

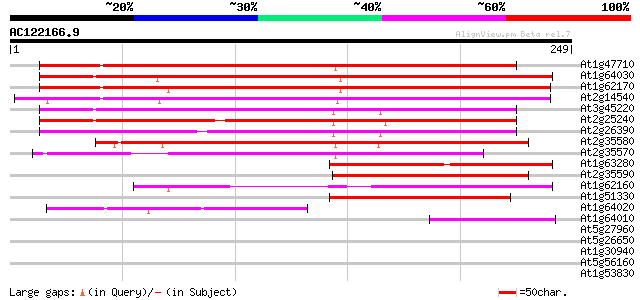
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC122166.9 + phase: 0 /pseudo
(249 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g47710 serpin, putative 218 2e-57
At1g64030 hypothetical protein 181 3e-46
At1g62170 hypothetical protein 180 7e-46
At2g14540 putative serpin 177 5e-45
At3g45220 serpin-like protein 174 3e-44
At2g25240 putative serpin 173 9e-44
At2g26390 putative serpin 169 1e-42
At2g35580 putative serpin 152 1e-37
At2g35570 putative serpin 147 5e-36
At1g63280 hypothetical protein 97 6e-21
At2g35590 putative serpin 90 1e-18
At1g62160 hypothetical protein 85 4e-17
At1g51330 hypothetical protein 81 6e-16
At1g64020 hypothetical protein 69 2e-12
At1g64010 hypothetical protein 52 3e-07
At5g27960 putative protein 31 0.71
At5g26650 putative protein 31 0.71
At1g30940 F17F8.19 31 0.71
At5g56160 putative protein 28 3.5
At1g53830 unknown protein 28 3.5
>At1g47710 serpin, putative
Length = 391
Score = 218 bits (556), Expect = 2e-57
Identities = 110/223 (49%), Positives = 153/223 (68%), Gaps = 12/223 (5%)
Query: 14 IGKSLTKFTNVSLNITKHLLSNQKLNEKNIVFSPLSLNTVLIMIATGSEGPTQNQLLSFL 73
+ +S++ VS+N+ KH+++ N N++FSP S+N VL +IA GS G T++Q+LSFL
Sbjct: 3 VRESISLQNQVSMNLAKHVITTVSQNS-NVIFSPASINVVLSIIAAGSAGATKDQILSFL 61
Query: 74 QSESTGDLKSLCSQVVSSVLSDGARAGGPCLSYVNGVWVEKSLPLQPSFKQLMTTDFEAT 133
+ ST L S S++VS+VL+DG+ GGP LS NG W++KSL +PSFKQL+ ++A
Sbjct: 62 KFSSTDQLNSFSSEIVSAVLADGSANGGPKLSVANGAWIDKSLSFKPSFKQLLEDSYKAA 121
Query: 134 LSAVDFVNKA-----------EKKTKGLIQDLLPHGSVNSLTSLIFANALYFKGVWKQKF 182
+ DF +KA EK+T GLI ++LP GS +S+T LIFANALYFKG W +KF
Sbjct: 122 SNQADFQSKAVEVIAEVNSWAEKETNGLITEVLPEGSADSMTKLIFANALYFKGTWNEKF 181
Query: 183 DTSKTKDYDFDLLNGKSVKVPFMTSKNDQFISSLDGFKVLGLP 225
D S T++ +F LL+G V PFMTSK Q++S+ DGFKVLGLP
Sbjct: 182 DESLTQEGEFHLLDGNKVTAPFMTSKKKQYVSAYDGFKVLGLP 224
>At1g64030 hypothetical protein
Length = 385
Score = 181 bits (459), Expect = 3e-46
Identities = 97/240 (40%), Positives = 148/240 (61%), Gaps = 13/240 (5%)
Query: 14 IGKSLTKFTNVSLNITKHLLSNQKLNEKNIVFSPLSLNTVLIMIATGSEGP-TQNQLLSF 72
+ +++ T+V++ ++ H+LS+ + N++FSP S+N+ + M A G G Q+LSF
