVVNALSEALEVTVWREGLEYTQTYSRGKPVTTLNCVMLPFENKDRQGTRIKF 150
G +G G + + + E + Q ++ V ++ ++ Q R+KF
Sbjct: 160 GTNGRGKQRSSDVKNPERYLLNPENPYFVQAVTKRNDVLYVSRPVV-------QSYRLKF 212
Query: 151 WPDKEVFT 158
P K T
Sbjct: 213 GPVKSTIT 220
>At3g29510 hypothetical protein
Length = 1158
Score = 30.0 bits (66), Expect = 4.0
Identities = 24/94 (25%), Positives = 41/94 (43%), Gaps = 5/94 (5%)
Query: 207 LAEYVQFLNTDKKAVHDVLSFRKEIDGITVDIALQWCEDAYSDTILGYANSIRTVDGGTH 266
+AE L D + H +L ++ +T+ A + I GY + + GTH
Sbjct: 1 MAEDKDILIVDSGSSHTILRDKRYFINLTLRNANISTIAGIASLIEGYGQAHVLLPNGTH 60
Query: 267 IDGMKASLTRTLNSLGKKSKVIKEKDITLSGEHV 300
++ ++ L S K ++ KDI L+G HV
Sbjct: 61 LE-----ISDDLYSPSSKRSLLSFKDIRLNGFHV 89
>At2g14410 hypothetical protein
Length = 679
Score = 30.0 bits (66), Expect = 4.0
Identities = 29/95 (30%), Positives = 40/95 (41%), Gaps = 20/95 (21%)
Query: 329 EVRKVVDQSIQEYLTEYLEL---HPDVLDSVLSKALNAFKAALAAKRARELVRQKSVLRS 385
+VR ++ + EY + L L H D D + + KA L KR R
Sbjct: 571 KVRAELESDVNEYESNLLLLDQTHED--DFSAERERDELKAVLEEKRNR----------L 618
Query: 386 SSLPGKLADCSSSNPEDCEIFIVEGDSAGGSAKQG 420
+SLP SS NP+ EI ++E D SA G
Sbjct: 619 ASLPS-----SSFNPQQFEIVVIEDDDGSDSAIPG 648
>At3g05140 putative serine/threonine protein kinase
Length = 460
Score = 29.6 bits (65), Expect = 5.2
Identities = 19/72 (26%), Positives = 31/72 (42%)
Query: 8 PRGLHHLVYEILDNAVDEAQAGFASKIDVVLHADDSVSIADNGRGIPTDMHPTTKKSALE 67
P G V + DE A F S++ ++ H D + G I MH + S L
Sbjct: 163 PEGKLIAVKRLTKGTPDEQTAEFLSELGIIAHVDHPNTAKFIGCCIEGGMHLVFRLSPLG 222
Query: 68 TVLTILHAGGKF 79
++ ++LH K+
Sbjct: 223 SLGSLLHGPSKY 234
>At1g80030 unknown protein
Length = 500
Score = 29.6 bits (65), Expect = 5.2
Identities = 35/133 (26%), Positives = 53/133 (39%), Gaps = 12/133 (9%)
Query: 230 EIDGITVDIALQWCEDAYSDTILGYANSIRTVDGGTHIDGMKASLTRTLNSLGKKSKVIK 289
E DGI + L +Y D ILG ++TV+G T + + + L KK V K
Sbjct: 339 ERDGINL---LSTLSISYLDAILGAVVKVKTVEGDTELQIPPGTQPGDVLVLAKKG-VPK 394
Query: 290 EKDITLSGEHVREGLTCVVSVKVPNPEFEGQTKTRLGNPEVRKVVDQSIQEYLTEYLELH 349
++ G+H+ V V VPN G+ + E+ + D S T
Sbjct: 395 LNRPSIRGDHL-----FTVKVSVPNQISAGERELL---EELASLKDTSSNRSRTRAKPQQ 446
Query: 350 PDVLDSVLSKALN 362
P L + S + N
Sbjct: 447 PSTLSTAPSGSEN 459
>At4g36280 unknown protein
Length = 626
Score = 29.3 bits (64), Expect = 6.8
Identities = 22/93 (23%), Positives = 39/93 (41%), Gaps = 8/93 (8%)
Query: 1 MYIGSTGPRGLHHLVYEILDNAVDEAQAGFA----SKIDVVLHADDSVSIADNGRGIPTD 56
++ +T + + E+LDNAVDE Q G KI++V ++ D+G G
Sbjct: 84 LHSNATSHKWAFGAIAELLDNAVDEIQNGATFVKIDKINIVKDNSPALVFQDDGGG---- 139
Query: 57 MHPTTKKSALETVLTILHAGGKFGGINSGYSVS 89
M P + + + + G +G+ S
Sbjct: 140 MDPAGLRKCMSLGYSSKKSNTTIGQYGNGFKTS 172
>At4g36270 putative protein
Length = 489
Score = 29.3 bits (64), Expect = 6.8
Identities = 16/46 (34%), Positives = 25/46 (53%), Gaps = 4/46 (8%)
Query: 15 VYEILDNAVDEAQAGFA----SKIDVVLHADDSVSIADNGRGIPTD 56
+ E++DNAVDE Q G KI++V ++ D+G G+ D
Sbjct: 87 IAELIDNAVDEIQNGATFVKIDKINIVKDNSPALVFQDDGGGMDPD 132
>At3g21010 unknown protein
Length = 438
Score = 29.3 bits (64), Expect = 6.8
Identities = 28/102 (27%), Positives = 45/102 (43%), Gaps = 20/102 (19%)
Query: 352 VLDSVLSKALNAFKAALAAKRARELVRQKSVLRSSSLPGKLADCSSSNPEDCEIFIVEGD 411
V DS+ ++++ L A R R + ++S + + K DCSSS P+ G
Sbjct: 195 VYDSITEESIHLM---LKANRLRSMAEEESCVLCNKNNHKQEDCSSSIPK-------AGK 244
Query: 412 SAGGSAKQGRDRRFQAILPLRGKILNIERRDEAAMYKNEEIQ 453
S+G A+Q + P +GK R A N++IQ
Sbjct: 245 SSG--ARQSK--------PKKGKCFQCGERGHKAKDCNKKIQ 276
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.136 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,443,918
Number of Sequences: 26719
Number of extensions: 578388
Number of successful extensions: 1721
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 1703
Number of HSP's gapped (non-prelim): 26
length of query: 622
length of database: 11,318,596
effective HSP length: 105
effective length of query: 517
effective length of database: 8,513,101
effective search space: 4401273217
effective search space used: 4401273217
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC122169.6