Sbjct: 3 VREAMKNQTHVAMILSGHVLSSAP-KDSNVIFSPASINSAITMHAAGPGGDLVSGQILSF 61
Query: 73 LQSESTGDLKSLCSQVVSSVLSDGARAGGPCLSYVNGVWVEKSLPLQPSFKQLMTTDFEA 132
L+S S +LK++ ++ S V +D + GGP ++ NG+W++KSLP P FK L F+A
Sbjct: 62 LRSSSIDELKTVFRELASVVYADRSATGGPKITAANGLWIDKSLPTDPKFKDLFENFFKA 121
Query: 133 TLSAVDFVNKAEK-----------KTKGLIQDLLPHGSVNSLTSLIFANALYFKGVWKQK 181
VDF ++AE+ T LI+DLLP GSV SLT+ I+ANAL FKG WK+
Sbjct: 122 VYVPVDFRSEAEEVRKEVNSWVEHHTNNLIKDLLPDGSVTSLTNKIYANALSFKGAWKRP 181
Query: 182 FDTSKTKDYDFDLLNGKSVKVPFMTSKNDQFISSLDGFKVLGLPINKAKMGVHSLFTFFF 241
F+ T+D DF L+NG SV VPFM+S +Q++ + DGFKVL LP + + F+ +F
Sbjct: 182 FEKYYTRDNDFYLVNGTSVSVPFMSSYENQYVRAYDGFKVLRLPYQRGSDDTNRKFSMYF 241
>At1g62170 hypothetical protein
Length = 433
Score = 180 bits (456), Expect = 7e-46
Identities = 100/242 (41%), Positives = 147/242 (60%), Gaps = 16/242 (6%)
Query: 14 IGKSLTKFTNVSLNITKHLLSNQKLNEKNIVFSPLSLNTVLIMIATGSEGPTQNQL---- 69
+G+++ K +V++ +T ++S+ N N VFSP S+N L M+A S G +L
Sbjct: 64 VGEAMKKQNDVAIFLTGIVISSVAKNS-NFVFSPASINAALTMVAASSGGEQGEELRSFI 122
Query: 70 LSFLQSESTGDLKSLCSQVVSSVLSDGARAGGPCLSYVNGVWVEKSLPLQPSFKQLMTTD 129
LSFL+S ST +L ++ ++ S VL DG++ GGP ++ VNG+W+++SL + P K L
Sbjct: 123 LSFLKSSSTDELNAIFREIASVVLVDGSKKGGPKIAVVNGMWMDQSLSVNPLSKDLFKNF 182
Query: 130 FEATLSAVDFVNKAEK-----------KTKGLIQDLLPHGSVNSLTSLIFANALYFKGVW 178
F A + VDF +KAE+ T GLI+DLLP GSV SLT ++ +ALYFKG W
Sbjct: 183 FSAAFAQVDFRSKAEEVRTEVNAWASSHTNGLIKDLLPRGSVTSLTDRVYGSALYFKGTW 242
Query: 179 KQKFDTSKTKDYDFDLLNGKSVKVPFMTSKNDQFISSLDGFKVLGLPINKAKMGVHSLFT 238
++K+ S TK F LLNG SV VPFM+S Q+I++ DGFKVL LP + + + F
Sbjct: 243 EEKYSKSMTKCKPFYLLNGTSVSVPFMSSFEKQYIAAYDGFKVLRLPYRQGRDNTNRNFA 302
Query: 239 FF 240
+
Sbjct: 303 MY 304
>At2g14540 putative serpin
Length = 407
Score = 177 bits (449), Expect = 5e-45
Identities = 103/252 (40%), Positives = 148/252 (57%), Gaps = 15/252 (5%)
Query: 3 PQKRKRTKKNLIG--KSLTKFTNVSLNITKHLLSNQKLNEKNIVFSPLSLNTVLIMIATG 60
P K KK I +++ VSL + ++S N N VFSP S+N VL + A
Sbjct: 18 PSLSKTNKKQKIDMQEAMKNQNEVSLLLVGKVISAVAKNS-NCVFSPASINAVLTVTAAN 76
Query: 61 SEGPT-QNQLLSFLQSESTGDLKSLCSQVVSSVLSDGARAGGPCLSYVNGVWVEKSLPLQ 119
++ T ++ +LSFL+S ST + ++ ++ S V DG+ GGP ++ VNGVW+E+SL
Sbjct: 77 TDNKTLRSFILSFLKSSSTEETNAIFHELASVVFKDGSETGGPKIAAVNGVWMEQSLSCN 136
Query: 120 PSFKQLMTTDFEATLSAVDFVNKAE-----------KKTKGLIQDLLPHGSVNSLTSLIF 168
P ++ L F+A+ + VDF +KAE + T LI+++LP GSV SLT+ I+
Sbjct: 137 PDWEDLFLNFFKASFAKVDFRHKAEEVRLDVNTWASRHTNDLIKEILPRGSVTSLTNWIY 196
Query: 169 ANALYFKGVWKQKFDTSKTKDYDFDLLNGKSVKVPFMTSKNDQFISSLDGFKVLGLPINK 228
NALYFKG W++ FD S T+D F LLNGKSV VPFM S QFI + DGFKVL LP +
Sbjct: 197 GNALYFKGAWEKAFDKSMTRDKPFHLLNGKSVSVPFMRSYEKQFIEAYDGFKVLRLPYRQ 256
Query: 229 AKMGVHSLFTFF 240
+ + F+ +
Sbjct: 257 GRDDTNREFSMY 268
>At3g45220 serpin-like protein
Length = 393
Score = 174 bits (442), Expect = 3e-44
Identities = 96/225 (42%), Positives = 136/225 (59%), Gaps = 14/225 (6%)
Query: 14 IGKSLTKFTNVSLNITKHLLSNQKLNEKNIVFSPLSLNTVLIMIATGSEGPTQNQLLSFL 73
+GKS+ T+V + + KH++ N N+VFSP+S+N +L +IA GS T+ Q+LSF+
Sbjct: 3 LGKSMENQTDVMVLLAKHVIPTVA-NGSNLVFSPMSINVLLCLIAAGSNCVTKEQILSFI 61
Query: 74 QSESTGDLKSLCSQVVSSVLSDGARAGGPCLSYVNGVWVEKSLPLQPSFKQLMTTDFEAT 133
S+ L ++ ++ VS L+DG LS GVW++KSL +PSFK L+ + AT
Sbjct: 62 MLPSSDYLNAVLAKTVSVALNDGMERSDLHLSTAYGVWIDKSLSFKPSFKDLLENSYNAT 121
Query: 134 LSAVDFVNK-----------AEKKTKGLIQDLLPHGSVNSL--TSLIFANALYFKGVWKQ 180
+ VDF K AE T GLI+++L S+ ++ + LI ANA+YFKG W +
Sbjct: 122 CNQVDFATKPAEVINEVNAWAEVHTNGLIKEILSDDSIKTIRESMLILANAVYFKGAWSK 181
Query: 181 KFDTSKTKDYDFDLLNGKSVKVPFMTSKNDQFISSLDGFKVLGLP 225
KFD TK YDF LL+G VKVPFMT+ Q++ DGFKVL LP
Sbjct: 182 KFDAKLTKSYDFHLLDGTMVKVPFMTNYKKQYLEYYDGFKVLRLP 226
>At2g25240 putative serpin
Length = 385
Score = 173 bits (438), Expect = 9e-44
Identities = 97/225 (43%), Positives = 136/225 (60%), Gaps = 18/225 (8%)
Query: 14 IGKSLTKFTNVSLNITKHLLSNQKLNEKNIVFSPLSLNTVLIMIATGSEGPTQNQLLSFL 73
+GKS+ +V + +TKH+++ N N+VFSP+S+N +L +IA GS T+ Q+LSFL
Sbjct: 3 LGKSIENHNDVVVRLTKHVIATVA-NGSNLVFSPISINVLLSLIAAGSCSVTKEQILSFL 61
Query: 74 QSESTGDLKSLCSQVVSSVLSDGARAGGPCLSYVNGVWVEKSLPLQPSFKQLMTTDFEAT 133
ST L + +Q++ G LS NGVW++K L+ SFK L+ ++AT
Sbjct: 62 MLPSTDHLNLVLAQIIDG----GTEKSDLRLSIANGVWIDKFFSLKLSFKDLLENSYKAT 117
Query: 134 LSAVDFVNK-----------AEKKTKGLIQDLLPHGSVNSLTS--LIFANALYFKGVWKQ 180
S VDF +K AE T GLI+ +L S++++ S L+ ANA+YFKG W
Sbjct: 118 CSQVDFASKPSEVIDEVNTWAEVHTNGLIKQILSRDSIDTIRSSTLVLANAVYFKGAWSS 177
Query: 181 KFDTSKTKDYDFDLLNGKSVKVPFMTSKNDQFISSLDGFKVLGLP 225
KFD + TK DF LL+G SVKVPFMT+ DQ++ S DGFKVL LP
Sbjct: 178 KFDANMTKKNDFHLLDGTSVKVPFMTNYEDQYLRSYDGFKVLRLP 222
>At2g26390 putative serpin
Length = 389
Score = 169 bits (428), Expect = 1e-42
Identities = 92/228 (40%), Positives = 133/228 (57%), Gaps = 20/228 (8%)
Query: 14 IGKSLTKFTNVSLNITKHLLSNQKLNEKNIVFSPLSLNTVLIMIATGSEGPTQNQLLSFL 73
+GKS+ NV + K ++ N N+VFSP+S+N +L +IA GS T+ ++LSFL
Sbjct: 3 LGKSIENQNNVVARLAKKVIETDVANGSNVVFSPMSINVLLSLIAAGSNPVTKEEILSFL 62
Query: 74 QSESTGDLKSLCSQVVSSVLSDGARAGGPCLSYVNGVWVEKSLPLQPSFKQLMTTDFEAT 133
S ST L + V++ + G CLS +GVW++KS L+PSFK+L+ ++A+
Sbjct: 63 MSPSTDHLNA----VLAKIADGGTERSDLCLSTAHGVWIDKSSYLKPSFKELLENSYKAS 118
Query: 134 LSAVDFVNK-----------AEKKTKGLIQDLLPHGSVNSL-----TSLIFANALYFKGV 177
S VDF K A+ T GLI+ +L +++ ++LI ANA+YFK
Sbjct: 119 CSQVDFATKPVEVIDEVNIWADVHTNGLIKQILSRDCTDTIKEIRNSTLILANAVYFKAA 178
Query: 178 WKQKFDTSKTKDYDFDLLNGKSVKVPFMTSKNDQFISSLDGFKVLGLP 225
W +KFD TKD DF LL+G +VKVPFM S DQ++ DGF+VL LP
Sbjct: 179 WSRKFDAKLTKDNDFHLLDGNTVKVPFMMSYKDQYLRGYDGFQVLRLP 226
>At2g35580 putative serpin
Length = 374
Score = 152 bits (385), Expect = 1e-37
Identities = 86/209 (41%), Positives = 130/209 (62%), Gaps = 18/209 (8%)
Query: 39 NEKNIVF---SPLSLNTVLIMIATGSEGPTQ--NQLLSFLQSESTGDLKSLCSQVVSSVL 93
N+ +IV +PL +N +L +IA S G T ++++S LQ+ ST L ++ S++V++VL
Sbjct: 23 NQNDIVLRLTAPL-INVILSIIAASSPGDTDTADKIVSLLQASSTDKLHAVSSEIVTTVL 81
Query: 94 SDGARAGGPCLSYVNGVWVEKSLPLQPSFKQLMTTDFEATLSAVDFVNKA---------- 143
+D +GGP +S NG+W+EK+L ++PSFK L+ ++A + VDF KA
Sbjct: 82 ADSTASGGPTISAANGLWIEKTLNVEPSFKDLLLNSYKAAFNRVDFRTKADEVNREVNSW 141
Query: 144 -EKKTKGLIQDLLPHGSVNS-LTSLIFANALYFKGVWKQKFDTSKTKDYDFDLLNGKSVK 201
EK+T GLI +LLP ++ LT IFANAL+F G W +FD S TKD DF LL+G V+
Sbjct: 142 VEKQTNGLITNLLPSNPKSAPLTDHIFANALFFNGRWDSQFDPSLTKDSDFHLLDGTKVR 201
Query: 202 VPFMTSKNDQFISSLDGFKVLGLPINKAK 230
VPFMT + ++ +GFKV+ L + +
Sbjct: 202 VPFMTGASCRYTHVYEGFKVINLQYRRGR 230
>At2g35570 putative serpin
Length = 213
Score = 147 bits (371), Expect = 5e-36
Identities = 81/211 (38%), Positives = 125/211 (58%), Gaps = 28/211 (13%)
Query: 11 KNLIGKSLTKFTNVSLNITKHLLSNQKLNEKNIVFSPLSLNTVLIMIATGSEGPTQNQLL 70
+NL G S+ ++ L +T+H++++ N+VFSP L+ +L
Sbjct: 15 ENLKG-SVGNLNDIILRLTQHVIASTAGKTSNLVFSPALLSVIL---------------- 57
Query: 71 SFLQSESTGDLKSLCSQVVSSVLSDGARAGGPCLSYVNGVWVEKSLPLQPSFKQLMTTDF 130
SF+ + S +L ++ S +V+++L+D +GGP +S NGVW+EK+L ++PSFK L+ +
Sbjct: 58 SFVAASSPDELNAVSSGIVTTILADSTPSGGPTISAANGVWIEKTLYVEPSFKDLLLNSY 117
Query: 131 EATLSAVDFVNKA-----------EKKTKGLIQDLLPHGSVNSLTSLIFANALYFKGVWK 179
+A + VDF KA E++T GLI DLLP S +S T LIFANAL+F G W
Sbjct: 118 KAAFNQVDFRTKADEVNKEVNSWVEEQTNGLITDLLPPNSASSWTDLIFANALFFNGRWD 177
Query: 180 QKFDTSKTKDYDFDLLNGKSVKVPFMTSKND 210
+F+ S TK+ DF LL+G V+VPFMT ++
Sbjct: 178 SQFNPSLTKESDFHLLDGTKVRVPFMTGAHE 208
>At1g63280 hypothetical protein
Length = 120
Score = 97.4 bits (241), Expect = 6e-21
Identities = 51/99 (51%), Positives = 60/99 (60%), Gaps = 2/99 (2%)
Query: 143 AEKKTKGLIQDLLPHGSVNSLTSLIFANALYFKGVWKQKFDTSKTKDYDFDLLNGKSVKV 202
A T GLI DLLP GSV S T ++ NALYFKG W+ KFD S TKD +F GK V V
Sbjct: 20 ASDHTNGLIIDLLPRGSVKSETVQVYGNALYFKGAWENKFDKSSTKDNEFH--QGKEVHV 77
Query: 203 PFMTSKNDQFISSLDGFKVLGLPINKAKMGVHSLFTFFF 241
PFM S Q+I + DGFKVLGLP + F+ +F
Sbjct: 78 PFMRSYESQYIMACDGFKVLGLPYQQGLDDTKRKFSIYF 116
>At2g35590 putative serpin
Length = 331
Score = 89.7 bits (221), Expect = 1e-18
Identities = 43/87 (49%), Positives = 58/87 (66%)
Query: 144 EKKTKGLIQDLLPHGSVNSLTSLIFANALYFKGVWKQKFDTSKTKDYDFDLLNGKSVKVP 203
E++T GLI DLLP S + LT LIFANAL+F G W +F+ S TK+ DF LL+G V+VP
Sbjct: 101 EEQTNGLITDLLPPNSASPLTDLIFANALFFNGRWDSQFNPSLTKESDFHLLDGTKVRVP 160
Query: 204 FMTSKNDQFISSLDGFKVLGLPINKAK 230
FMT ++ + + FKVL LP + +
Sbjct: 161 FMTGAHEDSLDVYEAFKVLNLPYREGR 187
>At1g62160 hypothetical protein
Length = 265
Score = 84.7 bits (208), Expect = 4e-17
Identities = 59/190 (31%), Positives = 85/190 (44%), Gaps = 57/190 (30%)
Query: 56 MIATGSEGPTQNQL----LSFLQSESTGDLKSLCSQVVSSVLSDGARAGGPCLSYVNGVW 111
M+A S ++L LSFL++ ST +L ++ ++ SSV DG++
Sbjct: 1 MVAASSGDEQDDELRFFILSFLKASSTDELNAVLRKIASSVFVDGSK------------- 47
Query: 112 VEKSLPLQPSFKQLMTTDFEATLSAVDFVNKAEKKTKGLIQDLLPHGSVNSLTSLIFANA 171
K K +G +LT I+ NA
Sbjct: 48 ------------------------------KGGPKMRGH----------KNLTKRIYGNA 67
Query: 172 LYFKGVWKQKFDTSKTKDYDFDLLNGKSVKVPFMTSKNDQFISSLDGFKVLGLPINKAKM 231
LYFKG W++KF S TK F L+NG SV VPFM+S DQ+I + DGFKVL LP + +
Sbjct: 68 LYFKGTWEKKFSKSMTKRKPFYLVNGISVSVPFMSSSKDQYIEAYDGFKVLRLPYRQGRD 127
Query: 232 GVHSLFTFFF 241
+ F+ +F
Sbjct: 128 NTNRNFSMYF 137
>At1g51330 hypothetical protein
Length = 193
Score = 80.9 bits (198), Expect = 6e-16
Identities = 41/80 (51%), Positives = 53/80 (66%)
Query: 143 AEKKTKGLIQDLLPHGSVNSLTSLIFANALYFKGVWKQKFDTSKTKDYDFDLLNGKSVKV 202
A + T GLI++LLP GSV + T I+ NALYFKG W+ KF S T F L+NGK V V
Sbjct: 43 ALRHTNGLIKNLLPPGSVTNQTIKIYGNALYFKGAWENKFGKSMTIHKPFHLVNGKQVLV 102
Query: 203 PFMTSKNDQFISSLDGFKVL 222
PFM S +++ + +GFKVL
Sbjct: 103 PFMKSYERKYMKAYNGFKVL 122
>At1g64020 hypothetical protein
Length = 121
Score = 69.3 bits (168), Expect = 2e-12
Identities = 40/117 (34%), Positives = 66/117 (56%), Gaps = 3/117 (2%)
Query: 17 SLTKFTNVSLNITKHLLSNQKLNEKNIVFSPLSLNTVLIMIATG-SEGPTQNQLLSFLQS 75
++ K V++ ++ HL S + N VFSP S+ V M+A+G +Q+LSFL S
Sbjct: 6 AMKKQNEVAMILSWHLFSTVAKHSNN-VFSPASITAVFTMMASGPGSSLISDQILSFLGS 64
Query: 76 ESTGDLKSLCSQVVSSVLSDGARAGGPCLSYVNGVWVEKSLPLQPSFKQLMTTDFEA 132
S +L S+ +V+++V +DG+ GGP + NG W+++S + S K L F+A
Sbjct: 65 SSIDELNSVF-RVITTVFADGSNIGGPTIKVANGAWIDQSFSIDSSSKNLFENFFKA 120
>At1g64010 hypothetical protein
Length = 185
Score = 52.0 bits (123), Expect = 3e-07
Identities = 26/56 (46%), Positives = 33/56 (58%)
Query: 187 TKDYDFDLLNGKSVKVPFMTSKNDQFISSLDGFKVLGLPINKAKMGVHSLFTFFFL 242
TKD DF L+NG SV V M+S DQ+I + DGFKVL LP + + F+L
Sbjct: 2 TKDRDFHLINGTSVSVSLMSSYKDQYIEAYDGFKVLKLPFRQGNDTSRNFSMHFYL 57
>At5g27960 putative protein
Length = 363
Score = 30.8 bits (68), Expect = 0.71
Identities = 14/45 (31%), Positives = 26/45 (57%)
Query: 4 QKRKRTKKNLIGKSLTKFTNVSLNITKHLLSNQKLNEKNIVFSPL 48
Q + + NL + +F N++LN+ +L SNQ LN++ +P+
Sbjct: 266 QVQYQAPNNLFNQIQREFYNINLNLNLNLNSNQYLNQQQSFMNPM 310
>At5g26650 putative protein
Length = 327
Score = 30.8 bits (68), Expect = 0.71
Identities = 14/45 (31%), Positives = 26/45 (57%)
Query: 4 QKRKRTKKNLIGKSLTKFTNVSLNITKHLLSNQKLNEKNIVFSPL 48
Q + + NL + +F N++LN+ +L SNQ LN++ +P+
Sbjct: 225 QVQYQAPNNLFNQIQREFYNINLNLNLNLNSNQYLNQQQSFMNPM 269
>At1g30940 F17F8.19
Length = 461
Score = 30.8 bits (68), Expect = 0.71
Identities = 31/112 (27%), Positives = 49/112 (43%), Gaps = 7/112 (6%)
Query: 89 VSSVLSDGARAGGPCLSYVNGVWVEKSLPLQPSFKQLMTTDFEATLSAVDFVNKAEKKTK 148
V SV +G A C+SY + L + + +QL ++ + + V K K +
Sbjct: 349 VPSVEVEGLGANRACMSYYAFADYVEDLSVNDAVRQLKSSPLQQ--QGQNIVTKRPKPRQ 406
Query: 149 GLIQDLLPHGSVNSLTSLIFANALYFKGVWKQKFDTSKTKDYDFDLLNGKSV 200
G QD H S S+ N +FK + K T T+D+ ++ L KSV
Sbjct: 407 G--QDT-SHDLSKSAPSV--KNNKWFKSINKDDVVTFTTEDFPWNTLRSKSV 453
>At5g56160 putative protein
Length = 592
Score = 28.5 bits (62), Expect = 3.5
Identities = 18/57 (31%), Positives = 27/57 (46%), Gaps = 2/57 (3%)
Query: 182 FDTSKTKDYDFDLLNGKSVKVPFMTSKNDQFISSLDGFKVLGLPINKAKMGVHSLFT 238
FD K + +F ++ K VK+P K SL+ K L L ++K K +H T
Sbjct: 516 FDRLKKMEKEFTEISRKQVKIPEANEK--LLAESLERIKSLELDLDKTKSVLHITLT 570
>At1g53830 unknown protein
Length = 587
Score = 28.5 bits (62), Expect = 3.5
Identities = 27/86 (31%), Positives = 41/86 (47%), Gaps = 7/86 (8%)
Query: 39 NEKNIVFSPLSLNTVLIM----IATGSEGPTQNQLLSFLQSESTGDLKSLCSQVVSSVL- 93
N K ++ S ++ +L+ IA + +NQ ++ L S S LKS+CS + L
Sbjct: 16 NNKKLILSSAAIALLLLASIVGIAATTTNQNKNQKITTLSSTSHAILKSVCSSTLYPELC 75
Query: 94 -SDGARAGGPCLSYVNGVWVEKSLPL 118
S A GG L+ V +E SL L
Sbjct: 76 FSAVAATGGKELTSQKEV-IEASLNL 100
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.133 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,377,227
Number of Sequences: 26719
Number of extensions: 213490
Number of successful extensions: 595
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 19
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 555
Number of HSP's gapped (non-prelim): 29
length of query: 249
length of database: 11,318,596
effective HSP length: 97
effective length of query: 152
effective length of database: 8,726,853
effective search space: 1326481656
effective search space used: 1326481656
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC122166.9


